Inter-Laboratory Comparison of RT-PCR-Based Methods for the Detection of Tomato Brown Rugose Fruit Virus on Tomato
Abstract
:1. Introduction
2. Results
2.1. Intra-Laboratory Evaluation
2.2. Test Performance Study
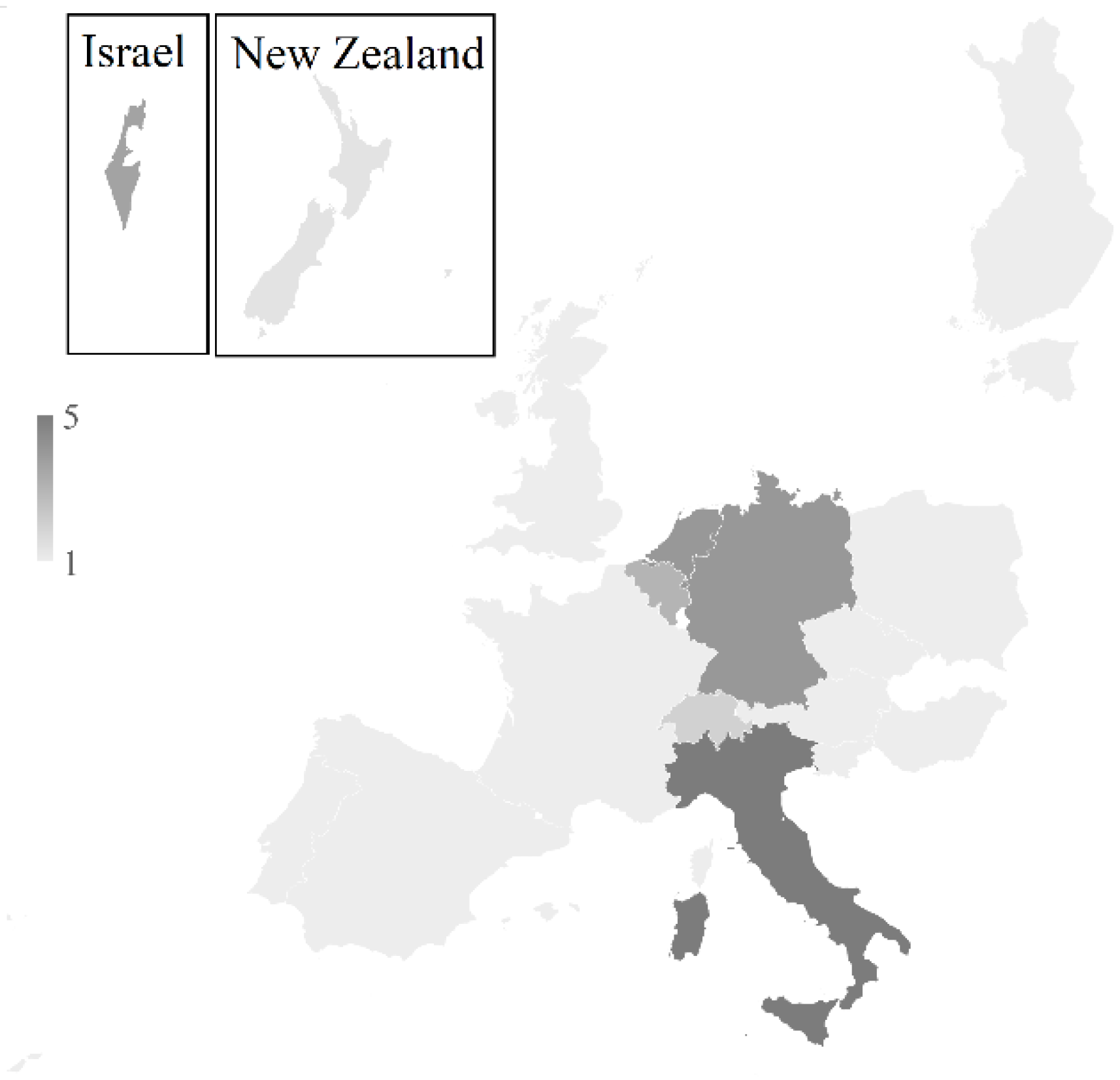
2.2.1. Participants
2.2.2. Data Set Evaluation
2.2.3. Repeatability and Reproducibility
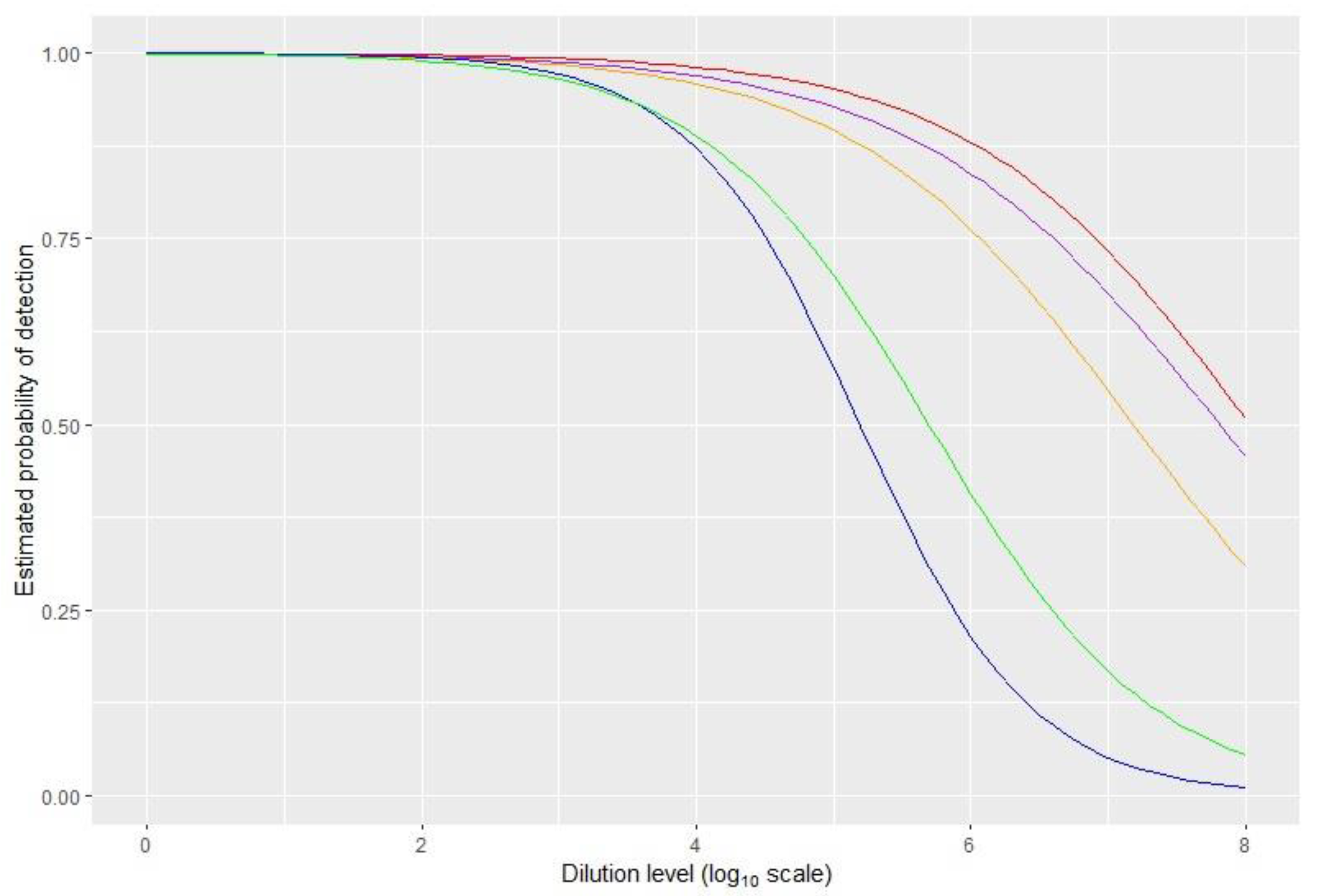
2.2.4. Analytical Sensitivity
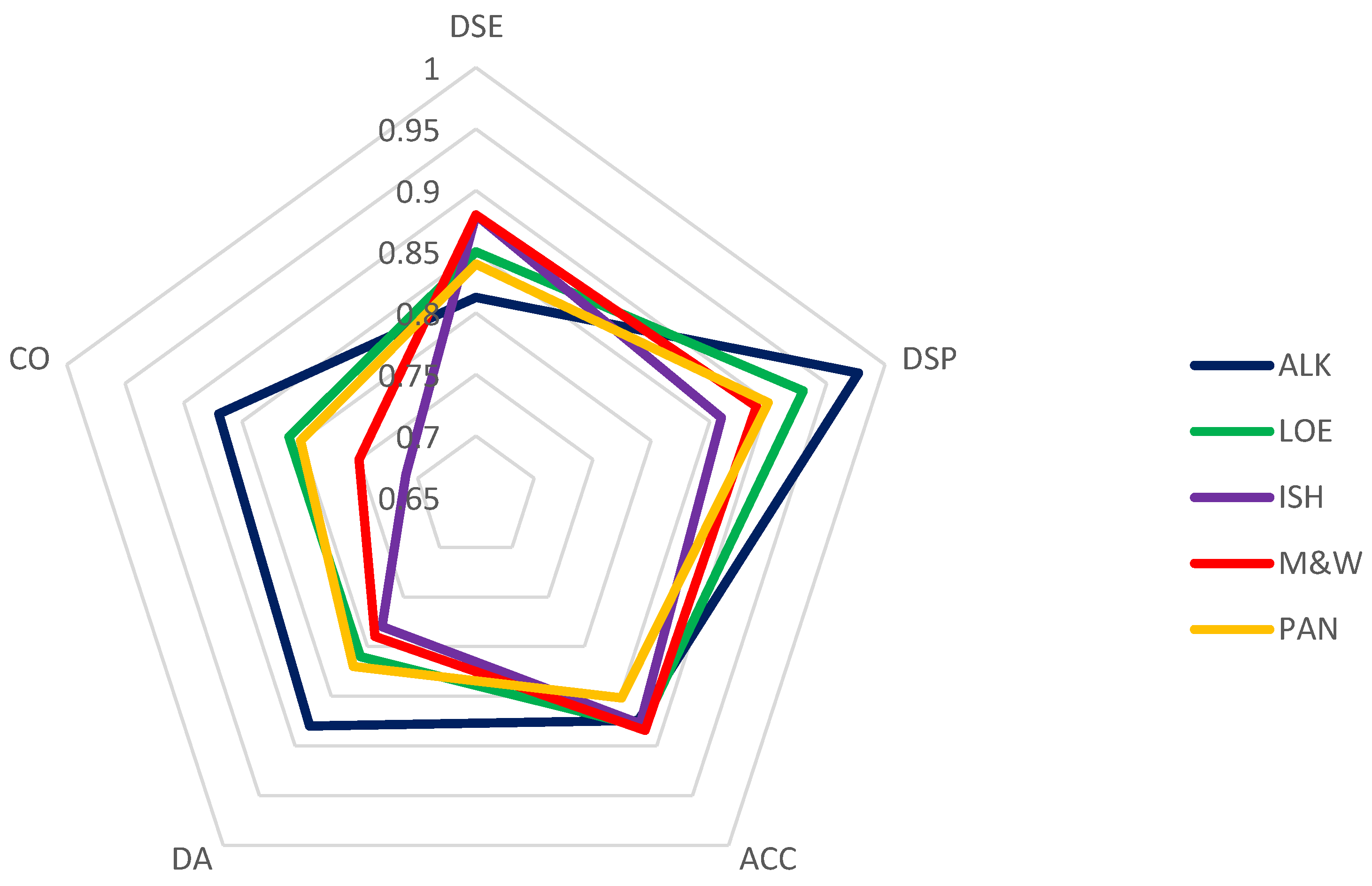
2.2.5. Evaluation of the Other Performance Criteria
2.2.6. Evaluation of the Deviations
3. Discussion
4. Materials and Methods
4.1. Sample Collection and Total RNA Extraction
4.2. Conventional RT-PCR Amplification
4.3. Real-Time RT-PCR
4.4. In-House Validation
4.5. TPS Participants
4.6. Panel of Test Items’ Composition and Preparation of the Shipping
4.7. Evaluation of the Performance Criteria
4.8. Outliers Results
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Luria, N.; Smith, E.; Reingold, V.; Bekelman, I.; Lapidot, M.; Levin, I.; Elad, N.; Tam, Y.; Sela, N.; Abu-Ras, A.; et al. A new Israeli Tobamovirus isolate infects tomato plants harboring Tm-22 resistance genes. PLoS ONE 2017, 12, e0170429. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Salem, N.; Mansour, A.; Ciuffo, M.; Falk, B.W.; Turina, M. A new tobamovirus infecting tomato crops in Jordan. Arch. Virol. 2016, 161, 503–506. [Google Scholar] [CrossRef] [PubMed]
- Cambrón-Crisantos, J.M.; Rodríguez-Mendoza, J.; Valencia-Luna, J.B.; Alcasio-Rangel, S.; García-Ávila, C.J.; López-Buenfil, J.A.; Ochoa-Martínez, D.L. First report of Tomato brown rugose fruit virus (ToBRFV) in Michoacan, Mexico. Rev. Mex. Fitopatol. 2018, 37, 1. [Google Scholar] [CrossRef] [Green Version]
- Ling, S.K.; Tian, T.; Gurung, S.; Salati, R.; Gilliard, A. First report of tomato brown rugose fruit virus infecting greenhouse tomato in the U.S. APS J. 2019, 103, 6. [Google Scholar] [CrossRef]
- Fidan, H.; Sarikaya, P.; Calis, O. First report of Tomato brown rugose fruit virus on tomato in Turkey. New Dis. Rep. 2019, 39, 18. [Google Scholar] [CrossRef] [Green Version]
- Yan, Z.-Y.; Ma, H.-Y.; Han, S.-L.; Geng, C.; Tian, Y.-P.; Li, X.-D. First report of Tomato brown rugose fruit virus infecting tomato in China. Plant Dis. 2019, 103, 2973. [Google Scholar] [CrossRef]
- Davino, S.; Caruso, A.G.; Bertacca, S.; Barone, S.; Panno, S. Tomato Brown Rugose Fruit Virus: Seed Transmission Rate and Efficacy of Different Seed Disinfection Treatments. Plants 2020, 9, 1615. [Google Scholar] [CrossRef] [PubMed]
- Salem, N.M.; Sulaiman, A.; Samarah, N.; Turina, M.; Vallino, M. Localization and mechanical transmission of tomato brown rugose fruit virus in tomato seeds. Plant Dis. 2021. [Google Scholar] [CrossRef]
- Levitzky, N.; Smith, E.; Lachman, O.; Luria, N.; Mizrahi, Y.; Bakelman, H.; Sela, N.; Laskar, O.; Milrot, E.; Dombrovsky, A. The bumblebee Bombus terrestris carries a primary inoculum of Tomato brown rugose fruit virus contributing to disease spread in tomatoes. PLoS ONE 2019, 14, e0210871. [Google Scholar] [CrossRef]
- EPPO Pest Risk Analysis for Tomato Brown Rugose Fruit Virus. EPPO. 2020. Paris. Available online: https://gd.eppo.int/taxon/TOBRFV/documents (accessed on 15 December 2021).
- Smith, E.; Dombrovsky, A. Chapter 3. In Plant Diseases—Current Threats and Management Trends; Topolovec-Pintaric, Ed.; IntechOpen: London, UK, 2019. [Google Scholar] [CrossRef] [Green Version]
- Alkowni, R.; Alabdallah, O.; Fadda, Z. Molecular identification of tomato brown rugose fruit virus in tomato in Palestine. J. Plant Pathol. 2019, 101, 719–723. [Google Scholar] [CrossRef]
- Panno, S.; Caruso, A.G.; Davino, S. First Report of Tomato Brown Rugose Fruit Virus on Tomato Crops in Italy. Plant Dis. 2019. [Google Scholar] [CrossRef]
- European and Mediterranean Plant Protection Organization. PM7/146 (1). EPPO Bull. 2021, 51, 178–197. [Google Scholar]
- European and Mediterranean Plant Protection Organization. PM7/98 (3). EPPO Bull. 2018, 48, 387–404. [Google Scholar]
- European and Mediterranean Plant Protection Organization. PM7/98 (4). EPPO Bull. 2019, 49, 530–563. [Google Scholar]
- Rodríguez-Mendoza, J.; de Jesús García-Ávila, C.; López-Buenfil, J.A.; Araujo-Ruiz, K.; Quezada-Salinas, A.; Cambrón-Crisantos, J.M.; Ochoa-Martínez, D.L. Identification of Tomato brown rugose fruit virus by RT-PCR from a coding region of replicase (RdRP). Rev. Mex Fis. 2019, 37. [Google Scholar] [CrossRef] [Green Version]
- ISF. Detection of Infectious Tomato Brown Rugose Fruit Virus (ToBRFV) in Tomato and Pepper Seed. 2020. Available online: https://www.worldseed.org/wp-content/uploads/2020/03/Tomato-ToBRFV_2020.03.pdf (accessed on 15 April 2020).
- Menzel, W.; Winter, S. Identification of novel and known tobamoviruses in tomato and other solanaceous crops using a new pair of generic primers and development of a specific RT-qPCR for ToBRFV. Acta Hortic. 2021, 1316, 143–148. [Google Scholar] [CrossRef]
- Panno, S.; Ruiz-Ruiz, S.; Caruso, A.G.; Alfaro-Fernandez, A.; Font San Ambrosio, M.I.; Davino, S. Real-time reverse transcription polymerase chain reaction development for rapid detection of Tomato brown rugose fruit virus and comparison with other techniques. Peer J. 2019, 7, e7928. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Doyle, J.J.; Doyle, J.L. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem. Bull. 1987, 19, 11–15. [Google Scholar]
- European and Mediterranean Plant Protection Organization. PM7/122 (1). EPPO Bull. 2014, 44, 390–399. [Google Scholar]
- Langton, S.D.; Chevennement, R.; Nagelkerke, N.; Lombard, B. Analysis collaborative trials for qualitative microbiological methods. Int. J. Food Microbiol. 2002, 79, 175–181. [Google Scholar] [CrossRef]

| Conventional RT-PCR | Real-time RT-PCR | |||||
|---|---|---|---|---|---|---|
| Test | ALK | LOE | ISH | M&W | PAN | |
| Ref. | [12] | [17] | [18] | [19] | [20] | |
| Conditions | Primers and/or probes final concentration (each) | 0.2 µM primer | - | 0.1 µM probe 0.15 µM primer | 0.3 µM probe 0.25 µM primer | 0.5 µM probe 0.25 µM primer |
| T annealing | 58 °C 30″ | 55 °C 20″ | 60 °C 1′ | 60 °C 1′ | 60 °C 1′ | |
| Analytical specificity | Inclusivity | 100% | 100% | 100% | 100% | 100% |
| Exclusivity | 100% | 100% | 100% | 100% | 100% | |
| Analytical sensitivity | Tomato | 10−3 | 10−5 | 10−7 | 10−7 | 10−7 |
| Pepper | 10−1 | 10−3 | 10−3 | 10−3 | 10−3 | |
| NIC | PIC | PAC | NAC | ||
|---|---|---|---|---|---|
| Results | Concordant (%) | 131 (87%) | 145 (97%) | 146 (97%) | 141 (94%) |
| Non-concordant (%) | 19 (13%) | 5 (3%) | 4 (3%) | 5 (3%) | |
| Untested (%) | 0 | 0 | 0 | 4 (3%) | |
| ISH | 36.28 ± 3.27 | 15.32 ± 3.05 | 19.94 ± 2.75 | 38.72 ± 3.02 | |
| M&W | 36.53 ± 3.17 | 15.68 ± 2.65 | 20.28 ± 3.43 | 39.23 ± 2.04 | |
| PAN | 36.81 ± 3.10 | 18.05 ± 3.79 | 22.58 ± 2.15 | 39.92 ± 0.37 |
| S1 | S2 | S3 | S4 | S5 | S6 | S7 | S8 | S9 | Total | |
|---|---|---|---|---|---|---|---|---|---|---|
| ALK | 100% | 95% | 97% | 72% | 76% | 100% | 100% | 91% | 61% | 88% |
| LOE | 95% | 79% | 91% | 50% | 75% | 100% | 100% | 91% | 49% | 81% |
| ISH | 80% | 71% | 50% | 72% | 100% | 100% | 100% | 54% | 75% | 78% |
| M&W | 88% | 73% | 49% | 71% | 100% | 100% | 100% | 53% | 78% | 79% |
| PAN | 95% | 70% | 63% | 65% | 100% | 100% | 90% | 72% | 79% | 82% |
| S1 | S2 | S3 | S4 | S5 | S6 | S7 | S8 | S9 | Total | |
|---|---|---|---|---|---|---|---|---|---|---|
| ALK | 100% | 95% | 97% | 69% | 76% | 100% | 100% | 91% | 57% | 87% |
| LOE | 95% | 79% | 91% | 47% | 75% | 100% | 100% | 91% | 49% | 81% |
| ISH | 79% | 71% | 49% | 63% | 88% | 88% | 96% | 54% | 66% | 73% |
| M&W | 88% | 73% | 48% | 65% | 92% | 92% | 100% | 52% | 72% | 76% |
| PAN | 95% | 70% | 62% | 65% | 100% | 100% | 90% | 70% | 79% | 81% |
| ALK | LOE | ISH | M&W | PAN | |
|---|---|---|---|---|---|
| Log dilution factor at 50% | 5.2 | 5.7 | 7.8 | N.A. | 7.2 |
| Log dilution factor at 95% | 3.4 | 3.3 | 4.6 | 5 | 4.2 |
| ALK | LOE | ISH | M&W | PAN | ||
|---|---|---|---|---|---|---|
| N. valid data set | 22 | 21 | 24 | 25 | 22 | |
| N. of samples | 352 | 336 | 384 | 389 | 344 | |
| TN (%) | (TN/N−%) | 129 (98%) | 118 (94%) | 79 (82%) | 88 (88%) | 78 (89%) |
| TP (%) | (TP/N+%) | 178 (81%) | 178 (85%) | 224 (85%) | 256 (85%) | 215 (81%) |
| FN (%) | (FN/N+%) | 41 (19%) | 32 (15%) | 32 (11%) | 34 (11%) | 42 (16%) |
| FP (%) | (FP/N−%) | 3 (2%) | 8 (6%) | 13 (14%) | 11 (11%) | 9 (10%) |
| INC (%) | (INC/N%) | 1 (0%) | 0 (0%) | 16 (4%) | 11 (3%) | 8 (2%) |
| ACC | (TP+TN)/ (TP+TN+FP+FN) | 87% | 88% | 88% | 88% | 85% |
| CIACC 95% | 66–100% | 73–100% | 66–100% | 66–100% | 54–94% | |
| p-Value Fisher ACC | NS | NS | NS | NS | NS | |
| DSE | TP/TP+FN | 81% | 85% | 88% | 88% | 84% |
| CIDSE 95% | 43–100% | 57–100% | 59–100% | 56–100% | 48–100% | |
| p-Value Fisher DSE | NS | NS | NS | NS | NS | |
| DSP | TN/TN+FP | 98% | 93% | 86% | 89% | 90% |
| CIDSP 95 % | 95–100% | 88–99% | 80–92% | 79–99% | 74–100% | |
| p-Value Fisher DSP | NS | NS | NS | NS | NS |
| ISH | M&W | PAN | ||||
|---|---|---|---|---|---|---|
| no-dev | dev | no-dev | dev | no-dev | dev | |
| N. valid dataset | 14 | 10 | 14 | 11 | 13 | 10 |
| Acc %Av | 91% | 87% | 92% | 87% | 87% | 84% |
| Acc % Min | 68% | 72% | 68% | 72% | 68% | 68% |
| Acc % Max | 100% | 100% | 100% | 100% | 100% | 95% |
| SAMPLE TYPE | SAMPLE ID | ISOLATE | HOST | SANITARY STATUS | EXPECTED OUTCOME (RT-PCR) | EXPECTED OUTCOME (REAL-TIME) |
|---|---|---|---|---|---|---|
| S1 | ToBRFV-M-1; M-2 | - | S. lycopersicum | Healthy | Negative | Negative |
| S2 | ToBRFV-M-3; M-4 | - | C. annuum | Healthy | Negative | Negative |
| S3 | ToBRFV-M-5; M-6; M-7 | ToB-SIC 21/19 | S. lycopersicum | 10−8 | Negative | Positive * |
| S4 | ToBRFV-M-8; M-9; M-10 | 10−6 | Positive * | Positive | ||
| S5 | ToBRFV-M-11; M-12; M-13 | 10−4 | Positive * | Positive | ||
| S6 | ToBRFV-M-14; M-15; M-16 | 10−2 | Positive | Positive | ||
| S7 | ToBRFV-M-17; M-18 | 100 | Positive | Positive | ||
| S8 | ToBRFV-M-19; M-20 | ToB-SIC 23/19 | S. lycopersicum | Low (10−6) | Positive * | Positive |
| S9 | ToBRFV-M-21; M-22 | ToB-SIC 25/19 | S. lycopersicum | Medium (10−4) | Positive * | Positive |
| NIC | ToBRFV-M-NIC | - ToB-SIC 24/19 | S. lycopersicum S. lycopersicum | 100 | Negative | Negative |
| PIC | ToBRFV-M-PIC | 100 | Positive | Positive | ||
| PAC | ToBRFV-M-PAC | ToB-SIC 22/19 | S. lycopersicum | 10−2 | Positive | Positive |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Luigi, M.; Manglli, A.; Tiberini, A.; Bertin, S.; Ferretti, L.; Taglienti, A.; Faggioli, F.; Tomassoli, L. Inter-Laboratory Comparison of RT-PCR-Based Methods for the Detection of Tomato Brown Rugose Fruit Virus on Tomato. Pathogens 2022, 11, 207. https://doi.org/10.3390/pathogens11020207
Luigi M, Manglli A, Tiberini A, Bertin S, Ferretti L, Taglienti A, Faggioli F, Tomassoli L. Inter-Laboratory Comparison of RT-PCR-Based Methods for the Detection of Tomato Brown Rugose Fruit Virus on Tomato. Pathogens. 2022; 11(2):207. https://doi.org/10.3390/pathogens11020207
Chicago/Turabian StyleLuigi, Marta, Ariana Manglli, Antonio Tiberini, Sabrina Bertin, Luca Ferretti, Anna Taglienti, Francesco Faggioli, and Laura Tomassoli. 2022. "Inter-Laboratory Comparison of RT-PCR-Based Methods for the Detection of Tomato Brown Rugose Fruit Virus on Tomato" Pathogens 11, no. 2: 207. https://doi.org/10.3390/pathogens11020207
APA StyleLuigi, M., Manglli, A., Tiberini, A., Bertin, S., Ferretti, L., Taglienti, A., Faggioli, F., & Tomassoli, L. (2022). Inter-Laboratory Comparison of RT-PCR-Based Methods for the Detection of Tomato Brown Rugose Fruit Virus on Tomato. Pathogens, 11(2), 207. https://doi.org/10.3390/pathogens11020207








