Wildlife Waterfowl as a Source of Pathogenic Campylobacter Strains
Abstract
1. Introduction
2. Results
2.1. Isolation and Identification of Bacterial Strains
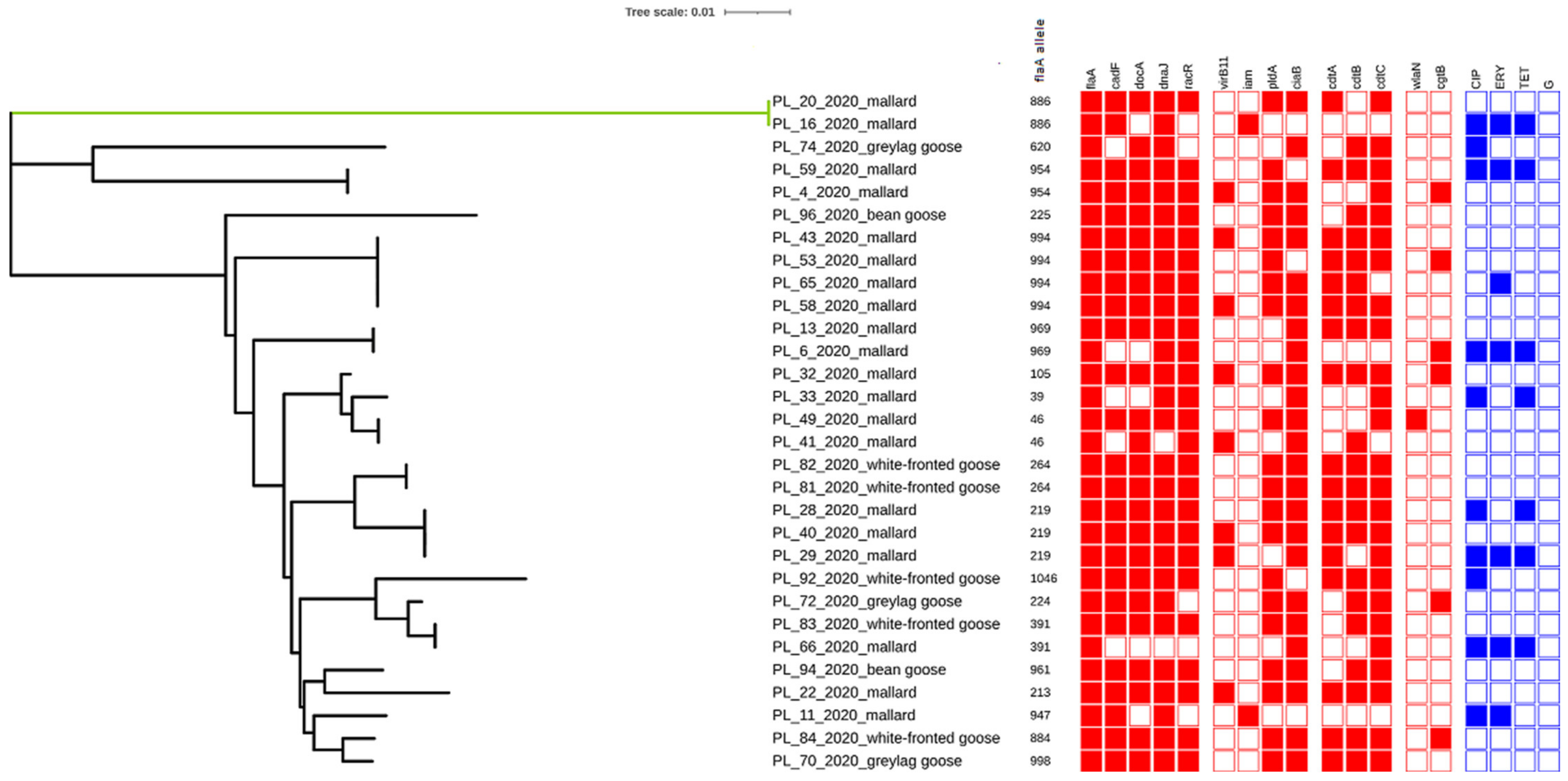
2.2. Detection of Virulence Genes
2.3. Sequencing of flaA-SVR
2.4. Antimicrobial Resistance
3. Discussion
4. Materials and Methods
4.1. Isolation and Identification of Bacterial Strains
4.2. Detection of Virulence Genes
4.3. Sequencing of flaA-SVR
4.4. Antimicrobial Resistance
4.5. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Tsiodras, S.; Kelesidis, T.; Kelesidis, I.; Bauchinger, U.; Falagas, M.E. Human infections associated with wild birds. J. Infect. 2008, 56, 83–98. [Google Scholar] [CrossRef] [PubMed]
- Reed, K.D.; Meece, J.K.; Henkel, J.S.; Shukla, S.K. Birds, migration and emerging zoonoses: West nile virus, lyme disease, influenza A and enteropathogens. Clin. Med. Res. 2003, 1, 5–12. [Google Scholar] [CrossRef] [PubMed]
- McKinney, M.L. Urbanization, Biodiversity, and Conservation: The impacts of urbanization on native species are poorly studied, but educating a highly urbanized human population about these impacts can greatly improve species conservation in all ecosystems. BioScience 2002, 52, 883–890. [Google Scholar] [CrossRef]
- Atterby, C.; Ramey, A.M.; Hall, G.G.; Järhult, J.; Börjesson, S.; Bonnedahl, J. Increased prevalence of antibiotic-resistant E. coli in gulls sampled in Southcentral Alaska is associated with urban environments. Infect. Ecol. Epidemiol. 2016, 6, 32334. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Smith, O.M.; Snyder, W.E.; Owen, J.P. Are we overestimating risk of enteric pathogen spillover from wild birds to humans? Biol. Rev. 2020, 95, 652–679. [Google Scholar] [CrossRef] [PubMed]
- European Centre for Disease Prevention and Control. Available online: https://www.ecdc.europa.eu/en/campylobacteriosis (accessed on 1 November 2021).
- Noormohamed, A.; Fakhr, M.K. Prevalence and Antimicrobial Susceptibility of Campylobacter spp. in Oklahoma Conventional and Organic Retail Poultry. Open Microbiol. J. 2014, 31, 130–137. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Kwon, Y.K.; Oh, J.Y.; Jeong, O.M.; Moon, O.K.; Kang, M.S.; Jung, B.Y.; An, B.K.; Youn, S.Y.; Kim, H.R.; Jang, I.; et al. Prevalence of Campylobacter species in wild birds South Korea. Avian Pathol. 2017, 46, 474–480. [Google Scholar] [CrossRef] [PubMed]
- Vogt, N.A.; Stevens, C.P.; Pearl, D.L.; Taboada, E.N.; Jardine, C.M. Generalizability and comparability of prevalence estimates in the wild bird literature: Methodological and epidemiological considerations. Anim. Health Res. Rev. 2020, 18, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Thomas, V.G.; Pain, D.J.; Kanstrup, N.; Green, R.E. Setting maximum levels for lead in game meat in EC regulations: An adjunct to replacement of lead ammunition. Ambio 2020, 49, 2026–2037. [Google Scholar] [CrossRef]
- Giacomelli, M.; Andrighetto, C.; Rossi, F.; Lombardi, A.; Rizzotti, L.; Martini, M.; Piccirillo, A. Molecular characterization and genotypic antimicrobial resistance analysis of Campylobacter jejuni and Campylobacter coli isolated from broiler flocks in northern Italy. Avian Pathol. 2012, 41, 579–588. [Google Scholar] [CrossRef]
- Marotta, F.; Garofolo, G.; Di Donato, G.; Aprea, G.; Platone, I.; Cianciavicchia, S.; Alessiani, A.; Di Giannatale, E. Population diversity of Campylobacter jejuni in poultry and its dynamic of contamination in chicken meat. BioMed Res. Int. 2015, 2015, 859845. [Google Scholar] [CrossRef] [PubMed]
- Hanage, W. Attack of the clones: What causes population structure in bacteria and how can we use it? In Proceedings of the APS March Meeting 2020, Denver, CO, USA, 2–6 March 2020. [Google Scholar]
- Reddy, S.; Zishiri, O.T. Genetic characterisation of virulence genes associated with adherence, invasion and cytotoxicity in Campylobacter spp. isolated from commercial chickens and human clinical cases. Onderstepoort J. Vet. Res. 2018, 85, e1–e9. [Google Scholar] [CrossRef] [PubMed]
- Guerry, P. Campylobacter flagella: Not just for motility. Trends Microbiol. 2007, 15, 456–461. [Google Scholar] [CrossRef] [PubMed]
- Sierra-Arguello, Y.M.; Perdoncini, G.; Rodrigues, L.B.; dos Santos, L.R.; Borges, K.A.; Furian, T.G.; Salle, C.T.P.; de Souza Moraes, H.L.; Gomes, M.J.P.; do Nascimento, V.P. Identification of pathogenic genes in Campylobacter jejuni isolated from broiler carcasses and broiler slaughterhouses. Sci. Rep. 2021, 11, 4588. [Google Scholar] [CrossRef]
- Krause-Gruszczynska, M.; van Alphen, L.B.; Oyarzabal, O.A.; Alter, T.; Hänel, I.; Schliephake, A.; König, W.; van Putten, J.P.M.; Konkel, M.E.; Backert, S. Expression patterns and role of the CadF protein in Campylobacter jejuni and Campylobacter coli. FEMS Microbiol. Lett. 2007, 274, 9–16. [Google Scholar] [CrossRef]
- Van der Stel, A.X.; van Mourik, A.; Łaniewski, P.; van Putten, J.P.; Jagusztyn-Krynicka, E.K.; Wösten, M.M. The Campylobacter jejuni RacRS two-component system activates the glutamate synthesis by directly upregulating γ-glutamyltranspeptidase (GGT). Front. Microbiol. 2015, 6, 567. [Google Scholar] [CrossRef]
- Hendrixson, D.R.; DiRita, V.J. Identification of Campylobacter jejuni genes involved in commensal colonization of the chick gastrointestinal tract. Mol. Microbiol. 2004, 52, 471–484. [Google Scholar] [CrossRef]
- Gharbi, M.; Béjaoui, A.; Ben Hamda, C.; Ghedira, K.; Ghram, A.; Maaroufi, A. Distribution of virulence and antibiotic resistance genes in Campylobacter jejuni and Campylobacter coli isolated from broiler chickens in Tunisia. J. Microbiol. Immunol. Infect. 2021, in press. [Google Scholar] [CrossRef]
- Pickett, C.L. Campylobacter Toxins and Their Role in Pathogenesis. In Campylobacter; Nachamkin, I., Blaser, M.J., Eds.; American Society for Microbiology: Washington, DC, USA, 2000; pp. 179–190. [Google Scholar]
- Guirado, P.; Paytubi, S.; Miró, E.; Iglesias-Torrens, Y.; Navarro, F.; Cerdà-Cuéllar, M.; Attolini, C.S.; Balsalobre, C.; Madrid, C. Differential Distribution of the wlaN and cgtB Genes, Associated with Guillain-Barré Syndrome, in Campylobacter jejuni Isolates from Humans, Broiler Chickens, and wild birds. Microorganisms 2020, 26, 325. [Google Scholar] [CrossRef] [PubMed]
- Bonnedahl, J.; Järhult, J.D. Antibiotic resistance in wild birds. Upsala J. Med. Sci. 2014, 119, 113–116. [Google Scholar] [CrossRef]
- Allen, H.K.; Donato, J.; Wang, H.H.; Cloud-Hansen, K.A.; Davies, J.; Handelsman, J. Call of the wild: Antibiotic resistance genes in natural environments. Nat. Rev. Microbiol. 2010, 8, 251–259. [Google Scholar] [CrossRef] [PubMed]
- Skurnik, D.; Ruimy, R.; Andremont, A.; Amorin, C.; Rouquet, P.; Picard, B.; Denamur, E. Effect of human vicinity on antimicrobial resistance and integrons in animal faecal Escherichia coli. J. Antimicrob. Chemother. 2006, 57, 1215–1219. [Google Scholar] [CrossRef] [PubMed]
- Elmberg, J.; Berg, C.; Lerner, H.; Waldenström, J.; Hessel, R. Potential disease transmission from wild geese and swans to livestock, poultry and humans: A review of the scientific literature from a One Health perspective. Infect. Ecol. Epidemiol. 2017, 7, 1300450. [Google Scholar] [CrossRef] [PubMed]
- French, N.P.; Midwinter, A.; Holland, B.; Collins-Emerson, J.; Pattison, R.; Colles, F.; Carter, P. Molecular epidemiology of Campylobacter jejuni isolates from wild-bird fecal material in children’s playgrounds. Appl. Environ. Microbiol. 2009, 75, 779–783. [Google Scholar] [CrossRef] [PubMed]
- Ramonaitė, S.; Novoslavskij, A.; Zakarienė, G.; Aksomaitienė, J.; Malakauskas, M. High Prevalence and Genetic Diversity of Campylobacter jejuni in Wild Crows and Pigeons. Curr. Microbiol. 2015, 71, 559–565. [Google Scholar] [CrossRef]
- Havelaar, A.H.; Mangen, M.-J.J.; de Koeijer, A.A.; Bogaardt, M.-J.; Evers, E.G.; Jacobs-Reitsma, W.E.; van Pelt, W.; Wagenaar, J.A.; de Wit, G.A.; van der Zee, H.; et al. Effectiveness and efficiency of controlling Campylobacter on broiler chicken meat. Risk Anal. 2007, 27, 831–844. [Google Scholar] [CrossRef] [PubMed]
- Wysok, B.; Wojtacka, J.; Kivistö, R. Pathogenicity of Campylobacter strains of poultry and human origin from Poland. Int. J. Food. Microbiol. 2020, 334, 108830. [Google Scholar] [CrossRef]
- Atterby, C.; Mourkas, E.; Méric, G.; Pascoe, B.; Wang, H.; Waldenström, J.; Sheppard, S.K.; Olsen, B.; Järhult, J.D.; Ellström, P. The Potential of Isolation Source to Predict Colonization in Avian Hosts: A Case Study in Campylobacter jejuni Strains from Three Bird Species. Front. Microbiol. 2018, 9, 591. [Google Scholar] [CrossRef] [PubMed]
- Colles, F.M.; Ali, J.S.; Sheppard, S.K.; McCarthy, N.D.; Maiden, M.C.J. Campylobacter populations in wild and domesticated Mallard ducks (Anas platyrhynchos). Environ. Microbiol. 2011, 3, 574–580. [Google Scholar] [CrossRef]
- Di Giannatale, E.; Garofolo, G.; Alessiani, A.; Di Donato, G.; Candeloro, L.; Vencia, W.; Decastelli, L.; Marotta, F. Tracing Back Clinical Campylobacter jejuni in the Northwest of Italy and Assessing Their Potential Source. Front. Microbiol. 2016, 7, 23. [Google Scholar] [CrossRef]
- Llarena, A.-K.; Zhang, J.; Vehkala, M.; Valimaki, N.; Hakkinen, M.; Hänninen, M.-L.; Roasto, M.; Mäesaar, M.; Taboada, E.N.; Barker, D.; et al. Monomorphic genotypes within a generalist lineage of Campylobacter jejuni show signs of global dispersion. Microb. Genom. 2016, 2, e000088. [Google Scholar] [CrossRef]
- Ehrlich, G.D.; Hiller, N.L.; Hu, F.Z. What makes pathogens pathogenic. Genome Biol. 2008, 9, 225. [Google Scholar] [CrossRef] [PubMed]
- Du, J.; Luo, J.; Huang, J.; Wang, C.; Li, M.; Wang, B.; Wang, B.; Chang, H.; Ji, J.; Sen, K.; et al. Emergence of Genetic Diversity and Multi-Drug Resistant Campylobacter jejuni From Wildlife waterfowl in Beijing, China. Front. Microbiol. 2019, 29, 2433. [Google Scholar] [CrossRef]
- Wei, B.; Kang, M.; Jang, H.K. Genetic characterization and epidemiological implications of Campylobacter isolates from wild birds in South Korea. Transbound. Emerg. Dis. 2019, 66, 56–65. [Google Scholar] [CrossRef] [PubMed]
- Shyaka, A.; Kusumoto, A.; Asakura, H.; Kawamoto, K. Whole-Genome Sequences of Eight Campylobacter jejuni Isolates from Wild Birds. Genome Announc. 2015, 3, e00315-15. [Google Scholar] [CrossRef]
- Wieczorek, K.; Wołkowicz, T.; Osek, J. Antimicrobial Resistance and Virulence-Associated Traits of Campylobacter jejuni Isolated from Poultry Food Chain and Humans with Diarrhea. Front. Microbiol. 2018, 9, 1508. [Google Scholar] [CrossRef]
- Wysok, B.; Wojtacka, J. Detection of virulence genes determining the ability to adhere and invade in Campylobacter spp. from cattle and swine in Poland. Microb. Pathog. 2018, 115, 257–263. [Google Scholar] [CrossRef] [PubMed]
- Gahamanyi, N.; Song, D.G.; Yoon, K.Y.; Mboera, L.E.G.; Matee, M.I.; Mutangana, D.; Amachawadi, R.G.; Komba, E.V.G.; Pan, C.H. Antimicrobial Resistance Profiles, Virulence Genes, and Genetic Diversity of Thermophilic Campylobacter Species Isolated from a Layer Poultry Farm in Korea. Front. Microbiol. 2021, 12, 554. [Google Scholar] [CrossRef]
- Melo, R.T.; Nalevaiko, P.C.; Mendonça, E.P.; Borges, L.W.; Fonseca, B.B.; Beletti, M.E.; Rossi, D.E. Campylobacter jejuni strains isolated from chicken meat harbour several virulence factors and represent a potential risk to humans. Food Control 2013, 33, 227–231. [Google Scholar] [CrossRef]
- Biswas, D.; Hannon, S.J.; Townsend, H.G.; Potter, A.; Allan, B.J. Genes coding for virulence determinants of Campylobacter jejuni in human clinical and cattle isolates from Alberta, Canada, and their potential role in colonization of poultry. Int. Microbiol. 2011, 14, 25–32. [Google Scholar] [CrossRef] [PubMed]
- Zhang, T.; Luo, Q.; Chen, Y.; Li, T.; Wen, G.; Zhang, R.; Luo, L.; Lu, Q.; Ai, D.; Wang, H.; et al. Molecular epidemiology, virulence determinants and antimicrobial resistance of Campylobacter spreading in retail chicken meat in Central China. Gut Pathog. 2016, 8, 48. [Google Scholar] [CrossRef] [PubMed]
- Bang, D.D.; Nielsen, E.M.; Scheutz, F.; Pedersen, K.; Handberg, K.; Madsen, M. PCR detection of seven virulence and toxin genes of Campylobacter jejuni and Campylobacter coli isolates from Danish pigs and cattle and cytolethal distending toxin production of the isolates. J. Appl. Microbiol. 2003, 94, 1003–1014. [Google Scholar] [CrossRef] [PubMed]
- González-Hein, G.; Huaracán, B.; García, P.; Figueroa, G. Prevalence of virulence genes in strains of Campylobacter jejuni isolated from human, bovine and broiler. Braz. J. Microbiol. 2014, 44, 1223–1229. [Google Scholar] [CrossRef] [PubMed]
- Tracz, D.M.; Keelan, M.; Ahmed-Bentley, J.; Gibreel, A.; Kowalewska-Grochowska, K.; Taylor, D.E. pVir and bloody diarrhea in Campylobacter jejuni enteritis. Emerg. Infect. Dis. 2005, 11, 838–843. [Google Scholar] [CrossRef] [PubMed]
- Sanad, Y.M.; Kassem, I.I.; Abley, M.; Gebreyes, W.; LeJeune, J.T.; Rajashekara, G. Genotypic and Phenotypic Properties of Cattle-Associated Campylobacter and Their Implications to Public Health in the USA. PLoS ONE 2011, 6, e25778. [Google Scholar] [CrossRef] [PubMed]
- Carvalho, A.F.; Silva, D.M.; Azevedo, S.S.; Piatti, R.M.; Genovez, M.E.; Scarcelli, E. Detection of CDT toxin genes in Campylobacter spp. strains isolated from broiler carcasses and vegetables in São Paulo, Brazil. Braz. J. Microbiol. 2014, 44, 693–699. [Google Scholar] [CrossRef]
- Wysok, B.; Uradziński, J.; Wojtacka, J. Determination of the cytotoxic activity of Campylobacter strains isolated from bovine and swine carcasses in north-eastern Poland. Pol. J. Vet. Sci. 2015, 18, 579–586. [Google Scholar] [CrossRef][Green Version]
- Cho, H.H.; Kim, S.H.; Min, W.; Ku, B.K.; Kim, Y.H. Prevalence of virulence and cytolethal distending toxin (CDT) genes in thermophilic Campylobacter spp. from dogs and humans in Gyeongnam and Busan, Korea. KJVR Korean J. Vet. Res. 2014, 54, 39–48. [Google Scholar] [CrossRef]
- Ang, C.W.; De Klerk, M.A.; Endtz, H.P.; Jacobs, B.C.; Laman, J.D.; van der Meche, F.G.A.; van Doorn, P.A. Guillain-Barre syndrome- and Miller Fisher syndrome-associated Campylobacter jejuni lipopolysaccharides induce anti-GM1 and anti-GQ1b antibodies in rabbits. Infect. Immun. 2001, 69, 2462–2469. [Google Scholar] [CrossRef]
- Datta, S.; Niwa, H.; Itoh, K. Prevalence of 11 pathogenic genes of Campylobacter jejuni by PCR in strains isolated from humans, poultry meat and broiler and bovine faeces. J. Med. Microbiol. 2003, 52, 345–348. [Google Scholar] [CrossRef]
- Wysok, B.; Wojtacka, J.; Wiszniewska-Łaszczych, A.; Szteyn, J. Antimicrobial Resistance and Virulence Properties of Campylobacter Spp. Originating from Domestic Geese in Poland. Animals 2020, 10, 742. [Google Scholar] [CrossRef] [PubMed]
- Casabonne, C.; Gonzalez, A.; Aquili, V.; Subils, T.; Balague, C. Prevalence of Seven Virulence Genes of Campylobacter jejuni Isolated from Patients with Diarrhea in Rosario, Argentina. Int. J. Infect. 2016, 3, e37727. [Google Scholar] [CrossRef]
- Oporto, B.; Juste, R.A.; López-Portolés, J.A.; Hurtado, A. Genetic diversity among Campylobacter jejuni isolates from healthy livestock and their links to human isolates in Spain. Zoonoses Public Health 2011, 58, 365–375. [Google Scholar] [CrossRef] [PubMed]
- Ma, F.; Xu, S.; Tang, Z.; Li, Z.; Zhang, L. Use of antimicrobials in food animals and impact of transmission of antimicrobial resistance on humans. Bios. Health 2021, 3, 32–38. [Google Scholar] [CrossRef]
- Wieczorek, K.; Osek, J. Antimicrobial resistance mechanisms among Campylobacter. BioMed Res. Int. 2013, 2013, 340605. [Google Scholar] [CrossRef] [PubMed]
- Hull, D.M.; Harrell, E.; van Vliet, A.H.M.; Correa, M.; Thakur, S. Antimicrobial resistance and interspecies gene transfer in Campylobacter coli and Campylobacter jejuni isolated from food animals, poultry processing, and retail meat in North Carolina, 2018-2019. PLoS ONE 2021, 16, e0246571. [Google Scholar] [CrossRef] [PubMed]
- García-Fernández, A.; Dionisi, A.M.; Arena, S.; Iglesias-Torrens, Y.; Carattoli, A.; Luzzi, I. Human Campylobacteriosis in Italy: Emergence of Multi-Drug Resistance to Ciprofloxacin, Tetracycline, and Erythromycin. Front. Microbiol. 2018, 9, 1906. [Google Scholar] [CrossRef]
- Marotta, F.; Garofolo, G.; Di Marcantonio, L.; Di Serafino, G.; Neri, D.; Romantini, R.; Sacchini, L.; Alessiani, A.; Di Donato, G.; Nuvoloni, R.; et al. Correction: Antimicrobial resistance genotypes and phenotypes of Campylobacter jejuni isolated in Italy from humans, birds from wild and urban habitats, and poultry. PLoS ONE 2019, 14, e0225231. [Google Scholar] [CrossRef]
- Wieczorek, K.; Osek, J. Multiplex PCR assays for simultaneous identification of Campylobacter jejuni and Campylobacter coli. Med. Weter. 2005, 61, 797–799. [Google Scholar]
- Meinersmann, R.J.; Helsel, L.O.; Fields, P.I.; Hiett, K.L. Discrimination of Campylobacter jejuni isolates by fla gene sequencing. J. Clin. Microbiol. 1997, 35, 2810–2814. [Google Scholar] [CrossRef] [PubMed]
- Müller, J.; Schulze, F.; Müller, W.; Hänel, I. PCR detection of virulence-associated genes in Campylobacter jejuni strains with differential ability to invade Caco-2 cells and to colonize the chick gut. Vet. Microbiol. 2006, 13, 123–129. [Google Scholar] [CrossRef] [PubMed]
- Chansiripornchai, N.; Sasipreeyajan, J. PCR detection of four virulence-associated genes of Campylobacter jejuni isolates from Thai broilers and their abilities of adhesion to and invasion of INT-407 cells. J. Vet. Med. Sci. 2009, 71, 839–844. [Google Scholar] [CrossRef] [PubMed]
- Konkel, M.E.; Kim, B.J.; Rivera-Amill, V.; Garvis, S.G. Identification of proteins required for the internalization of Campylobacter jejuni into cultured mammalian cells. Adv. Exp. Med. Biol. 1999, 473, 215–224. [Google Scholar] [PubMed]
- Carvalho, A.C.; Ruiz Palacios, G.M.; Ramos Cervantes, P.; Cervantes, L.E.; Jiang, X.; Pickering, L.K. Molecular characterization of invasive and noninvasive Campylobacter jejuni and Campylobacter coli isolates. J. Clin. Microbiol. 2001, 39, 1353–1359. [Google Scholar] [CrossRef] [PubMed]
- Linton, D.; Gilbert, M.; Hitchen, P.G.; Dell, A.; Morris, H.R.; Wakarchuk, W.W.; Gregson, N.A.; Wren, B.W. Phase variation of a beta-1,3 galactosyltransferase involved in generation of the ganglioside GM1-like lipo-oligosaccharide of Campylobacter jejuni. Mol. Microbiol. 2000, 37, 501–514. [Google Scholar] [CrossRef] [PubMed]
- Hunter, P.R.; Gaston, M.A. Numerical index of the discriminatory ability of typing systems: An application of Simpson’s index of diversity. J. Clin. Microbiol. 1988, 26, 2465–2466. [Google Scholar] [CrossRef] [PubMed]
- Carriço, J.A.; Silva-Costa, C.; Melo-Cristino, J.; Pinto, F.R.; de Lencastre, H.; Almeida, J.S.; Ramirez, M. Illustration of a common framework for relating multiple typing methods by application to macrolide-resistant Streptococcus pyogenes. J. Clin. Microbiol. 2006, 44, 2524–2532. [Google Scholar] [CrossRef]
- EUCAST. European Committee on Antimicrobial Susceptibility Testing. Available online: http://www.eucast.org/clinical_breakpoints (accessed on 1 November 2021).
- CLSI. Methods for Antimicrobial Dilution and Disc Susceptibility Testing of Infrequently Isolated or Fastidious Bacteria; CLSI Guideline M45-A3; Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2016. [Google Scholar]
- Magiorakos, A.P.; Srinivasan, A.; Carey, R.B.; Carmeli, Y.; Falagas, M.E.; Giske, C.G.; Harbarth, S.; Hindler, J.F.; Kahlmeter, G.; Olsson-Liljequist, B.; et al. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: An international expert proposal for interim standard definitions for acquired resistance. Clin. Microbiol. Infect. 2012, 18, 268–281. [Google Scholar] [CrossRef] [PubMed]
| Source | No. of Samples | No. of Positive Samples (%) | ||
|---|---|---|---|---|
| Common Name | Latin Name | C. jejuni | C. coli | |
| Mallard duck | Anas platyrhynchos | 61 | 18 (29.5%) | 2 (3.3%) |
| White-fronted goose | Anser albifrons | 11 | 5 (45.5%) | 0 |
| Greylag goose | Anser anser | 8 | 3 (37.5%) | 0 |
| Eurasian teal | Anas crecca | 6 | 0 | 0 |
| Bean goose | Anser fabalis | 5 | 2 (40%) | 0 |
| Target Gene | Sequences (5′–3′) | Product Size (bp) | Annealing Temperature °C | References |
|---|---|---|---|---|
| 16S rRNA for Campylobacter spp. | F-ATCTAATGGCTTAACCATTAAAC | 857 | 58 | [62] |
| R GGACGGTAACTAGTTTAGTATT | ||||
| mapA for C. jejuni | F-CTATTTTATTTTTGAGTGCTTGTG | 589 | 58 | [62] |
| R-GCTTTATTTGCCATTTGTTTTATTA | ||||
| ceuE for C. coli | F-AATTGAAAATTGCTCCAACTATG | 462 | 58 | [62] |
| R-TGATTTTATTATTTGTAGCAGCG | ||||
| flaA-SVR | F-CTA TGG ATG AGC AAT T(AT)A AAA T | 383 | 53 | [63] |
| R-CAA G(AT)C CTG TTC C(AT)A CTG AAG | ||||
| flaA | F-AATAAAAATGCTGATAAAACAGGTG | 855 | 53 | [53] |
| R-TACCGAACCAATGTCTGCTCTGATT | ||||
| flhA | F-GGAAGCGGCACTTGGTTTGC | 735 | 53 | [64] |
| R-GCTGTGAGTGAGATTATAGCAG | ||||
| dnaJ | F-ATTGATTTTGCTGCGGGTAG | 177 | 50 | [65] |
| R-ATCCGCAAAAGCTTCAAAAA | ||||
| cadF | F-TTGAAGGTAATTTAGATATG | 400 | 45 | [66] |
| R-CTAATACCTAAAGTTGAAAC | ||||
| virB11 | F-TCTTGTGAGTTGCCTTACCCCTTTT | 494 | 53 | [53] |
| R-CCTGCGTGTCCTGTGTTATTTACCC | ||||
| docA | F-ATAAGGTGCGGTTTTGGC | 725 | 50 | [64] |
| R-GTCTTTGCAGTAGATATG | ||||
| iam | F-GCGCAAAATATTATCACCC | 518 | 52 | [67] |
| R-TTCACGACTACTATGCGG | ||||
| ciaB | F-TGCGAGATTTTTCGAGAATG | 527 | 54 | [65] |
| R-TGCCCGCCTTAGAACTTACA | ||||
| racR | F-GATGATCCTGACTTTG | 584 | 45 | [53] |
| R-TCTCCTATTTTTACCC | ||||
| pldA | F-AAGCTTATGCGTTTTT | 913 | 45 | [53] |
| R-TATAAGGCTTTCTCCA | ||||
| cdtA | F-CCTTGTGATGCAAGCAATC | 370 | 49 | [53] |
| R-ACACTCCATTTGCTTTCTG | ||||
| cdtB | F-CAGAAAGCAAATGGAGTGTT | 620 | 51 | [53] |
| R-AGCTAAAAGCGGTGGAGTAT | ||||
| cdtC | F-CGATGAGTTAAAACAAAAAGATA | 182 | 47 | [53] |
| R-TTGGCATTATAGAAAATACAGTT | ||||
| wlaN | F-TGCTGGGTATACAAAGGTTGTG | 330 | 55 | [64] |
| R-ATTTTGGATATGGGTGGGG | ||||
| cgtB | F-TAAGAGCAAGATATGAAGGTG | 561 | 52 | [68] |
| R-GCACATAGAGAACGCTACAA |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wysok, B.; Sołtysiuk, M.; Stenzel, T. Wildlife Waterfowl as a Source of Pathogenic Campylobacter Strains. Pathogens 2022, 11, 113. https://doi.org/10.3390/pathogens11020113
Wysok B, Sołtysiuk M, Stenzel T. Wildlife Waterfowl as a Source of Pathogenic Campylobacter Strains. Pathogens. 2022; 11(2):113. https://doi.org/10.3390/pathogens11020113
Chicago/Turabian StyleWysok, Beata, Marta Sołtysiuk, and Tomasz Stenzel. 2022. "Wildlife Waterfowl as a Source of Pathogenic Campylobacter Strains" Pathogens 11, no. 2: 113. https://doi.org/10.3390/pathogens11020113
APA StyleWysok, B., Sołtysiuk, M., & Stenzel, T. (2022). Wildlife Waterfowl as a Source of Pathogenic Campylobacter Strains. Pathogens, 11(2), 113. https://doi.org/10.3390/pathogens11020113






