Molecular Mimicry Analyses Unveiled the Human Herpes Simplex and Poxvirus Epitopes as Possible Candidates to Incite Autoimmunity
Abstract
:1. Introduction
2. Results
2.1. Non-Paralogous Viral and Human Proteome Sets
2.2. Sequence Similarity Search
2.3. Pathway Enrichment Analysis
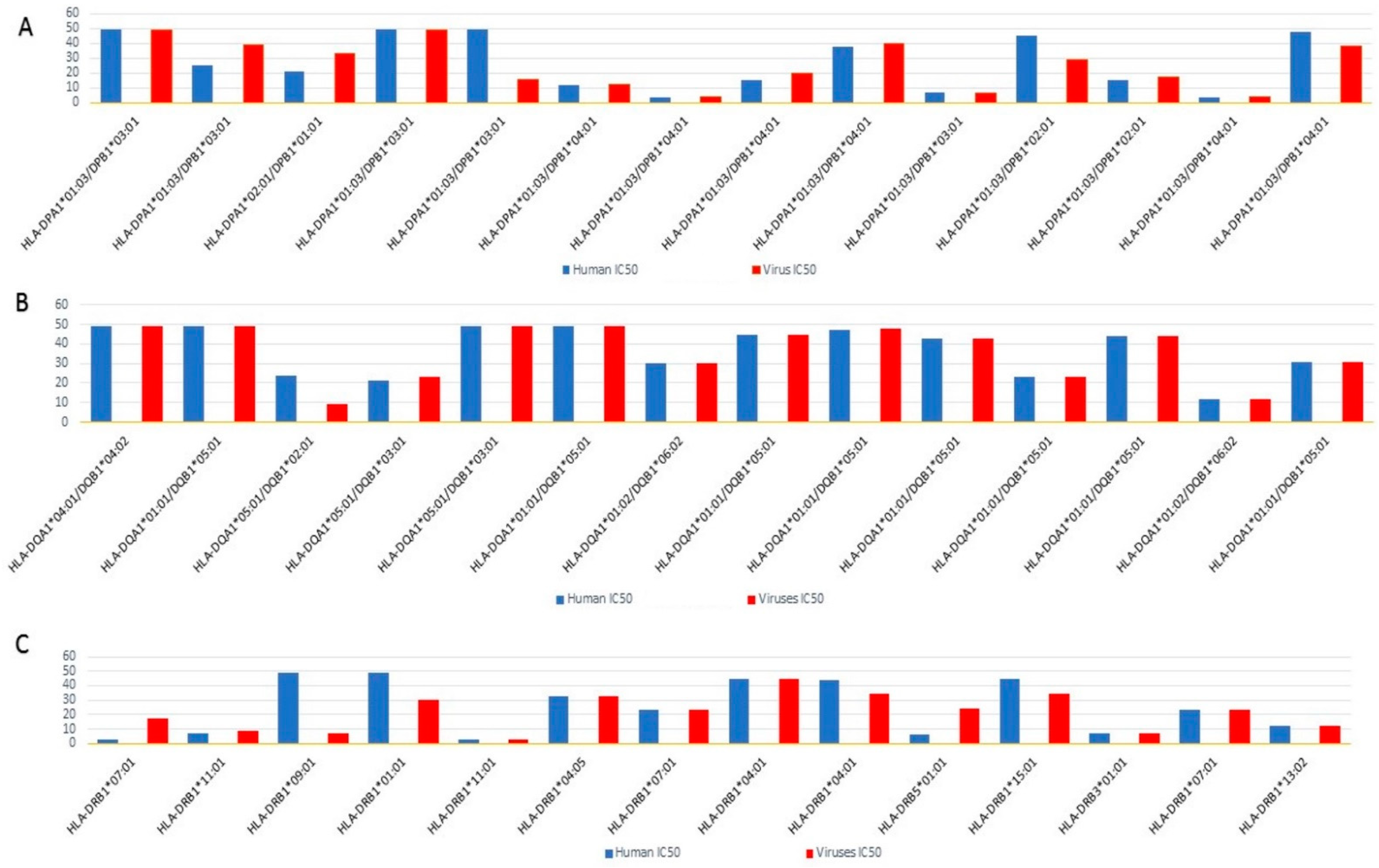
2.4. Epitope Prediction
2.5. Molecular Mimicry Prediction of Viral-Human Homolog Epitopes
2.6. Molecular Docking
3. Discussion
4. Materials and Methods
4.1. Non-Paralogous Viral and Human Proteome Sequence Retrieval
4.2. Sequence Similarity Search
4.3. Metabolic Pathway Enrichment
4.4. Epitope Candidate Prediction
4.5. Molecular Modeling and Docking Analyses
4.6. Structural Mimicry Prediction
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ercolini, A.M.; Miller, S.D. The role of infections in autoimmune disease. Clin. Exp. Immuno. 2009, 155, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Smatti, M.K.; Cyprian, F.S.; Nasrallah, G.K.; Al Thani, A.A.; Almishal, R.O.; Yassine, H.M. Viruses and Autoimmunity: A Review on the Potential Interaction and Molecular Mechanisms. Viruses 2019, 11, 762. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Selgrade, M.K.; Cooper, G.S.; Germolec, D.R.; Heindel, J.J. Linking environmental agents and autoimmune disease: An agenda for future research. Environ. Health Perspect. 1999, 107, 811–813. [Google Scholar]
- Casiraghi, C.; Shanina, I.; Cho, S.; Freeman, M.L.; Blackman, M.A.; Horwitz, M.S. Gammaherpesvirus Latency Accentuates EAE Pathogenesis: Relevance to Epstein-Barr Virus and Multiple Sclerosis. PLoS Pathogens. 2012, 8, e1002715. [Google Scholar] [CrossRef] [Green Version]
- Pahari, S.; Chatterjee, D.; Negi, S.; Kaur, J.; Singh, B.; Agrewala, J.N. Morbid Sequences Suggest Molecular Mimicry between Microbial Peptides and Self-Antigens: A Possibility of Inciting Autoimmunity. Front. Microbiol. 2017, 8, 1938. [Google Scholar] [CrossRef] [Green Version]
- Arleevskaya, M.I.; Manukyan, G.; Inoue, R.; Aminov, R. Editorial: Microbial and Environmental Factors in Autoimmune and Inflammatory Diseases. Front. Immunol. 2017, 8, 243. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Finnegan, A.; Needleman, B.W.; Hodes, R.J. Function of Autoreactive T Cells in Immune Responses. Immunol. Rev. 1990, 116, 15–31. [Google Scholar] [CrossRef] [PubMed]
- Wang, P.; Zheng, S.G. Regulatory T cells and B cells: Implication on autoimmune diseases. Int. J. Clin. Exp. Pathol. 2013, 6, 2668–2674. [Google Scholar]
- Curran, S.A.; Hollan, I.; Erridge, C.; Lappin, D.F.; Murray, C.A.; Sturfelt, G.; Mikkelsen, K.; Førre, O.T.; Almdahl, S.M.; Fagerhol, M.K.; et al. Bacteria in the Adventitia of Cardiovascular Disease Patients with and without Rheumatoid Arthritis. PLoS ONE 2014, 9, e98627. [Google Scholar] [CrossRef]
- Rioux, J.D.; Abbas, A.K. Paths to understanding the genetic basis of autoimmune disease. Nature 2005, 435, 584–589. [Google Scholar] [CrossRef]
- Mohammed, J.P.; Fusakio, M.E.; Rainbow, D.B.; Moule, C.; Fraser, H.I.; Clark, J.; Todd, J.A.; Peterson, L.B.; Savage, P.B.; Wills-Karp, M.; et al. Identification of Cd101 as a susceptibility gene for Novosphingobium aromaticivorans-induced liver autoimmunity. J. Immunol. 2011, 187, 337. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lleo, A.; Bowlus, C.L.; Yang, G.X.; Invernizzi, P.; Podda, M.; Van de Water, J.; Ansari, A.A.; Coppel, R.L.; Worman, H.J.; Gores, G.J.; et al. Biliary apotopes and anti-mitochondrial antibodies activate innate immune responses in primary biliary cirrhosis. Hepatology 2010, 52, 987–998. [Google Scholar] [CrossRef] [PubMed]
- Kim, B.; Kaistha, S.D.; Rouse, B.T. Viruses and autoimmunity. Autoimmunity 2006, 39, 71–77. [Google Scholar] [CrossRef]
- Rojas, M.; Restrepo-Jiménez, P.; Monsalve, D.M.; Pacheco, Y.; Acosta-Ampudia, Y.; Ramírez-Santana, C.; Leung, P.S.C.; Ansari, A.A.; Gershwin, M.E.; Anaya, J.M. Molecular mimicry and autoimmunity. J. Autoimmun. 2018, 95, 100–123. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Z.S.; Granucci, F.; Yeh, L.; Schaffer, P.A.; Cantor, H. Molecular mimicry by herpes simplex virus-type 1: Autoimmune disease after viral infection. Science 1998, 279, 1344–1347. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Coppieters, K.T.; Wiberg, A.; von Herrath, M.G. Viral infections and molecular mimicry in type 1 diabetes. Apmis 2012, 120, 941–949. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gauntt, C.J.; Arizpe, H.M.; Higdon, A.L.; Wood, H.J.; Bowers, D.F.; Rozek, M.M.; Crawley, R. Molecular mimicry, anti-coxsackievirus B3 neutralizing monoclonal antibodies, and myocarditis. J. Immunol. 1995, 154, 2983–2995. [Google Scholar]
- Getts, D.R.; Chastain, E.M.; Terry, R.L.; Miller, S.D. Virus infection, antiviral immunity, and autoimmunity. Immunol. Rev. 2013, 255, 197–209. [Google Scholar] [CrossRef] [Green Version]
- Fujinami, R.S.; von Herrath, M.G.; Christen, U.; Whitton, J.L. Molecular mimicry, bystander activation, or viral persistence: Infections and autoimmune disease. Clin. Microbiol. Rev. 2006, 19, 80–94. [Google Scholar] [CrossRef] [Green Version]
- Kivity, S.; Agmon-Levin, N.; Blank, M.; Shoenfeld, Y. Infections and autoimmunity—Friends or foes? Trends Immunol. 2009, 30, 409–414. [Google Scholar] [CrossRef]
- Vanderlugt, C.L.; Miller, S.D. Epitope spreading in immune-mediated diseases: Implications for immunotherapy. Nat. Rev. Immunol. 2002, 2, 85–95. [Google Scholar] [CrossRef] [PubMed]
- Olson, J.K.; Croxford, J.L.; Miller, S.D. Virus-Induced Autoimmunity: Potential Role of Viruses in Initiation, Perpetuation, and Progression of T-Cell–Mediated Autoimmune Disease. Viral Immunol. 2001, 14, 227–250. [Google Scholar] [CrossRef] [PubMed]
- Garcia, A.F.; Yamaga, K.M.; Shafer, L.A.; Bollt, O.; Tam, E.K.; Cunningham, M.W.; Kurahara, D.K. Cardiac Myosin Epitopes Recognized by Autoantibody in Acute and Convalescent Rheumatic Fever. Pediatr. Infect. Dis. J. 2016, 35, 1021–1026. [Google Scholar] [CrossRef] [PubMed]
- Constantinescu, C.S.; Farooqi, N.; O’Brien, K.; Gran, B. Experimental autoimmune encephalomyelitis (EAE) as a model for multiple sclerosis (MS). Br. J. Pharmacol. 2011, 164, 1079–1106. [Google Scholar] [CrossRef] [Green Version]
- Leis, A.A.; Szatmary, G.; Ross, M.A.; Stokic, D.S. West nile virus infection and myasthenia gravis. Muscle Nerve 2014, 49, 26–29. [Google Scholar] [CrossRef]
- Miller, S.D.; Vanderlugt, C.L.; Begolka, W.S.; Pao, W.; Yauch, R.L.; Neville, K.L.; Katz-Levy, Y.; Carrizosa, A.; Kim, B.S. Persistent infection with Theiler’s virus leads to CNS autoimmunity via epitope spreading. Nat. Med. 1997, 3, 1133–1136. [Google Scholar] [CrossRef]
- Liebert, U.G.; Linington, C.; ter Meulen, V. Induction of autoimmune reactions to myelin basic protein in measles virus encephalitis in Lewis rats. J. Neuroimmunol. 1988, 17, 103–118. [Google Scholar] [CrossRef]
- Martinsen, V.; Kursula, P. Multiple sclerosis and myelin basic protein: Insights into protein disorder and disease. Amino Acids 2022, 54, 99–109. [Google Scholar] [CrossRef]
- Filippi, C.M.; von Herrath, M.G. Viral trigger for type 1 diabetes: Pros and cons. Diabetes 2008, 57, 2863–2871. [Google Scholar] [CrossRef] [Green Version]
- Kaufman, D.L.; Erlander, M.G.; Clare-Salzler, M.; Atkinson, M.A.; Maclaren, N.K.; Tobin, A.J. Autoimmunity to two forms of glutamate decarboxylase in insulin-dependent diabetes mellitus. J. Clin. Investig. 1992, 89, 283–292. [Google Scholar] [CrossRef]
- Pane, J.A.; Coulson, B.S. Lessons from the mouse: Potential contribution of bystander lymphocyte activation by viruses to human type 1 diabetes. Diabetologia 2015, 58, 1149–1159. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Honeyman, M.C.; Coulson, B.S.; Stone, N.L.; Gellert, S.A.; Goldwater, P.N.; Steele, C.E.; Couper, J.J.; Tait, B.D.; Colman, P.G.; Harrison, L.C. Association between rotavirus infection and pancreatic islet autoimmunity in children at risk of developing type 1 diabetes. Diabetes 2000, 49, 1319–1324. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pane, J.A.; Webster, N.L.; Coulson, B.S. Rotavirus activates lymphocytes from non-obese diabetic mice by triggering toll-like receptor 7 signaling and interferon production in plasmacytoid dendritic cells. PLoS Pathog. 2014, 10, e1003998. [Google Scholar] [CrossRef] [PubMed]
- Roman, L.M.; Simons, L.F.; Hammer, R.E.; Sambrook, J.F.; Gething, M.J. The expression of influenza virus hemagglutinin in the pancreatic beta cells of transgenic mice results in autoimmune diabetes. Cell 1990, 61, 383–396. [Google Scholar] [CrossRef]
- Tan, H.; Wang, C.; Yu, Y. H1N1 influenza: The trigger of diabetic ketoacidosis in a young woman with ketosis-prone diabetes. Am. J. Med. Sci. 2012, 343, 180–183. [Google Scholar] [CrossRef]
- Capua, I.; Mercalli, A.; Pizzuto, M.S.; Romero-Tejeda, A.; Kasloff, S.; De Battisti, C.; Bonfante, F.; Patrono, L.V.; Vicenzi, E.; Zappulli, V.; et al. Influenza A viruses grow in human pancreatic cells and cause pancreatitis and diabetes in an animal model. J. Virol. 2013, 87, 597–610. [Google Scholar] [CrossRef] [Green Version]
- Oughton, M.; Dascal, A.; Laporta, D.; Charest, H.; Afilalo, M.; Miller, M. Evidence of viremia in 2 cases of severe pandemic influenza A H1N1/09. Diagn. Microbiol. Infect. Dis. 2011, 70, 213–217. [Google Scholar] [CrossRef]
- Lönnrot, M.; Lynch, K.F.; Elding Larsson, H.; Lernmark, Å.; Rewers, M.J.; Törn, C.; Burkhardt, B.R.; Briese, T.; Hagopian, W.A.; She, J.X.; et al. Respiratory infections are temporally associated with initiation of type 1 diabetes autoimmunity: The TEDDY study. Diabetologia 2017, 60, 1931–1940. [Google Scholar] [CrossRef] [Green Version]
- Luo, G.; Ambati, A.; Lin, L.; Bonvalet, M.; Partinen, M.; Ji, X.; Maecker, H.T.; Mignot, E.J. Autoimmunity to hypocretin and molecular mimicry to flu in type 1 narcolepsy. Proc. Natl. Acad. Sci. USA 2018, 115, e12323–e12332. [Google Scholar] [CrossRef] [Green Version]
- Draborg, A.H.; Duus, K.; Houen, G. Epstein-Barr virus and systemic lupus erythematosus. Clin. Dev. Immunol. 2012, 2012, 370516. [Google Scholar] [CrossRef] [Green Version]
- Numazaki, K.; Goldman, H.; Seemayer, T.A.; Wong, I.; Wainberg, M.A. Infection by human cytomegalovirus and rubella virus of cultured human fetal islets of Langerhans. In Vivo 1990, 4, 49–54. [Google Scholar]
- Ramondetti, F.; Sacco, S.; Comelli, M.; Bruno, G.; Falorni, A.; Iannilli, A.; d’Annunzio, G.; Iafusco, D.; Songini, M.; Toni, S.; et al. Type 1 diabetes and measles, mumps and rubella childhood infections within the Italian Insulin-dependent Diabetes Registry. Diabet Med. 2012, 29, 761–766. [Google Scholar] [CrossRef] [PubMed]
- Christen, U.; von Herrath, M.G. Do viral infections protect from or enhance type 1 diabetes and how can we tell the difference? Cell Mol. Immunol. 2011, 8, 193–198. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Korber, B.; LaBute, M.; Yusim, K. Immunoinformatics comes of age. PLoS Comput. Biol. 2006, 2, e71. [Google Scholar] [CrossRef] [PubMed]
- Tomar, N.; De, R.K. Immunoinformatics: An integrated scenario. Immunology 2010, 131, 153–168. [Google Scholar] [CrossRef] [PubMed]
- NCBI Resource Coordinators. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2013, 41, D8–D20. [Google Scholar]
- Rahmati, S.; Abovsky, M.; Pastrello, C.; Kotlyar, M.; Lu, R.; Cumbaa, C.A.; Rahman, P.; Chandran, V.; Jurisica, I. pathDIP 4: An extended pathway annotations and enrichment analysis resource for human, model organisms and domesticated species. Nucleic Acids Res. 2020, 48, D479–D488. [Google Scholar] [CrossRef]
- Ahmed, R.K.S.; Maeurer, M.J. T-cell epitope mapping. Methods Mol. Biol. 2009, 524, 427–438. [Google Scholar]
- Bernecker, C.; Ostapczuk, M.; Vordenbäumen, S.; Ehlers, M.; Thiel, A.; Schinner, S.; Willenberg, H.; Scherbaum, W.A.; Schott, M. HLA-A2 phenotype may be protective against Graves’ disease but not against Hashimoto’s thyroiditis in Caucasians. Horm. Metab. Res. 2013, 45, 74–77. [Google Scholar] [CrossRef]
- Jiang, L.; Jiang, D.; Han, Y.; Shi, X.; Ren, C. Association of HLA-DPB1 polymorphisms with rheumatoid arthritis: A systemic review and meta-analysis. Int. J. Surg. 2018, 52, 98–104. [Google Scholar] [CrossRef]
- Venigalla, S.S.K.; Premakumar, S.; Janakiraman, V. A possible role for autoimmunity through molecular mimicry in alphavirus mediated arthritis. Sci Rep. 2020, 10, 938. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zanelli, E.; Breedveld, F.C.; de Vries, R.R. HLA association with autoimmune disease: A failure to protect? Rheumatology 2000, 39, 1060–1066. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Du, Z.; Su, H.; Wang, W.; Ye, L.; Wei, H.; Peng, Z.; Anishchenko, I.; Baker, D.; Yang, J. The trRosetta server for fast and accurate protein structure prediction. Nat. Protoc. 2021, 16, 5634–5651. [Google Scholar] [CrossRef] [PubMed]
- Kozakov, D.; Hall, D.R.; Xia, B.; Porter, K.A.; Padhorny, D.; Yueh, C.; Beglov, D.; Vajda, S. The ClusPro web server for protein–protein docking. Nat. Protoc. 2017, 12, 255–278. [Google Scholar] [CrossRef]
- Cusick, M.F.; Libbey, J.E.; Fujinami, R.S. Molecular mimicry as a mechanism of autoimmune disease. Clin. Rev. Allergy Immunol. 2012, 42, 102–111. [Google Scholar]
- Sanders, V.J.; Waddell, A.E.; Felisan, S.L.; Li, X.; Conrad, A.J.; Tourtellotte, W.W. Herpes simplex virus in postmortem multiple sclerosis brain tissue. Arch. Neurol. 1996, 53, 125–133. [Google Scholar] [CrossRef]
- Chucair-Elliott, A.J.; Conrady, C.; Zheng, M.; Kroll, C.M.; Lane, T.E.; Carr, D.J. Microglia-induced IL-6 protects against neuronal loss following HSV-1 infection of neural progenitor cells. Glia 2014, 62, 1418–1434. [Google Scholar] [CrossRef]
- Bradshaw, M.J.; Pawate, S.; Lennon, V.A.; Bloch, K.C.; Brown, K.M. Herpes simplex virus 1 encephalitis associated with voltage-gated calcium channel autoimmunity. Neurology 2015, 85, 2176–2177. [Google Scholar] [CrossRef] [Green Version]
- Yu, S.F.; Wu, H.C.; Tsai, W.C.; Yen, J.H.; Chiang, W.; You, C.Y.; Lu, S.N.; Chiang, L.C.; Chen, C.J. Detecting Epstein-Barr virus DNA from peripheral blood mononuclear cells in adult patients with systemic lupus erythematosus in Taiwan. Med. Microbiol. Immunol. 2005, 194, 115–120. [Google Scholar] [CrossRef]
- Yoon, S.I.; Logsdon, N.J.; Sheikh, F.; Donnelly, R.P.; Walter, M.R. Conformational changes mediate interleukin-10 receptor 2 (IL-10R2) binding to IL-10 and assembly of the signaling complex. J. Biol. Chem. 2006, 281, 35088–35096. [Google Scholar] [CrossRef] [Green Version]
- Sun, L.; Louie, M.C.; Vannella, K.M.; Wilke, C.A.; LeVine, A.M.; Moore, B.B.; Shanley, T.P. New concepts of IL-10-induced lung fibrosis: Fibrocyte recruitment and M2 activation in a CCL2/CCR2 axis. Am. J. Physiol. Lung Cell Mol. Physiol. 2011, 300, L341–L353. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Moudgil, K.D.; Choubey, D. Cytokines in autoimmunity: Role in induction, regulation, and treatment. J. Interferon. Cytokine Res. 2011, 31, 695–703. [Google Scholar] [CrossRef] [PubMed]
- Nicholas, J.; Ruvolo, V.; Zong, J.; Ciufo, D.; Guo, H.G.; Reitz, M.S.; Hayward, G.S. A single 13-kilobase divergent locus in the Kaposi sarcoma-associated herpesvirus (human herpesvirus 8) genome contains nine open reading frames that are homologous to or related to cellular proteins. J. Virol. 1997, 71, 1963–1974. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Balada, E.; Ramentol, M.; Felip, L.; Ordi-Ros, J.; Selva-O’Callaghan, A.; Simeón-Aznar, C.P.; Solans-Laqué, R.; Vilardell-Tarrés, M. Prevalence of HHV-8 in systemic autoimmune diseases. J. Clin. Virol. 2015, 62, 84–88. [Google Scholar] [CrossRef] [PubMed]
- Levin, G.; Duffin, K.L.; Obukowicz, M.G.; Hummert, S.L.; Fujiwara, H.; Needleman, P.; Raz, A. Differential metabolism of dihomo-gamma-linolenic acid and arachidonic acid by cyclo-oxygenase-1 and cyclo-oxygenase-2: Implications for cellular synthesis of prostaglandin E1 and prostaglandin E2. Biochem. J. 2002, 365, 489–496. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rotondi, M.; Chiovato, L.; Romagnani, S.; Serio, M.; Romagnani, P. Role of chemokines in endocrine autoimmune diseases. Endocr. Rev. 2007, 28, 492–520. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Seet, B.T.; Johnston, J.B.; Brunetti, C.R.; Barrett, J.W.; Everett, H.; Cameron, C.; Sypula, J.; Nazarian, S.H.; Lucas, A.; McFadden, G. Poxviruses and immune evasion. Annu. Rev. Immunol. 2003, 21, 377–423. [Google Scholar] [CrossRef] [PubMed]
- Huang, N.; Perl, A. Metabolism as a Target for Modulation in Autoimmune Diseases. Trends Immunol. 2018, 39, 562–576. [Google Scholar] [CrossRef]
- Pickett, B.E.; Sadat, E.L.; Zhang, Y.; Noronha, J.M.; Squires, R.B.; Hunt, V.; Liu, M.; Kumar, S.; Zaremba, S.; Gu, Z.; et al. ViPR: An open bioinformatics database and analysis resource for virology research. Nucleic Acids Res. 2012, 40, D593–D598. [Google Scholar] [CrossRef]
- Huang, Y.; Niu, B.; Gao, Y.; Fu, L.; Li, W. CD-HIT Suite: A web server for clustering and comparing biological sequences. Bioinformatics 2010, 26, 680–682. [Google Scholar] [CrossRef]
- Johnson, M.; Zaretskaya, I.; Raytselis, Y.; Merezhuk, Y.; McGinnis, S.; Madden, T.L. NCBI BLAST: A better web interface. Nucleic Acids Res. 2008, 36, W5–W9. [Google Scholar] [CrossRef] [PubMed]
- Fleri, W.; Paul, S.; Dhanda, S.K.; Mahajan, S.; Xu, X.; Peters, B.; Sette, A. The Immune Epitope Database and Analysis Resource in Epitope Discovery and Synthetic Vaccine Design. Front. Immunol. 2017, 8, 278. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Couch, G.S.; Greenblatt, D.M.; Meng, E.C.; Ferrin, T.E. UCSF Chimera--a visualization system for exploratory research and analysis. J. Comput. Chem. 2004, 25, 1605–1612. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rahman, N.; Begum, S.; Khan, A.; Afridi, S.G.; Khayam Sahibzada, M.U.; Atwah, B.; Alhindi, Z.; Khan, H. An insight in Salmonella typhi associated autoimmunity candidates’ prediction by molecular mimicry. Comput. Biol. Med. 2022, 148, 105865. [Google Scholar] [CrossRef] [PubMed]

| S. No | Query Sequence IDs | Subject Sequence IDs | Score | Query Coverage | Percent-Identity | E-Value |
|---|---|---|---|---|---|---|
| 1 | gb:AXN75085 | sp|P23921|RIR1_HUMAN | 3148 | 98 | 75.297 | 0 |
| 2 | gb:AST09466 | sp|P23921|RIR1_HUMAN | 3130 | 100 | 73.106 | 0 |
| 3 | gb:AEV80548 | sp|P35354|PGH2_HUMAN | 2452 | 98 | 73.729 | 0 |
| 4 | gb:AAY97564 | sp|P49916|DNLI3_HUMAN | 1700 | 98 | 55.124 | 0 |
| 5 | gb:AST09563 | sp|P49916|DNLI3_HUMAN | 1594 | 99 | 51.852 | 0 |
| 6 | gb:AZY90656 | sp|P31350|RIR2_HUMAN | 1412 | 95 | 80.625 | 0 |
| 7 | gb:AST09433 | sp|P31350|RIR2_HUMAN | 1399 | 99 | 80.312 | 0 |
| 8 | gb:QCA43223 | sp|P04818|TYSY_HUMAN | 1162 | 95 | 71.579 | 2.94 × 10−161 |
| 9 | gb:BBA90853 | sp|P04818|TYSY_HUMAN | 1153 | 89 | 69.333 | 2.63 × 10−159 |
| 10 | gb:AQY16903 | sp|Q9H2F3|3BHS7_HUMAN | 834 | 99 | 50.559 | 9.70 × 10−110 |
| 11 | gb:ADZ29327 | sp|Q9HC24|LFG4_HUMAN | 833 | 100 | 71.849 | 1.56 × 10−113 |
| 12 | gb:AZT86284 | sp|P07203|GPX1_HUMAN | 831 | 83 | 84.153 | 6.52 × 10−114 |
| 13 | gb:AXN75107 | sp|P04183|KITH_HUMAN | 656 | 97 | 69.006 | 1.45 × 10−87 |
| 14 | gb:AST09487 | sp|P04183|KITH_HUMAN | 625 | 98 | 64.205 | 6.60 × 10−83 |
| 15 | gb:QCF48225 | sp|P22301|IL10_HUMAN | 599 | 95 | 81.287 | 6.42 × 10−80 |
| 16 | gb:ABD28857 | sp|P00374|DYR_HUMAN | 498 | 86 | 50 | 9.21 × 10−64 |
| 17 | gb:AAY97032 | tr|H0YNW5|H0YNW5_HUMAN | 477 | 90 | 64.964 | 7.74 × 10−62 |
| 18 | gb:AUL80434 | tr|A0A0C4DGL3|A0A0C4DGL3_HUMAN | 394 | 92 | 55.882 | 1.42 × 10−49 |
| 19 | gb:AUL80132 | sp|Q8IV08|PLD3_HUMAN | 377 | 85 | 50.595 | 6.87 × 10−43 |
| 20 | gb:AAY97407 | sp|Q9HC24|LFG4_HUMAN | 367 | 97 | 63.248 | 9.72 × 10−45 |
| 21 | gb:AUL80484 | sp|P23921|RIR1_HUMAN | 306 | 86 | 57.143 | 2.35 × 10−33 |
| 22 | gb:AUL80431 | tr|A0A3B3ITT3|A0A3B3ITT3_HUMAN | 235 | 86 | 54.167 | 2.36 × 10−25 |
| 23 | gb:AEV80662 | sp|P09341|GROA_HUMAN | 208 | 71 | 65.079 | 1.00 × 10−22 |
| 24 | gb:AEV80661 | sp|P19875|CXCL2_HUMAN | 180 | 60 | 55.932 | 2.11 × 10−18 |
| S. No | Human Proteins | Human Peptides | Docking Score in Human HLA | Docking Score in Human TLR4 | Virus Proteins | Virus Peptides | Docking Score in Human HLA | Docking Score in Human TLR4 | Structural Mimicry | RMSD Value |
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | Ribonucleoside-diphosphate reductase subunit M2 | IFFSGSFASIFWLKK | −1105.4 | −829.4 | CPXV051 | IFFSGSFASIFWLKK | −1105.4 | −829.4 | IFFSGSFASIFWLKK ::::::::::::::: IFFSGSFASIFWLKK | 0.00 |
| 2 | Thymidine kinase, cytosolic | STELMRRVRRFQIAQ | −864 | −689 | thymidine kinase | STELIRRVRRYQIAQ | −853 | −596.4 | STELMRRVRRFQIAQ ::::::::::::::: STELIRRVRRYQIAQ | 0.1 |
| 3 | Ribonucleoside-diphosphate reductase large subunit | RDFSYNYFGFKTLER | −854 | −659.7 | ribonucleotide reductase large subunit | RDFSYNYFGFKTLER | −854 | −659.7 | RDFSYNYFGFKTLER ::::::::::::::: RDFSYNYFGFKTLEK | 0.1 |
| 4 | Prostaglandin G/H synthase 2 | MFAFFAQHFTHQFFK | −962 | −742.8 | prostaglandin G/H synthase 2 | MFAFFAQHFTHQFFK | −962 | −742.8 | MFAFFAQHFTHQFFK ::::::::::::::: MFAFFGQHFTHQFFR | 0.1 |
| 5 | Thymidylate synthase | TKRVFWKGVLEELLW | −962.9 | −747 | ORF13 | TKRVFWRAVVEELLW | −117.6 | −738 | TKRVFWKGVLEELLW ::::::::::::::: TKRVFWRAVVEELLW | 0.2 |
| 6 | Thymidylate synthase | VPFNIASYALLTYMI | −874.7 | −613 | ORF70 | VPFNIASYSLLTYML | −867 | −745.6 | VPFNIASYALLTYMI :::::::::::: VPFNIASYSLLTYML | 0.3 |
| 7 | Dihydrofolate reductase | RPLKGRINLVLSREL | −823 | −1070 | ORF2 | RPLAGRINVVLSRTL | −1087 | −782 | RPLKGRINLVLSREL ::::::::::::::: RPLAGRINVVLSRTL | 0.8 |
| 8 | DNA ligase 3 | FVFDCIYFNDVSLMD | −951.7 | −861 | ATP-dependent DNA ligase | FVFDCIYFNDVSLMD | −951.7 | −861 | FVFDCIYFNDVSLMD : :::::::::: FVFDCIYFNDVSLMD | 0.9 |
| 9 | thymidine kinase | LMRRVRRFQIAQYKC | −863.6 | −691 | thymidine kinase | LIRRVKRYQIAKYDC | −907 | −652 | LMRRVRRFQIAQYKC ::::::::::::::: LIRRVKRYQIAKYDC | 1 |
| 10 | Ribonucleoside-diphosphate reductase large subunit | AGRRAAGASVATELR | −875.3 | −781.9 | ribonucleotide reductas | LMSLIAYCQSATELR | −917 | −806.5 | AGRRAAGASVATELR :::::::::: LMSLIAYCQSATELR | 1.0 |
| 11 | Growth-regulated alpha protein | IIYDRDFSYNYFGFK | −661 | −602.4 | chemokine vCXCL7 | IINDRDFSYNYFGFK | −789.5 | −580.5 | IIYDRDFSYNYFGFK ::::::::: IINDRDFSYNYFGFK | 1.0 |
| 12 | C-X-C motif chemokine 2 | LLLVAASRRAAGAPL | −763 | −781 | chemokine vCXCL6 | SRLLVATLLGTLLAC | −1006.9 | −598 | LLLVAASRRAAGAPL ::::::::: SRLLVATLLGTLLAC | 1.5 |
| 13 | DNA ligase 3 | CLFVFDCIYFNDVSL | −820.4 | −742.3 | DNA ligase | CLFVFDCLYFDGFDM | −942 | −878 | CLFVFDCIYFNDVSL ::: ::::::::: CLFVFDCLYFDGFDM | 2.1 |
| S/No | HLA-DP | HLA-DQ | HLA-DR |
|---|---|---|---|
| 1 | DPA1*01:03-DPB1*02:01 | DQA1*01:02–DQB1*06:02 | DRB1*03:01 |
| 2 | DPA1*02:01–DPB1*05:01 | -DQA1*04:01–DQB1*04:02 | DRB1*04:04 |
| 3 | DPA1*03:01–DPB1*04:02 | DQA1*05:01–DQB1*03:01 | DRB1*07:01 |
| 4 | DPA1*01:03–DPB1*04:01 | DQA1*01:01–DQB1*05:01 | DRB1*11:01 |
| 5 | DPA1*01:03-DPB1*03:01-DPB1*04:01 | DQA1*05:01–DQB1*03:01 | DRB1*13:02 |
| 6 | DPA1*02:01-DPB1*01:01 | DQA1*03:01–QB1*03:02 | DRB3*01:01 |
| DRB5*01:01 | |||
| DRB1*01:01 | |||
| DRB4*01:01 | |||
| DRB1*04:01 | |||
| DRB1*15:01 | |||
| DRB1*04:05 | |||
| DRB1*11:01 | |||
| DRB1*08:02 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Begum, S.; Aiman, S.; Ahmad, S.; Samad, A.; Almehmadi, M.; Allahyani, M.; Aljuaid, A.; Afridi, S.G.; Khan, A. Molecular Mimicry Analyses Unveiled the Human Herpes Simplex and Poxvirus Epitopes as Possible Candidates to Incite Autoimmunity. Pathogens 2022, 11, 1362. https://doi.org/10.3390/pathogens11111362
Begum S, Aiman S, Ahmad S, Samad A, Almehmadi M, Allahyani M, Aljuaid A, Afridi SG, Khan A. Molecular Mimicry Analyses Unveiled the Human Herpes Simplex and Poxvirus Epitopes as Possible Candidates to Incite Autoimmunity. Pathogens. 2022; 11(11):1362. https://doi.org/10.3390/pathogens11111362
Chicago/Turabian StyleBegum, Sara, Sara Aiman, Shujaat Ahmad, Abdus Samad, Mazen Almehmadi, Mamdouh Allahyani, Abdulelah Aljuaid, Sahib Gul Afridi, and Asifullah Khan. 2022. "Molecular Mimicry Analyses Unveiled the Human Herpes Simplex and Poxvirus Epitopes as Possible Candidates to Incite Autoimmunity" Pathogens 11, no. 11: 1362. https://doi.org/10.3390/pathogens11111362
APA StyleBegum, S., Aiman, S., Ahmad, S., Samad, A., Almehmadi, M., Allahyani, M., Aljuaid, A., Afridi, S. G., & Khan, A. (2022). Molecular Mimicry Analyses Unveiled the Human Herpes Simplex and Poxvirus Epitopes as Possible Candidates to Incite Autoimmunity. Pathogens, 11(11), 1362. https://doi.org/10.3390/pathogens11111362







