Phylogenetic Inference of the 2022 Highly Pathogenic H7N3 Avian Influenza Outbreak in Northern Mexico
Abstract
:1. Introduction
2. Results
2.1. Phylogenetic Relationships
2.2. Phylogenetic Transmission Network
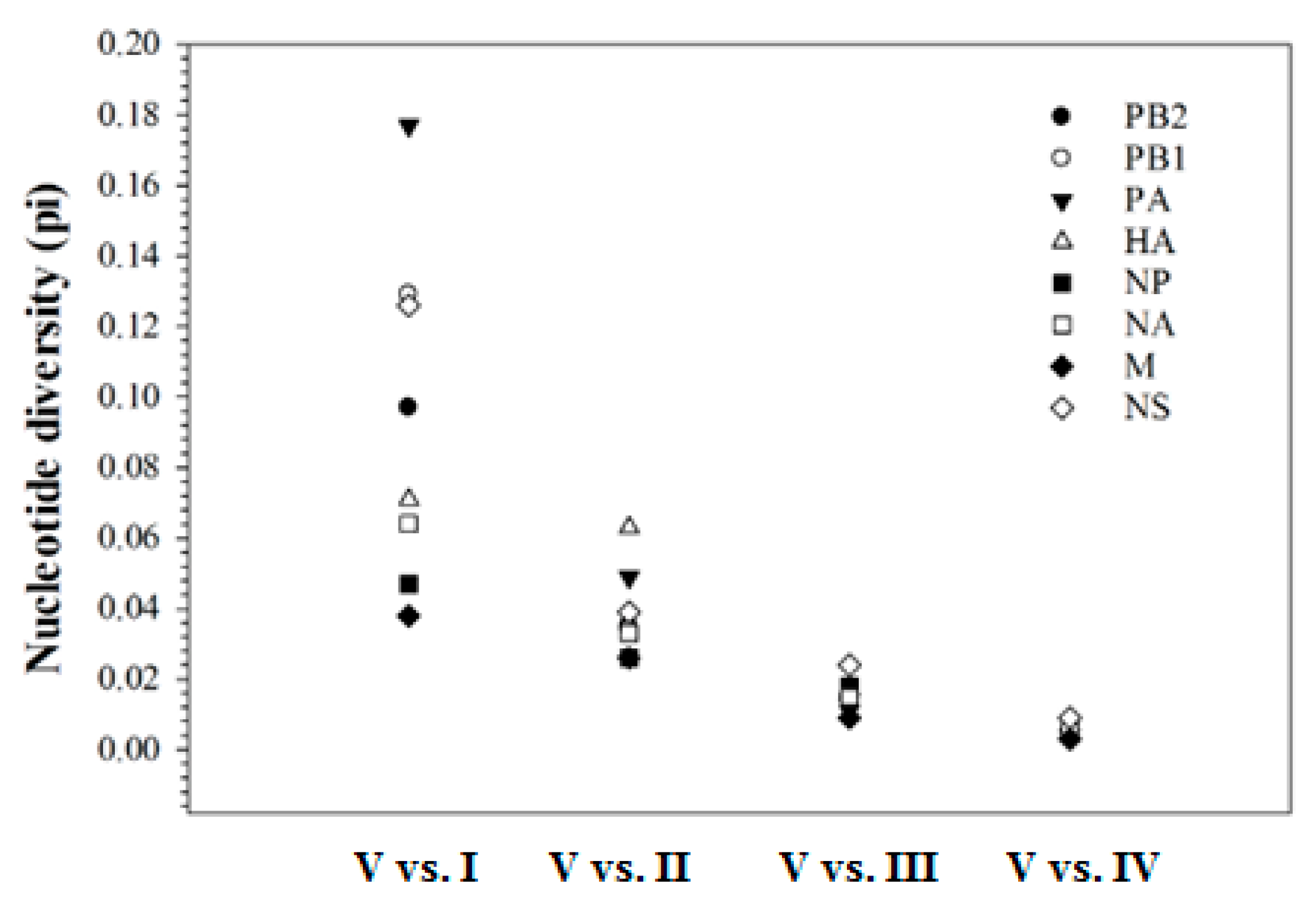
2.3. Genetic Diversity in Mexican H7N3 HPAIVs
2.4. Selection Pressures Acting on the Viral Genes
2.5. Antigenic Evolution of HA Gene
3. Discussion
4. Materials and Methods
4.1. Virus Isolation and Genome Sequencing
4.2. Nucleotide Sequences Used in the Study
4.3. Sequence Analyses
4.4. Transmission Network Construction
4.5. Analysis of Selection Pressure
4.6. 3D Structural Analyses
4.7. Antigenic and Glycosylation Analyses of HA Protein
4.8. Nucleotide Sequence Accession Numbers
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Webby, R.J.; Webster, R.G. Emergence of Influenza A Viruses. Philos. Trans. R. Soc. Lond. Ser. B Biol. Sci. 2001, 356, 1817–1828. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Deshpande, K.L.; Fried, V.A.; Ando, M.; Webster, R.G. Glycosylation Affects Cleavage of an H5N2 Influenza Virus Hemagglutinin and Regulates Virulence. Proc. Natl. Acad. Sci. USA 1987, 84, 36–40. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, L.M.; Blixt, O.; Stevens, J.; Lipatov, A.S.; Davis, C.T.; Collins, B.E.; Cox, N.J.; Paulson, J.C.; Donis, R.O. In Vitro Evolution of H5N1 Avian Influenza Virus Toward Human-Type Receptor Specificity. Virology 2012, 422, 105–113. [Google Scholar] [CrossRef] [Green Version]
- Bonfanti, L.; Monne, I.; Tamba, M.; Santucci, U.; Massi, P.; Patregnani, T.; Loli Piccolomini, L.; Natalini, S.; Ferri, G.; Cattoli, G.; et al. Highly Pathogenic H7N7 Avian Influenza in Italy. Vet. Rec. 2014, 174, 382. [Google Scholar] [CrossRef] [PubMed]
- Belser, J.A.; Lash, R.R.; Garg, S.; Tumpey, T.M.; Maines, T.R. The Eyes have it: Influenza Virus Infection Beyond the Respiratory Tract. Lancet Infect. Dis. 2018, 18, e220–e227. [Google Scholar] [CrossRef]
- Shu, Y.; McCauley, J. GISAID: Global Initiative on Sharing all Influenza Data—From Vision to Reality. Eurosurveillance 2017, 22, 30494. [Google Scholar] [CrossRef] [Green Version]
- Banks, J.; Speidel, E.C.; Harris, P.A.; Alexander, D.J. Phylogenetic Analysis of Influenza A Viruses of H9 Haemagglutinin Subtype. Avian Pathol. 2000, 29, 353–359. [Google Scholar] [CrossRef]
- Escalera-Zamudio, M.; Golden, M.; Gutierrez, B.; Theze, J.; Keown, J.R.; Carrique, L.; Bowden, T.A.; Pybus, O.G. Publisher Correction: Parallel Evolution in the Emergence of Highly Pathogenic Avian Influenza A Viruses. Nat. Commun. 2020, 11, 5511. [Google Scholar] [CrossRef]
- Lopez-Martinez, I.; Balish, A.; Barrera-Badillo, G.; Jones, J.; Nunez-Garcia, T.E.; Jang, Y.; Aparicio-Antonio, R.; Azziz-Baumgartner, E.; Belser, J.A.; Ramirez-Gonzalez, J.E.; et al. Highly Pathogenic Avian Influenza A(H7N3) Virus in Poultry Workers, Mexico, 2012. Emerg. Infect. Dis. 2013, 19, 1531–1534. [Google Scholar] [CrossRef]
- Lu, L.; Lycett, S.J.; Leigh Brown, A.J. Determining the Phylogenetic and Phylogeographic Origin of Highly Pathogenic Avian Influenza (H7N3) in Mexico. PLoS ONE 2014, 9, e107330. [Google Scholar] [CrossRef]
- Maurer-Stroh, S.; Lee, R.T.; Gunalan, V.; Eisenhaber, F. The Highly Pathogenic H7N3 Avian Influenza Strain from July 2012 in Mexico Acquired an Extended Cleavage Site through Recombination with Host 28S rRNA. Virol. J. 2013, 10, 139. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Youk, S.; Lee, D.H.; Ferreira, H.L.; Afonso, C.L.; Absalon, A.E.; Swayne, D.E.; Suarez, D.L.; Pantin-Jackwood, M.J. Rapid Evolution of Mexican H7N3 Highly Pathogenic Avian Influenza Viruses in Poultry. PLoS ONE 2019, 14, e0222457. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Trovao, N.S.; Talavera, G.A.; Nelson, M.I.; Perez de la Rosa, J.D. Evolution of Highly Pathogenic H7N3 Avian Influenza Viruses in Mexico. Zoonoses Public Health 2020, 67, 318–323. [Google Scholar] [CrossRef] [PubMed]
- Gao, W.; Zu, Z.; Liu, J.; Song, J.; Wang, X.; Wang, C.; Liu, L.; Tong, Q.; Wang, M.; Sun, H.; et al. Prevailing I292V PB2 Mutation in Avian Influenza H9N2 Virus Increases Viral Polymerase Function and Attenuates IFN-Beta Induction in Human Cells. J. Gen. Virol. 2019, 100, 1273–1281. [Google Scholar] [CrossRef]
- Hu, M.; Yuan, S.; Zhang, K.; Singh, K.; Ma, Q.; Zhou, J.; Chu, H.; Zheng, B.J. PB2 Substitutions V598T/I Increase the Virulence of H7N9 Influenza A Virus in Mammals. Virology 2017, 501, 92–101. [Google Scholar] [CrossRef]
- Song, W.; Wang, P.; Mok, B.W.; Lau, S.Y.; Huang, X.; Wu, W.L.; Zheng, M.; Wen, X.; Yang, S.; Chen, Y.; et al. The K526R Substitution in Viral Protein PB2 Enhances the Effects of E627K on Influenza Virus Replication. Nat. Commun. 2014, 5, 5509. [Google Scholar] [CrossRef] [Green Version]
- Schat, K.A.; Bingham, J.; Butler, J.M.; Chen, L.M.; Lowther, S.; Crowley, T.M.; Moore, R.J.; Donis, R.O.; Lowenthal, J.W. Role of Position 627 of PB2 and the Multibasic Cleavage Site of the Hemagglutinin in the Virulence of H5N1 Avian Influenza Virus in Chickens and Ducks. PLoS ONE 2012, 7, e30960. [Google Scholar] [CrossRef] [Green Version]
- Sun, H.; Cui, P.; Song, Y.; Qi, Y.; Li, X.; Qi, W.; Xu, C.; Jiao, P.; Liao, M. PB2 Segment Promotes High-Pathogenicity of H5N1 Avian Influenza Viruses in Mice. Front. Microbiol. 2015, 6, 73. [Google Scholar] [CrossRef] [Green Version]
- Li, J.; Ishaq, M.; Prudence, M.; Xi, X.; Hu, T.; Liu, Q.; Guo, D. Single Mutation at the Amino Acid Position 627 of PB2 that Leads to Increased Virulence of an H5N1 Avian Influenza Virus during Adaptation in Mice can be Compensated by Multiple Mutations at Other Sites of PB2. Virus Res. 2009, 144, 123–129. [Google Scholar] [CrossRef]
- Elgendy, E.M.; Arai, Y.; Kawashita, N.; Daidoji, T.; Takagi, T.; Ibrahim, M.S.; Nakaya, T.; Watanabe, Y. Identification of Polymerase Gene Mutations that Affect Viral Replication in H5N1 Influenza Viruses Isolated from Pigeons. J. Gen. Virol. 2017, 98, 6–17. [Google Scholar] [CrossRef]
- Feng, X.; Wang, Z.; Shi, J.; Deng, G.; Kong, H.; Tao, S.; Li, C.; Liu, L.; Guan, Y.; Chen, H. Glycine at Position 622 in PB1 Contributes to the Virulence of H5N1 Avian Influenza Virus in Mice. J. Virol. 2015, 90, 1872–1879. [Google Scholar] [CrossRef] [Green Version]
- Gabriel, G.; Abram, M.; Keiner, B.; Wagner, R.; Klenk, H.D.; Stech, J. Differential Polymerase Activity in Avian and Mammalian Cells Determines Host Range of Influenza Virus. J. Virol. 2007, 81, 9601–9604. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schmolke, M.; Manicassamy, B.; Pena, L.; Sutton, T.; Hai, R.; Varga, Z.T.; Hale, B.G.; Steel, J.; Perez, D.R.; Garcia-Sastre, A. Differential Contribution of PB1-F2 to the Virulence of Highly Pathogenic H5N1 Influenza A Virus in Mammalian and Avian Species. PLoS Pathog. 2011, 7, e1002186. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Conenello, G.M.; Zamarin, D.; Perrone, L.A.; Tumpey, T.; Palese, P. A Single Mutation in the PB1-F2 of H5N1 (HK/97) and 1918 Influenza A Viruses Contributes to Increased Virulence. PLoS Pathog. 2007, 3, 1414–1421. [Google Scholar] [CrossRef] [PubMed]
- Yamayoshi, S.; Yamada, S.; Fukuyama, S.; Murakami, S.; Zhao, D.; Uraki, R.; Watanabe, T.; Tomita, Y.; Macken, C.; Neumann, G.; et al. Virulence-Affecting Amino Acid Changes in the PA Protein of H7N9 Influenza A Viruses. J. Virol. 2014, 88, 3127–3134. [Google Scholar] [CrossRef] [Green Version]
- Hu, M.; Chu, H.; Zhang, K.; Singh, K.; Li, C.; Yuan, S.; Chow, B.K.; Song, W.; Zhou, J.; Zheng, B.J. Amino Acid Substitutions V63I Or A37S/I61T/V63I/V100A in the PA N-Terminal Domain Increase the Virulence of H7N7 Influenza A Virus. Sci. Rep. 2016, 6, 37800. [Google Scholar] [CrossRef] [Green Version]
- Hu, M.; Yuan, S.; Ye, Z.W.; Singh, K.; Li, C.; Shuai, H.; Fai, N.; Chow, B.K.C.; Chu, H.; Zheng, B.J. PAN Substitutions A37S, A37S/I61T and A37S/V63I Attenuate the Replication of H7N7 Influenza A Virus by Impairing the Polymerase and Endonuclease Activities. J. Gen. Virol. 2017, 98, 364–373. [Google Scholar] [CrossRef]
- DesRochers, B.L.; Chen, R.E.; Gounder, A.P.; Pinto, A.K.; Bricker, T.; Linton, C.N.; Rogers, C.D.; Williams, G.D.; Webby, R.J.; Boon, A.C. Residues in the PB2 and PA Genes Contribute to the Pathogenicity of Avian H7N3 Influenza A Virus in DBA/2 Mice. Virology 2016, 494, 89–99. [Google Scholar] [CrossRef]
- Song, J.; Feng, H.; Xu, J.; Zhao, D.; Shi, J.; Li, Y.; Deng, G.; Jiang, Y.; Li, X.; Zhu, P.; et al. The PA Protein Directly Contributes to the Virulence of H5N1 Avian Influenza Viruses in Domestic Ducks. J. Virol. 2011, 85, 2180–2188. [Google Scholar] [CrossRef] [Green Version]
- Song, J.; Xu, J.; Shi, J.; Li, Y.; Chen, H. Synergistic Effect of S224P and N383D Substitutions in the PA of H5N1 Avian Influenza Virus Contributes to Mammalian Adaptation. Sci. Rep. 2015, 5, 10510. [Google Scholar] [CrossRef]
- Yamayoshi, S.; Kiso, M.; Yasuhara, A.; Ito, M.; Shu, Y.; Kawaoka, Y. Enhanced Replication of Highly Pathogenic Influenza A(H7N9) Virus in Humans. Emerg. Infect. Dis. 2018, 24, 746–750. [Google Scholar] [CrossRef] [Green Version]
- Wang, W.; Lu, B.; Zhou, H.; Suguitan, A.L., Jr.; Cheng, X.; Subbarao, K.; Kemble, G.; Jin, H. Glycosylation at 158N of the Hemagglutinin Protein and Receptor Binding Specificity Synergistically Affect the Antigenicity and Immunogenicity of a Live Attenuated H5N1 A/Vietnam/1203/2004 Vaccine Virus in Ferrets. J. Virol. 2010, 84, 6570–6577. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Watanabe, Y.; Ibrahim, M.S.; Ellakany, H.F.; Kawashita, N.; Mizuike, R.; Hiramatsu, H.; Sriwilaijaroen, N.; Takagi, T.; Suzuki, Y.; Ikuta, K. Acquisition of Human-Type Receptor Binding Specificity by New H5N1 Influenza Virus Sublineages during their Emergence in Birds in Egypt. PLoS Pathog. 2011, 7, e1002068. [Google Scholar] [CrossRef] [PubMed]
- Zhu, W.; Zhou, J.; Li, Z.; Yang, L.; Li, X.; Huang, W.; Zou, S.; Chen, W.; Wei, H.; Tang, J.; et al. Biological Characterisation of the Emerged Highly Pathogenic Avian Influenza (HPAI) A(H7N9) Viruses in Humans, in Mainland China, 2016 to 2017. Eurosurveillance 2017, 22, 30533. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sun, X.; Belser, J.A.; Pulit-Penaloza, J.A.; Zeng, H.; Lewis, A.; Shieh, W.J.; Tumpey, T.M.; Maines, T.R. Pathogenesis and Transmission Assessments of Two H7N8 Influenza A Viruses Recently Isolated from Turkey Farms in Indiana using Mouse and Ferret Models. J. Virol. 2016, 90, 10936–10944. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Abdelwhab, E.M.; Veits, J.; Ulrich, R.; Kasbohm, E.; Teifke, J.P.; Mettenleiter, T.C. Composition of the Hemagglutinin Polybasic Proteolytic Cleavage Motif Mediates Variable Virulence of H7N7 Avian Influenza Viruses. Sci. Rep. 2016, 6, 39505. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sun, X.; Belser, J.A.; Yang, H.; Pulit-Penaloza, J.A.; Pappas, C.; Brock, N.; Zeng, H.; Creager, H.M.; Stevens, J.; Maines, T.R. Identification of Key Hemagglutinin Residues Responsible for Cleavage, Acid Stability, and Virulence of Fifth-Wave Highly Pathogenic Avian Influenza A(H7N9) Viruses. Virology 2019, 535, 232–240. [Google Scholar] [CrossRef]
- Wasilenko, J.L.; Sarmento, L.; Pantin-Jackwood, M.J. A Single Substitution in Amino Acid 184 of the NP Protein Alters the Replication and Pathogenicity of H5N1 Avian Influenza Viruses in Chickens. Arch. Virol. 2009, 154, 969–979. [Google Scholar] [CrossRef]
- Kode, S.S.; Pawar, S.D.; Tare, D.S.; Keng, S.S.; Hurt, A.C.; Mullick, J. A Novel I117T Substitution in Neuraminidase of Highly Pathogenic Avian Influenza H5N1 Virus Conferring Reduced Susceptibility to Oseltamivir and Zanamivir. Vet. Microbiol. 2019, 235, 21–24. [Google Scholar] [CrossRef]
- Fan, S.; Deng, G.; Song, J.; Tian, G.; Suo, Y.; Jiang, Y.; Guan, Y.; Bu, Z.; Kawaoka, Y.; Chen, H. Two Amino Acid Residues in the Matrix Protein M1 Contribute to the Virulence Difference of H5N1 Avian Influenza Viruses in Mice. Virology 2009, 384, 28–32. [Google Scholar] [CrossRef]
- Nao, N.; Kajihara, M.; Manzoor, R.; Maruyama, J.; Yoshida, R.; Muramatsu, M.; Miyamoto, H.; Igarashi, M.; Eguchi, N.; Sato, M.; et al. A Single Amino Acid in the M1 Protein Responsible for the Different Pathogenic Potentials of H5N1 Highly Pathogenic Avian Influenza Virus Strains. PLoS ONE 2015, 10, e0137989. [Google Scholar] [CrossRef] [Green Version]
- Lan, Y.; Zhang, Y.; Dong, L.; Wang, D.; Huang, W.; Xin, L.; Yang, L.; Zhao, X.; Li, Z.; Wang, W.; et al. A Comprehensive Surveillance of Adamantane Resistance among Human Influenza A Virus Isolated from Mainland China between 1956 and 2009. Antivir. Ther. 2010, 15, 853–859. [Google Scholar] [CrossRef]
- Cheung, C.L.; Rayner, J.M.; Smith, G.J.; Wang, P.; Naipospos, T.S.; Zhang, J.; Yuen, K.Y.; Webster, R.G.; Peiris, J.S.; Guan, Y.; et al. Distribution of Amantadine-Resistant H5N1 Avian Influenza Variants in Asia. J. Infect. Dis. 2006, 193, 1626–1629. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bean, W.J.; Threlkeld, S.C.; Webster, R.G. Biologic Potential of Amantadine-Resistant Influenza A Virus in an Avian Model. J. Infect. Dis. 1989, 159, 1050–1056. [Google Scholar] [CrossRef]
- Ilyushina, N.A.; Govorkova, E.A.; Webster, R.G. Detection of Amantadine-Resistant Variants among Avian Influenza Viruses Isolated in North America and Asia. Virology 2005, 341, 102–106. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- He, G.; Qiao, J.; Dong, C.; He, C.; Zhao, L.; Tian, Y. Amantadine-Resistance among H5N1 Avian Influenza Viruses Isolated in Northern China. Antivir. Res. 2008, 77, 72–76. [Google Scholar] [CrossRef]
- Puthavathana, P.; Auewarakul, P.; Charoenying, P.C.; Sangsiriwut, K.; Pooruk, P.; Boonnak, K.; Khanyok, R.; Thawachsupa, P.; Kijphati, R.; Sawanpanyalert, P. Molecular Characterization of the Complete Genome of Human Influenza H5N1 Virus Isolates from Thailand. J. Gen. Virol. 2005, 86, 423–433. [Google Scholar] [CrossRef] [PubMed]
- Buranathai, C.; Amonsin, A.; Chaisigh, A.; Theamboonlers, A.; Pariyothorn, N.; Poovorawan, Y. Surveillance Activities and Molecular Analysis of H5N1 Highly Pathogenic Avian Influenza Viruses from Thailand, 2004–2005. Avian Dis. 2007, 51, 194–200. [Google Scholar] [CrossRef]
- Jiao, P.; Tian, G.; Li, Y.; Deng, G.; Jiang, Y.; Liu, C.; Liu, W.; Bu, Z.; Kawaoka, Y.; Chen, H. A Single-Amino-Acid Substitution in the NS1 Protein Changes the Pathogenicity of H5N1 Avian Influenza Viruses in Mice. J. Virol. 2008, 82, 1146–1154. [Google Scholar] [CrossRef] [Green Version]
- Ayllon, J.; Domingues, P.; Rajsbaum, R.; Miorin, L.; Schmolke, M.; Hale, B.G.; Garcia-Sastre, A. A Single Amino Acid Substitution in the Novel H7N9 Influenza A Virus NS1 Protein Increases CPSF30 Binding and Virulence. J. Virol. 2014, 88, 12146–12151. [Google Scholar] [CrossRef]
- Li, J.; Zhang, K.; Chen, Q.; Zhang, X.; Sun, Y.; Bi, Y.; Zhang, S.; Gu, J.; Li, J.; Liu, D.; et al. Three Amino Acid Substitutions in the NS1 Protein Change the Virus Replication of H5N1 Influenza Virus in Human Cells. Virology 2018, 519, 64–73. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Jiang, Y.; Jiao, P.; Wang, A.; Zhao, F.; Tian, G.; Wang, X.; Yu, K.; Bu, Z.; Chen, H. The NS1 Gene Contributes to the Virulence of H5N1 Avian Influenza Viruses. J. Virol. 2006, 80, 11115–11123. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kuo, R.L.; Krug, R.M. Influenza a Virus Polymerase is an Integral Component of the CPSF30-NS1A Protein Complex in Infected Cells. J. Virol. 2009, 83, 1611–1616. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Spesock, A.; Malur, M.; Hossain, M.J.; Chen, L.M.; Njaa, B.L.; Davis, C.T.; Lipatov, A.S.; York, I.A.; Krug, R.M.; Donis, R.O. The Virulence of 1997 H5N1 Influenza Viruses in the Mouse Model is Increased by Correcting a Defect in their NS1 Proteins. J. Virol. 2011, 85, 7048–7058. [Google Scholar] [CrossRef] [Green Version]
- Kanrai, P.; Mostafa, A.; Madhugiri, R.; Lechner, M.; Wilk, E.; Schughart, K.; Ylosmaki, L.; Saksela, K.; Ziebuhr, J.; Pleschka, S. Identification of Specific Residues in Avian Influenza A Virus NS1 that Enhance Viral Replication and Pathogenicity in Mammalian Systems. J. Gen. Virol. 2016, 97, 2135–2148. [Google Scholar] [CrossRef]
- Jackson, D.; Hossain, M.J.; Hickman, D.; Perez, D.R.; Lamb, R.A. A New Influenza Virus Virulence Determinant: The NS1 Protein Four C-Terminal Residues Modulate Pathogenicity. Proc. Natl. Acad. Sci. USA 2008, 105, 4381–4386. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Richard, M.; Fouchier, R.; Monne, I.; Kuiken, T. Mechanisms and Risk Factors for Mutation from Low to Highly Pathogenic Avian Influenza Virus 2017. EFSA Support. Publ. 2017, 14, 1287E. [Google Scholar]
- Rohm, C.; Horimoto, T.; Kawaoka, Y.; Suss, J.; Webster, R.G. Do Hemagglutinin Genes of Highly Pathogenic Avian Influenza Viruses Constitute Unique Phylogenetic Lineages? Virology 1995, 209, 664–670. [Google Scholar] [CrossRef] [Green Version]
- Bataille, A.; van der Meer, F.; Stegeman, A.; Koch, G. Evolutionary Analysis of Inter-Farm Transmission Dynamics in a Highly Pathogenic Avian Influenza Epidemic. PLoS Pathog. 2011, 7, e1002094. [Google Scholar] [CrossRef] [Green Version]
- Xu, W.; Berhane, Y.; Dube, C.; Liang, B.; Pasick, J.; VanDomselaar, G.; Alexandersen, S. Epidemiological and Evolutionary Inference of the Transmission Network of the 2014 Highly Pathogenic Avian Influenza H5N2 Outbreak in British Columbia, Canada. Sci. Rep. 2016, 6, 30858. [Google Scholar] [CrossRef] [Green Version]
- Bos, M.E.; Van Boven, M.; Nielen, M.; Bouma, A.; Elbers, A.R.; Nodelijk, G.; Koch, G.; Stegeman, A.; De Jong, M.C. Estimating the Day of Highly Pathogenic Avian Influenza (H7N7) Virus Introduction into a Poultry Flock Based on Mortality Data. Vet. Res. 2007, 38, 493–504. [Google Scholar] [CrossRef] [PubMed]
- Xu, W.; Navarro-Lopez, R.; Solis-Hernandez, M.; Liljehult-Fuentes, F.; Molina-Montiel, M.; Lagunas-Ayala, M.; Rocha-Martinez, M.; Ferrara-Tijera, E.; Perez de la Rosa, J.; Berhane, Y. Evolutionary Dynamics of Mexican Lineage H5N2 Avian Influenza Viruses. Viruses 2022, 14, 958. [Google Scholar] [CrossRef] [PubMed]
- Zhou, B.; Donnelly, M.E.; Scholes, D.T.; St George, K.; Hatta, M.; Kawaoka, Y.; Wentworth, D.E. Single-Reaction Genomic Amplification Accelerates Sequencing and Vaccine Production for Classical and Swine Origin Human Influenza a Viruses. J. Virol. 2009, 83, 10309–10313. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Stamatakis, A. RAxML Version 8: A Tool for Phylogenetic Analysis and Post-Analysis of Large Phylogenies. Bioinformatics 2014, 30, 1312–1313. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bouckaert, R.; Vaughan, T.G.; Barido-Sottani, J.; Duchene, S.; Fourment, M.; Gavryushkina, A.; Heled, J.; Jones, G.; Kuhnert, D.; De Maio, N.; et al. BEAST 2.5: An Advanced Software Platform for Bayesian Evolutionary Analysis. PLoS Comput. Biol. 2019, 15, e1006650. [Google Scholar] [CrossRef] [Green Version]
- Kosakovsky Pond, S.L.; Frost, S.D. Not so Different After all: A Comparison of Methods for Detecting Amino Acid Sites Under Selection. Mol. Biol. Evol. 2005, 22, 1208–1222. [Google Scholar] [CrossRef] [Green Version]
- Murrell, B.; Wertheim, J.O.; Moola, S.; Weighill, T.; Scheffler, K.; Kosakovsky Pond, S.L. Detecting Individual Sites Subject to Episodic Diversifying Selection. PLoS Genet. 2012, 8, e1002764. [Google Scholar] [CrossRef] [Green Version]
- Murrell, B.; Moola, S.; Mabona, A.; Weighill, T.; Sheward, D.; Kosakovsky Pond, S.L.; Scheffler, K. FUBAR: A Fast, Unconstrained Bayesian Approximation for Inferring Selection. Mol. Biol. Evol. 2013, 30, 1196–1205. [Google Scholar]
- Delport, W.; Poon, A.F.; Frost, S.D.; Kosakovsky Pond, S.L. Datamonkey 2010: A Suite of Phylogenetic Analysis Tools for Evolutionary Biology. Bioinformatics 2010, 26, 2455–2457. [Google Scholar] [CrossRef] [Green Version]
- Xiong, X.; Martin, S.R.; Haire, L.F.; Wharton, S.A.; Daniels, R.S.; Bennett, M.S.; McCauley, J.W.; Collins, P.J.; Walker, P.A.; Skehel, J.J.; et al. Receptor Binding by an H7N9 Influenza Virus from Humans. Nature 2013, 499, 496–499. [Google Scholar] [CrossRef] [PubMed]
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Couch, G.S.; Greenblatt, D.M.; Meng, E.C.; Ferrin, T.E. UCSF Chimera—A Visualization System for Exploratory Research and Analysis. J. Comput. Chem. 2004, 25, 1605–1612. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An Integrative Toolkit Developed for Interactive Analyses of Big Biological Data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef] [PubMed]

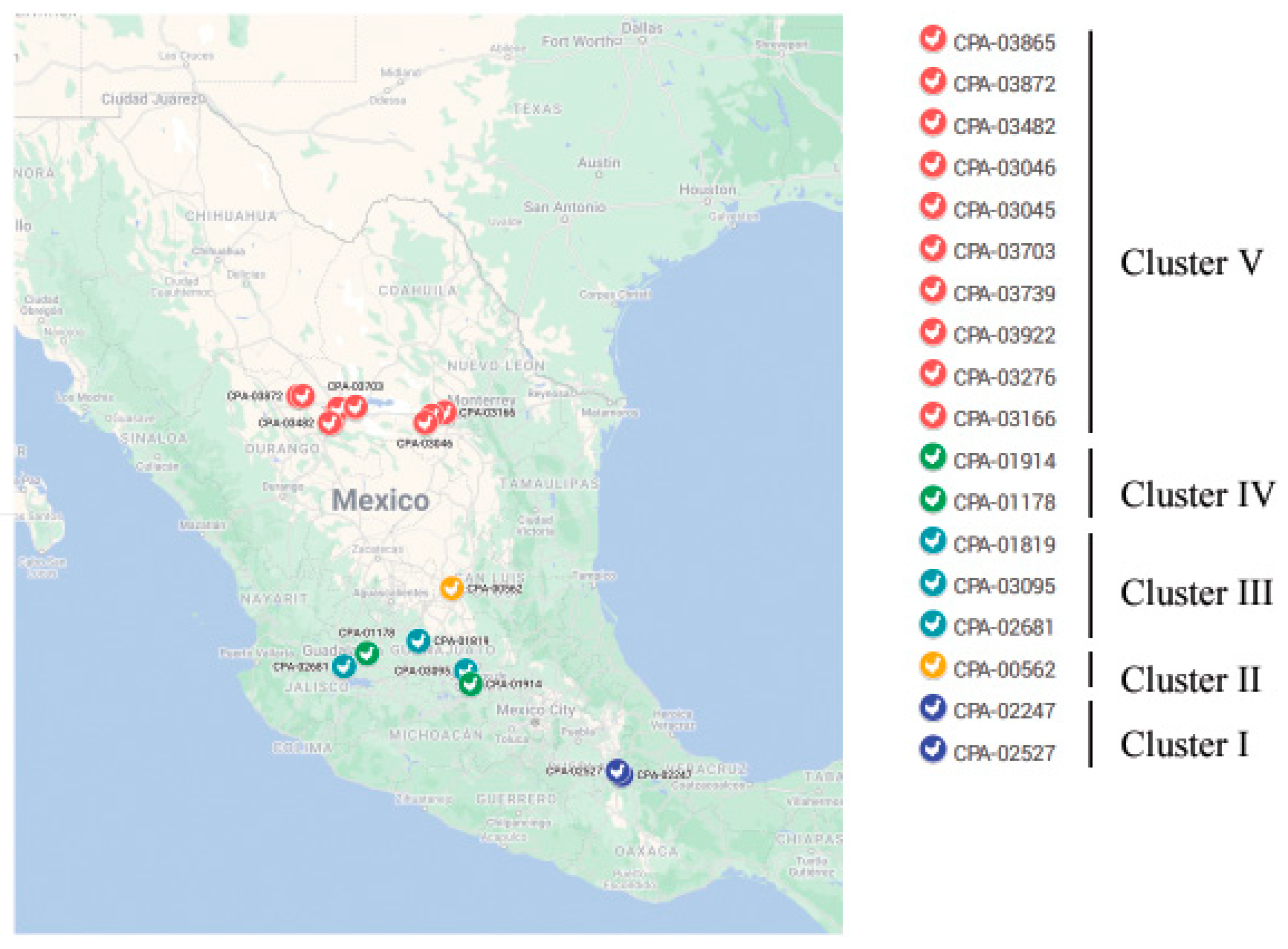



| Strain | Collection Date (y/m/d) | Type of Surveillance | Type of Sample Bird | Farm ID | Network ID | HA Cleavage Site Motif |
|---|---|---|---|---|---|---|
| A/chicken/San Luis Potosí/CPA-00562-22/2022 | 2022-02-25 | Passive | Sick | Farm A | CPA-00562 | PENPKGRKSLHRKTR/G |
| A/chicken/Jalisco/CPA-01178-22/2022 | 2022-03-12 | Passive | Dead corpse | Farm B | CPA-01178 | PENPKSRKSQHRKTR/G |
| A/chicken/Coahuila/CPA-03045-22/2022 | 2022-04-21 | Passive | Dead corpse | Farm H | CPA-03045 | PENPKSRKSQHRKTR/G |
| A/chicken/Coahuila/CPA-03046-22/2022 | 2022-04-21 | Passive | Dead corpse | Farm I | CPA-03046 | PENPKSRKSQHRKTR/G |
| A/chicken/Puebla/CPA-02247-22/2022 | 2022-04-05 | Passive | Dead corpse | Farm E | CPA-02247 | PENPKGGKSRHRKTR/G |
| A/turkey/Puebla/CPA-02527-22/2022 | 2022-04-11 | Passive | Dead corpse | Farm F | CPA-02527 | PENPKGGKSRHRKTR/G |
| A/chicken/Guanajuato/CPA-01819-22/2022 | 2022-03-28 | Active | Healthy | Farm C | CPA-01819 | PENPKSRKSRHRKTR/G |
| A/chicken/Guanajuato/CPA-01914-22/2022 | 2022-03-28 | Active | Healthy | Farm D | CPA-01914 | PENPKSRKSQHRKTR/G |
| A/chicken/Jalisco/CPA-02681-22/2022 | 2022-04-18 | Passive | Sick | Farm G | CPA-02681 | PENPKSRKSRHRKTR/G |
| A/chicken/Guanajuato/CPA-03095-22/2022 | 2022-04-24 | Passive | Sick | Farm J | CPA-03095 | PENPKSRKSRHRKTR/G |
| A/chicken/Coahuila/CPA-03166-22/2022 | 2022-04-24 | Active | Dead corpse | Farm K | CPA-03166 | PENPKSRKSQHRKTR/G |
| A/chicken/Durango/CPA-03482-22/2022 | 2022-04-26 | Active | Dead corpse | Farm M | CPA-03482 | PENPKSRKSQHRKTR/G |
| A/Chicken/Durango/CPA-03865-22/2022 | 2022-04-28 | Active | Healthy | Farm P | CPA-03865 | PENPKSRKSQHRKTR/G |
| A/chicken/Coahuila/CPA-03703-22/2022 | 2022-04-27 | Active | Healthy | Farm N | CPA-03703 | PENPKSRKSQHRKTR/G |
| A/chicken/Durango/CPA-03739-22/2022 | 2022-04-27 | Active | Dead corpse | Farm O | CPA-03739 | PENPKSRKSQHRKTR/G |
| A/chicken/Durango/CPA-03872-22/2022 | 2022-04-28 | Active | Dead corpse | Farm Q | CPA-03872 | PENPKSRKSQHRKTR/G |
| A/chicken/Durango/CPA-03922-22/2022 | 2022-04-28 | Active | Dead corpse | Farm R | CPA-03922 | PENPKSRKSQHRKTR/G |
| A/chicken/Durango/CPA-03276-22/2022 | 2022-04-25 | Active | Sick | Farm L | CPA-03276 | PENPKSRKSQHRKTR/G |
| Gene | Position a/Residue/Motif (%) b | Phenotype [Reference Number] |
|---|---|---|
| PB2 | 292V (89) | Increased polymerase activity in mammalian cell line and increased virulence in mice [14] |
| 389R (100) | Increased polymerase activity and replication in mammalian cell line [15] | |
| 526R (6) | Increased polymerase activity in mammalian cell line [16] | |
| 598T (100) | Increased polymerase activity and replication in mammalian cells and increased virulence in mice [15] | |
| 627E (100) | Increased virulence in chickens [17] | |
| 715N (89) | Decreased virulence in mice [18] | |
| 89V, 309D (11) | Increased polymerase activity in mammalian cell line and increased virulence in mice [19] | |
| PB1 | 3V (100) | Increased polymerase activity and viral replication in avian and mammalian cell lines [20] |
| 622G (100) | Increased polymerase activity and virulence in mice [21] | |
| 678N (6) | Increased replication in avian and mammalian cell lines [22] | |
| PB1-F2 | 66S (100) | Enhanced replication, virulence and antiviral response in mice [23,24] |
| PA | 37A (100) | Increased polymerase activity in mammalian cell line [25] |
| 63I (89) | Increased polymerase activity and enhanced replication in mammalian cell line, increased virulence in mice [26,27] | |
| 190S (100) | Decreased virulence in mice [28] | |
| 383D (100) | Increased polymerase activity in mammalian and avian cell lines [29,30] | |
| 400P (89) | Decreased virulence in mice [28] | |
| 409S (100) | Increased polymerase activity and replication in mammalian cell line [25] | |
| 497R (6) | Increased polymerase activity in mammalian cell line [31] | |
| HA | 126N (6) | Increased virus binding to α2-6 [32] |
| 214I (6) | Increased virus binding to α2-6 [33] | |
| 326 to 329 (100) | Polybasic cleavage motif sequence required for high pathogenicity avian influenza viruses [34,35,36] | |
| 393E (100) | Increased pH of fusion, decreased HA stability, decreased virulence in mice [37] | |
| NP | 184K (100) | Increased replication in avian cells and virulence in chickens enhanced IFN response [38] |
| NA | 117T (100) | Reduced susceptibility to oseltamivir and zanamivir [39] |
| M1 | 30D (100) | Increased virulence in mice [40] |
| 43M (100) | Increased virulence in mice, chickens, and ducks [41] | |
| 215A (100) | Increased virulence in mice [40] | |
| M2 | 31N (100) | Increased resistance to amantadine and rimantadine [42,43,44,45,46,47,48] |
| NS1 | 42S (100) | Increased virulence and decreased antiviral response in mice [49] |
| 106M (100) | Increased viral replication in mammalian cells and increased virulence in mice [50] | |
| 138F (100) | Increased replication in mammalian cells, decreased interferon response [51] | |
| 149A (100) | Increased virulence and decreased interferon response in chicken [52] | |
| 103F, 106M (100) | Increased virulence in mice [53,54] | |
| 3S, 41K (11) | Enhanced replication in mammalian cells and pathogenicity in mice [55] | |
| 55E, 66E, 138F (100) | Enhanced replication in mammalian cells, decreased IF response [51] | |
| ESEV (227-230, PDZ domain) (100) | Increased virulence in mice Decreased viral replication in mammalian and avian cell lines Increased viral replication and virulence in mice decreased viral replication in human and duck cell lines [56] |
| Amino Acid Position a | Structural Location | Sequons b | % of Isolates |
|---|---|---|---|
| 30 (22) | stalk | NGTK | 44 |
| 46 (38) | stalk | NATE | 100 |
| 134 (126) | RBD | NYSG * | 6 |
| 141 (133) | RBD | NGTS | 100 |
| 166/167 (n/a) | RBD | NVTF | 83 |
| 171/172/173 (162/163/164) | RBD | NMTL(R) | 83 |
| 174 (165) | RBD | NLSY * | 17 |
| 248/249/250 (239/240/241) | RBD | NDTV(I) | 100 |
| 500/501/502 (482/483/484) | stalk | NNTY | 100 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Navarro-Lopez, R.; Xu, W.; Gomez-Romero, N.; Velazquez-Salinas, L.; Berhane, Y. Phylogenetic Inference of the 2022 Highly Pathogenic H7N3 Avian Influenza Outbreak in Northern Mexico. Pathogens 2022, 11, 1284. https://doi.org/10.3390/pathogens11111284
Navarro-Lopez R, Xu W, Gomez-Romero N, Velazquez-Salinas L, Berhane Y. Phylogenetic Inference of the 2022 Highly Pathogenic H7N3 Avian Influenza Outbreak in Northern Mexico. Pathogens. 2022; 11(11):1284. https://doi.org/10.3390/pathogens11111284
Chicago/Turabian StyleNavarro-Lopez, Roberto, Wanhong Xu, Ninnet Gomez-Romero, Lauro Velazquez-Salinas, and Yohannes Berhane. 2022. "Phylogenetic Inference of the 2022 Highly Pathogenic H7N3 Avian Influenza Outbreak in Northern Mexico" Pathogens 11, no. 11: 1284. https://doi.org/10.3390/pathogens11111284
APA StyleNavarro-Lopez, R., Xu, W., Gomez-Romero, N., Velazquez-Salinas, L., & Berhane, Y. (2022). Phylogenetic Inference of the 2022 Highly Pathogenic H7N3 Avian Influenza Outbreak in Northern Mexico. Pathogens, 11(11), 1284. https://doi.org/10.3390/pathogens11111284







