Recombinase Polymerase Amplification Combined with Fluorescence Immunochromatography Assay for On-Site and Ultrasensitive Detection of SARS-CoV-2
Abstract
:1. Introduction
2. Experimental Section
2.1. Reagents and Materials
2.2. Nucleic Acid Release
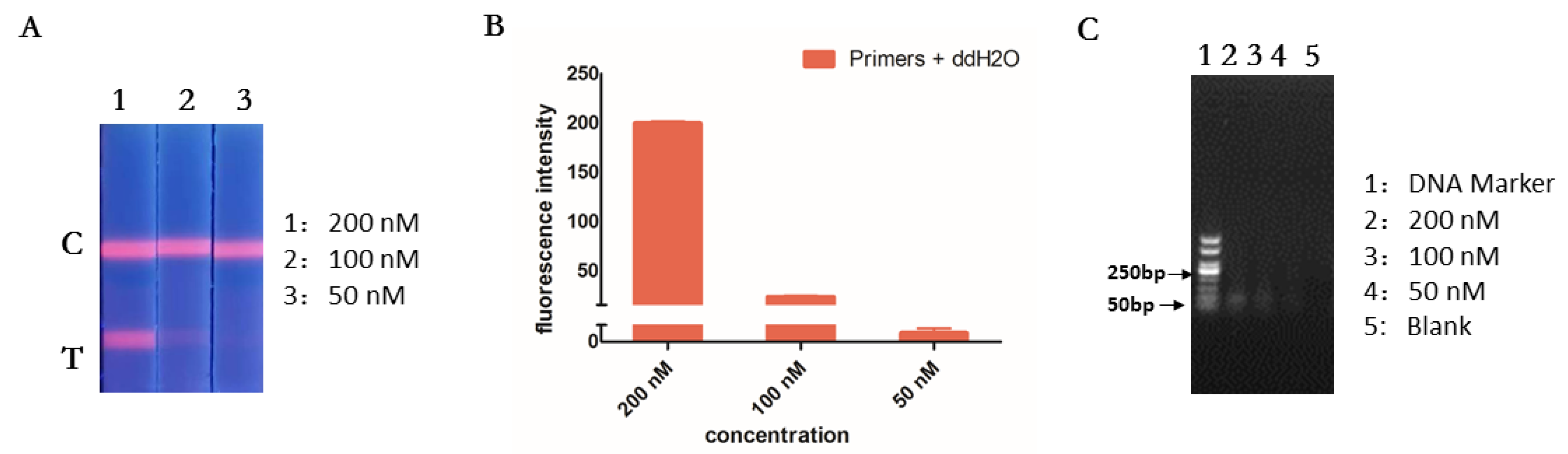
2.3. Primer Design
2.4. RT-RPA Assay
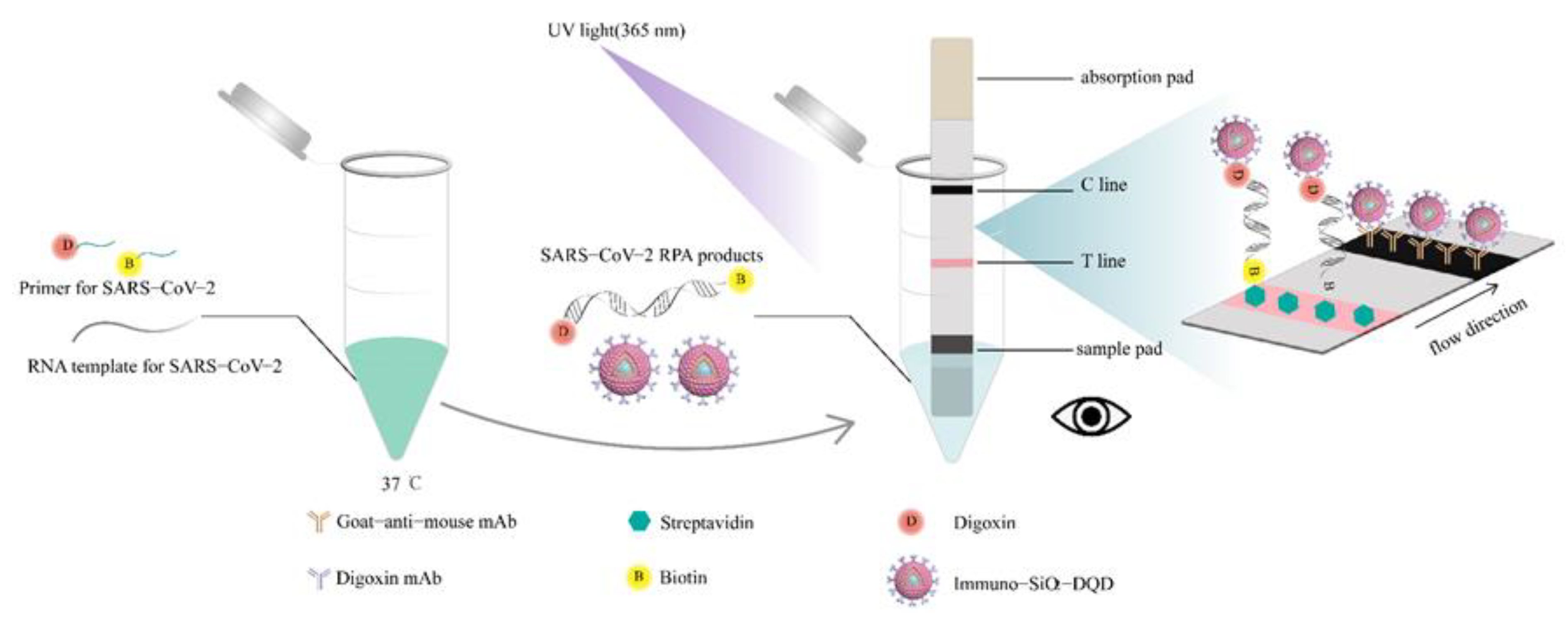
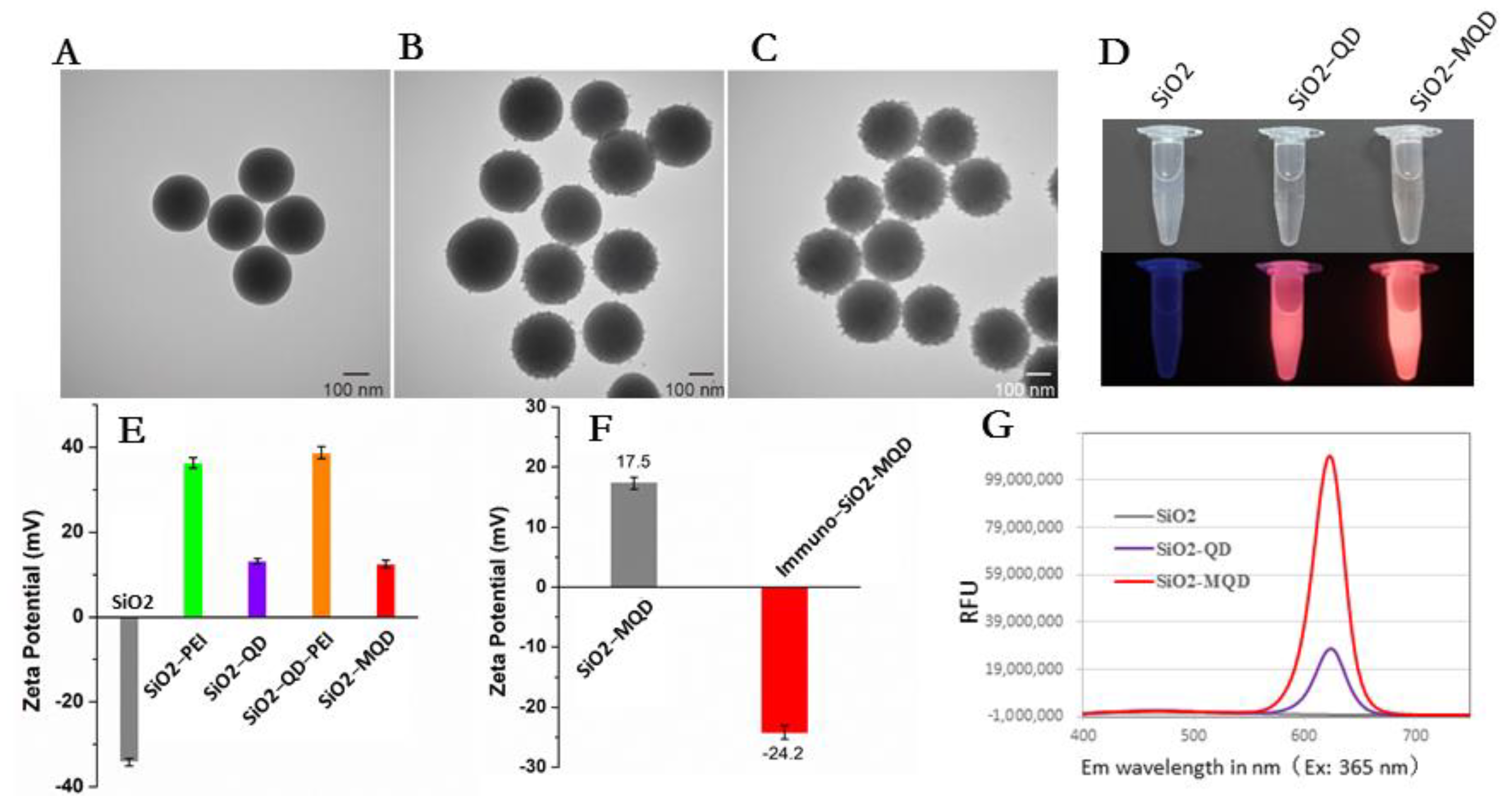
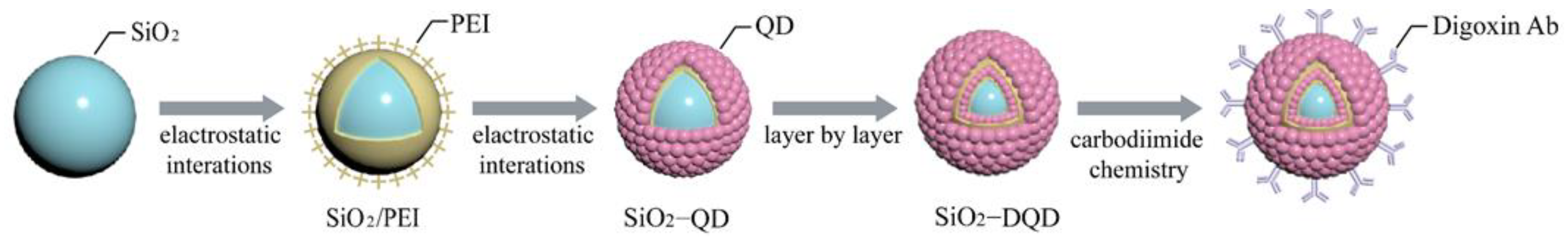
2.5. Synthesis of SiO2-MQDs Nanocomposite
2.6. Preparation of Immuno-SiO2-MQDs Labels
2.7. Preparation of the ICA Strip
2.8. Analytical Procedure for Detection of SARS-CoV-2
2.9. Optimization of Experimental Procedures and Methodological Investigation
3. Results and Discussion
3.1. Construction of Immuno-MQD Labels and MQD-Based ICA
3.2. Optimization of RT-RPA and ICA
3.3. Specificity of the RT-RPA-Combined ICA
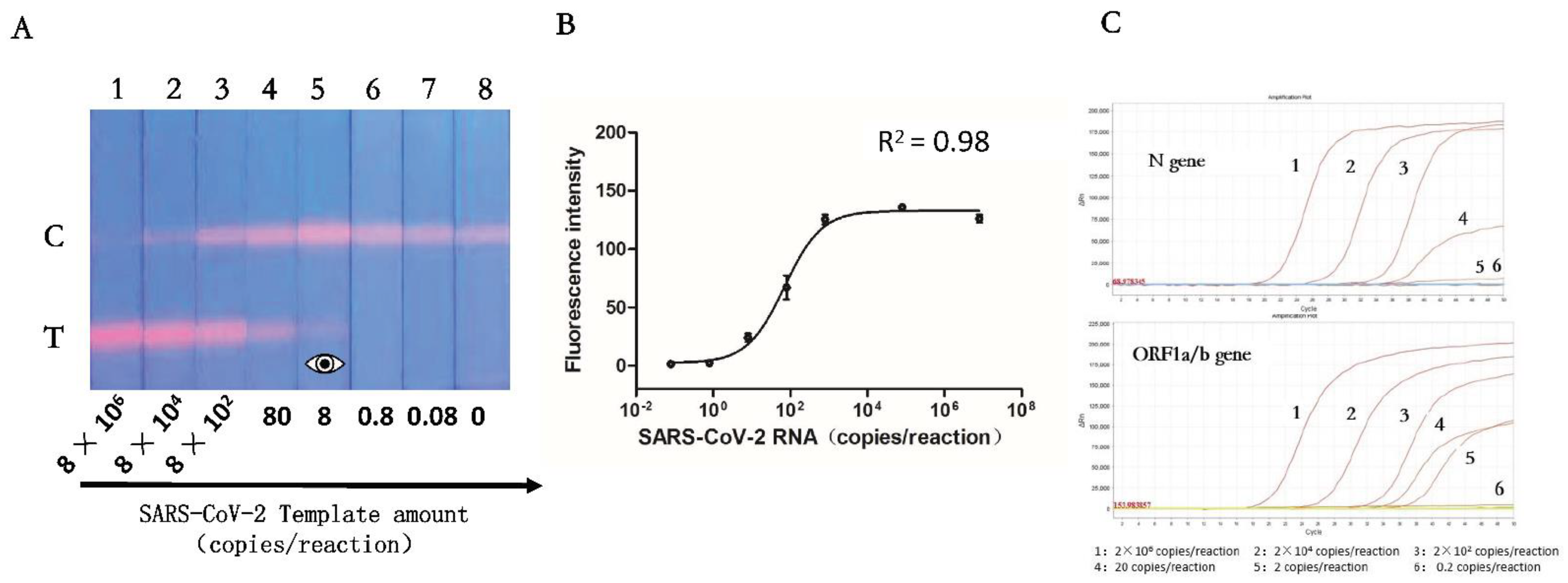
3.4. Sensitivity of the RT-RPA-Combined ICA
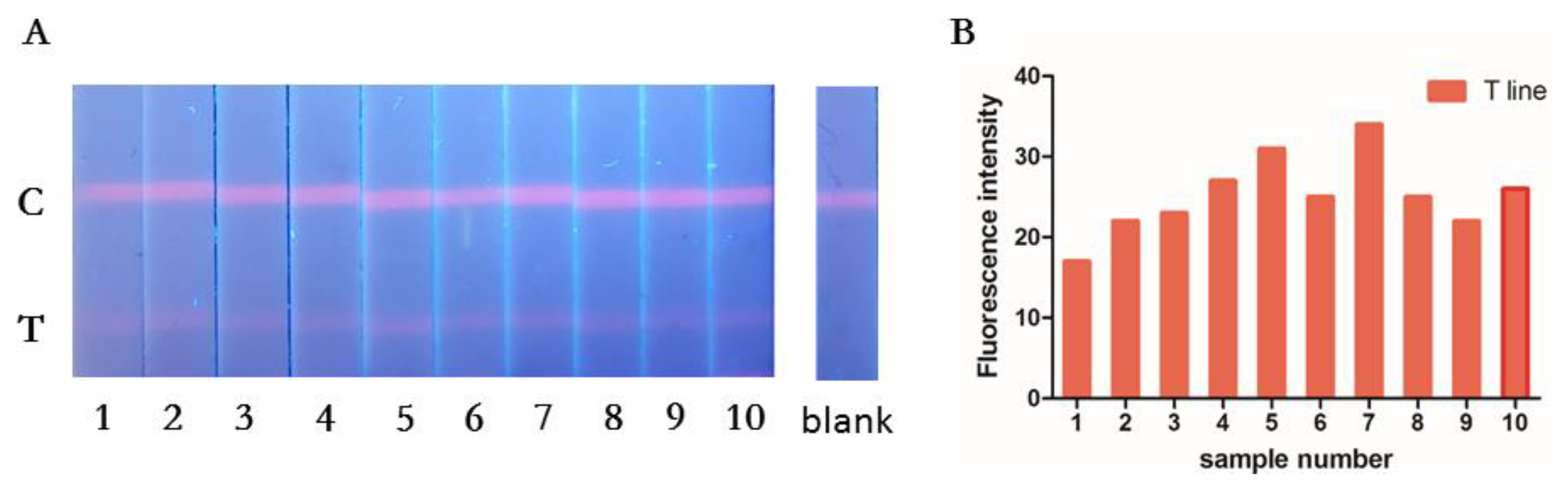
3.5. Repeatability of the RT-RPA-Combined ICA
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Yang, X.; Yu, Y.; Xu, J.; Shu, H.; Xia, J.; Liu, H.; Wu, Y.; Zhang, L.; Yu, Z.; Fang, M.; et al. Clinical course and outcomes of critically ill patients with SARS-CoV-2 pneumonia in Wuhan, China: A single-centered, retrospective, observational study. Lancet Respir. Med. 2020, 8, 475–481. [Google Scholar] [CrossRef] [Green Version]
- Wu, F.; Zhao, S.; Yu, B.; Chen, Y.-M.; Wang, W.; Song, Z.-G.; Hu, Y.; Tao, Z.-W.; Tian, J.-H.; Pei, Y.-Y.; et al. A new coronavirus associated with human respiratory disease in China. Nature 2020, 579, 265–269. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chan, J.F.-W.; Yuan, S.; Kok, K.-H.; To, K.K.-W.; Chu, H.; Yang, J.; Xing, F.; Liu, J.; Yip, C.C.-Y.; Poon, R.W.-S.; et al. A familial cluster of pneumonia associated with the 2019 novel coronavirus indicating person-to-person transmission: A study of a family cluster. Lancet 2020, 395, 514–523. [Google Scholar] [CrossRef] [Green Version]
- Zhu, N.; Zhang, D.; Wang, W.; Li, X.; Yang, B.; Song, J.; Zhao, X.; Huang, B.; Shi, W.; Lu, R.; et al. A Novel Coronavirus from Patients with Pneumonia in China, 2019. N. Engl. J. Med. 2020, 382, 727–733. [Google Scholar] [CrossRef] [PubMed]
- Mohit, E.; Rostami, Z.; Vahidi, H. A comparative review of immunoassays for COVID-19 detection. Expert Rev. Clin. Immunol. 2021, 17, 573–599. [Google Scholar] [CrossRef]
- Yoo, H.; Kim, I.-H.; Kim, S. Nucleic Acid Testing of SARS-CoV-2. Int. J. Mol. Sci. 2021, 22, 6150. [Google Scholar] [CrossRef] [PubMed]
- Dutta, D.; Naiyer, S.; Mansuri, S.; Soni, N.; Singh, V.; Bhat, K.H.; Singh, N.; Arora, G.; Mansuri, M.S. COVID-19 Diagnosis: A Comprehensive Review of the RT-qPCR Method for Detection of SARS-CoV-2. Diagnostics 2022, 12, 1503. [Google Scholar] [CrossRef]
- CDC 2019-Novel Coronavirus (2019-nCoV) Real-Time RT-PCR Diagnostic Panel; Division of Viral Diseases: Atlanta, GA, USA, 2021.
- Abbasi, H.; Tabaraei, A.; Hosseini, S.M.; Khosravi, A.; Nikoo, H.R. Real-time PCR Ct value in SARS-CoV-2 detection: RdRp or N gene? Infection 2021, 50, 537–540. [Google Scholar] [CrossRef]
- Adams, E.R.; Ainsworth, M.; Anand, R.; Andersson, M.I.; Auckland, K.; Baillie, J.K.; Barnes, E.; Beer, S.; Bell, J.I.; Berry, T.; et al. Antibody testing for COVID-19: A report from the National COVID Scientific Advisory Panel. Wellcome Open Res. 2020, 5, 139. [Google Scholar] [CrossRef]
- Flower, B.; Brown, J.C.; Simmons, B.; Moshe, M.; Frise, R.; Penn, R.; Kugathasan, R.; Petersen, C.; Daunt, A.; Ashby, D.; et al. Clinical and laboratory evaluation of SARS-CoV-2 lateral flow assays for use in a national COVID-19 seroprevalence survey. Thorax 2020, 75, 1082–1088. [Google Scholar] [CrossRef]
- Serrano, M.M.; Rodríguez, D.N.; Palop, N.T.; Arenas, R.O.; Córdoba, M.M.; Mochón, M.D.O.; Cardona, C.G. Comparison of commercial lateral flow immunoassays and ELISA for SARS-CoV-2 antibody detection. J. Clin. Virol. 2020, 129, 104529. [Google Scholar] [CrossRef] [PubMed]
- Theel, E.S.; Slev, P.; Wheeler, S.; Couturier, M.R.; Wong, S.J.; Kadkhoda, K. The Role of Antibody Testing for SARS-CoV-2: Is There One? J. Clin. Microbiol. 2020, 58, e00797-20. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.-B.; Du, X.-J.; Zang, Y.-X.; Li, P.; Wang, S. SERS-Based Lateral Flow Strip Biosensor for Simultaneous Detection of Listeria monocytogenes and Salmonella enterica Serotype Enteritidis. J. Agric. Food Chem. 2017, 65, 10290–10299. [Google Scholar] [CrossRef]
- Cha, H.; Kim, H.; Joung, Y.; Kang, H.; Moon, J.; Jang, H.; Park, S.; Kwon, H.-J.; Lee, I.-C.; Kim, S.; et al. Surface-enhanced Raman scattering-based immunoassay for severe acute respiratory syndrome coronavirus 2. Biosens. Bioelectron. 2022, 202, 114008. [Google Scholar] [CrossRef] [PubMed]
- Hang, Y.; Boryczka, J.; Wu, N. Visible-light and near-infrared fluorescence and surface-enhanced Raman scattering point-of-care sensing and bio-imaging: A review. Chem. Soc. Rev. 2021, 51, 329–375. [Google Scholar] [CrossRef]
- Li, H.; Dong, B.; Dou, L.; Yu, W.; Yu, X.; Wen, K.; Ke, Y.; Shen, J.; Wang, Z. Fluorescent lateral flow immunoassay for highly sensitive detection of eight anticoagulant rodenticides based on cadmium-free quantum dot-encapsulated nanospheres. Sensors Actuators B Chem. 2020, 324, 128771. [Google Scholar] [CrossRef]
- Rong, Z.; Wang, Q.; Sun, N.; Jia, X.; Wang, K.; Xiao, R.; Wang, S. Smartphone-based fluorescent lateral flow immunoassay platform for highly sensitive point-of-care detection of Zika virus nonstructural protein 1. Anal. Chim. Acta 2018, 1055, 140–147. [Google Scholar] [CrossRef]
- Shao, Y.; Duan, H.; Guo, L.; Leng, Y.; Lai, W.; Xiong, Y. Quantum dot nanobead-based multiplexed immunochromatographic assay for simultaneous detection of aflatoxin B1 and zearalenone. Anal. Chim. Acta 2018, 1025, 163–171. [Google Scholar] [CrossRef]
- Hu, J.; Zhang, Z.-L.; Wen, C.-Y.; Tang, M.; Wu, L.-L.; Liu, C.; Zhu, L.; Pang, D.-W. Sensitive and Quantitative Detection of C-Reaction Protein Based on Immunofluorescent Nanospheres Coupled with Lateral Flow Test Strip. Anal. Chem. 2016, 88, 6577–6584. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Lv, Y.; Li, N.; Wu, R.; Xing, M.; Shen, H.; Li, L.S.; Chen, X. Robust synthesis of bright multiple quantum dot-embedded nanobeads and its application to quantitative immunoassay. Chem. Eng. J. 2019, 361, 499–507. [Google Scholar] [CrossRef]
- Huang, L.; Liao, T.; Wang, J.; Ao, L.; Su, W.; Hu, J. Brilliant pitaya-type silica colloids with central–radial and high-density quantum dots incorporation for ultrasensitive fluorescence immunoassays. Adv. Funct. Mater. 2018, 28, 1705380. [Google Scholar] [CrossRef]
- Zheng, S.; Wu, T.; Li, J.; Qin, J.; Xiao, R.; Wang, S.; Wang, C. Difunctional immunochromatographic assay based on magnetic quantum dot for ultrasensitive and simultaneous detection of multiple mycotoxins in foods. Sens. Actuators B Chem. 2022, 359, 131528. [Google Scholar] [CrossRef]
- Cheng, X.; Zheng, S.; Wang, W.; Han, H.; Yang, X.; Shen, W.; Wang, C.; Wang, S. Synthesis of two-dimensional graphene oxide-fluorescent nanoprobe for ultrasensitive and multiplex immunochromatographic detection of respiratory bacteria. Chem. Eng. J. 2021, 426, 131836. [Google Scholar] [CrossRef]
- Wang, C.; Cheng, X.; Liu, L.; Zhang, X.; Yang, X.; Zheng, S.; Rong, Z.; Wang, S. Ultrasensitive and Simultaneous Detection of Two Specific SARS-CoV-2 Antigens in Human Specimens Using Direct/Enrichment Dual-Mode Fluorescence Lateral Flow Immunoassay. ACS Appl. Mater. Interfaces 2021, 13, 40342–40353. [Google Scholar] [CrossRef]
- Wang, C.; Yang, X.; Gu, B.; Liu, H.; Zhou, Z.; Shi, L.; Cheng, X.; Wang, S. Sensitive and simultaneous detection of SARS-CoV-2-specific IgM/IgG using lateral flow immunoassay based on dual-mode quantum dot nanobeads. Anal. Chem. 2020, 92, 15542–15549. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Xiao, R.; Wang, S.; Yang, X.; Bai, Z.; Li, X.; Rong, Z.; Shen, B.; Wang, S. Magnetic quantum dot based lateral flow assay biosensor for multiplex and sensitive detection of protein toxins in food samples. Biosens. Bioelectron. 2019, 146, 111754. [Google Scholar] [CrossRef]
- Chen, H.; Wang, Y.; Wei, H.; Rong, Z.; Wang, S. A rapid water bath PCR combined with lateral flow assay for the simultaneous detection of SARS-CoV-2 and influenza B virus. RSC Adv. 2022, 12, 3437–3444. [Google Scholar] [CrossRef]
- Li, J.; Macdonald, J.; von Stetten, F. Review: A comprehensive summary of a decade development of the recombinase polymerase amplification. Analyst 2018, 144, 31–67. [Google Scholar] [CrossRef] [Green Version]
- Lobato, I.M.; O’Sullivan, C.K. Recombinase polymerase amplification: Basics, applications and recent advances. TrAC Trends Anal. Chem. 2018, 98, 19–35. [Google Scholar] [CrossRef]
- Daher, R.K.; Stewart, G.; Boissinot, M.; Bergeron, M.G. Recombinase Polymerase Amplification for Diagnostic Applications. Clin. Chem. 2016, 62, 947–958. [Google Scholar] [CrossRef]
- Mota, D.S.; Guimaraes, J.M.; Gandarilla, A.M.D.; Filho, J.C.B.S.; Brito , W.R.; Mariuba , L.A.M.; Mota, D.S. Recombinase polymerase amplification in the molecular diagnosis of microbiological targets and its applications. Can J. Microbiol. 2022, 68, 383–402. [Google Scholar] [CrossRef]
- Wang, C.; Yang, X.; Zheng, S.; Cheng, X.; Xiao, R.; Li, Q.; Wang, W.; Liu, X.; Wang, S. Development of an ultrasensitive fluorescent immunochromatographic assay based on multilayer quantum dot nanobead for simultaneous detection of SARS-CoV-2 antigen and influenza A virus. Sens. Actuators B Chem. 2021, 345, 130372. [Google Scholar] [CrossRef]
- Havasi, A.; Visan, S.; Cainap, C.; Cainap, S.S.; Mihaila, A.A.; Pop, L.-A. Influenza A, Influenza B, and SARS-CoV-2 Similarities and Differences—A Focus on Diagnosis. Front. Microbiol. 2022, 13, 908525. [Google Scholar] [CrossRef]
- Wang, X.; Choi, N.; Cheng, Z.; Ko, J.; Chen, L.; Choo, J. Simultaneous Detection of Dual Nucleic Acids Using a SERS-Based Lateral Flow Assay Biosensor. Anal. Chem. 2016, 89, 1163–1169. [Google Scholar] [CrossRef]
- Zhang, D.; Huang, L.; Liu, B.; Ni, H.; Sun, L.; Su, E.; Chen, H.; Gu, Z.; Zhao, X. Quantitative and ultrasensitive detection of multiplex cardiac biomarkers in lateral flow assay with core-shell SERS nanotags. Biosens. Bioelectron. 2018, 106, 204–211. [Google Scholar] [CrossRef]
- WHO. Clinical Management of Severe Acute Respiratory Infection when Novel Coronavirus (nCoV) Infection Is Suspected. Interim Guidance; WHO: Geneva, Switzerland, 2020. [Google Scholar]
- Hu, J.; Jiang, Y.-Z.; Wu, L.-L.; Wu, Z.; Bi, Y.; Wong, G.; Qiu, X.; Chen, J.; Pang, D.-W.; Zhang, Z.-L. Dual-Signal Readout Nanospheres for Rapid Point-of-Care Detection of Ebola Virus Glycoprotein. Anal. Chem. 2017, 89, 13105–13111. [Google Scholar] [CrossRef]



Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, G.; Yang, X.; Dong, H.; Tu, Z.; Zhou, Y.; Rong, Z.; Wang, S. Recombinase Polymerase Amplification Combined with Fluorescence Immunochromatography Assay for On-Site and Ultrasensitive Detection of SARS-CoV-2. Pathogens 2022, 11, 1252. https://doi.org/10.3390/pathogens11111252
Wang G, Yang X, Dong H, Tu Z, Zhou Y, Rong Z, Wang S. Recombinase Polymerase Amplification Combined with Fluorescence Immunochromatography Assay for On-Site and Ultrasensitive Detection of SARS-CoV-2. Pathogens. 2022; 11(11):1252. https://doi.org/10.3390/pathogens11111252
Chicago/Turabian StyleWang, Guangyu, Xingsheng Yang, Hao Dong, Zhijie Tu, Yong Zhou, Zhen Rong, and Shengqi Wang. 2022. "Recombinase Polymerase Amplification Combined with Fluorescence Immunochromatography Assay for On-Site and Ultrasensitive Detection of SARS-CoV-2" Pathogens 11, no. 11: 1252. https://doi.org/10.3390/pathogens11111252



