The Detection of Periodic Reemergence Events of SARS-CoV-2 Delta Strain in Communities Dominated by Omicron
Abstract
:1. Introduction
2. Materials and Methods
2.1. Wastewater Sampling and Preparation
2.2. cDNA and Library Synthesis
2.3. Next-Generation Sequencing and Data Analysis
2.4. Accession and Analysis of City of Louisville KY Public Health Data
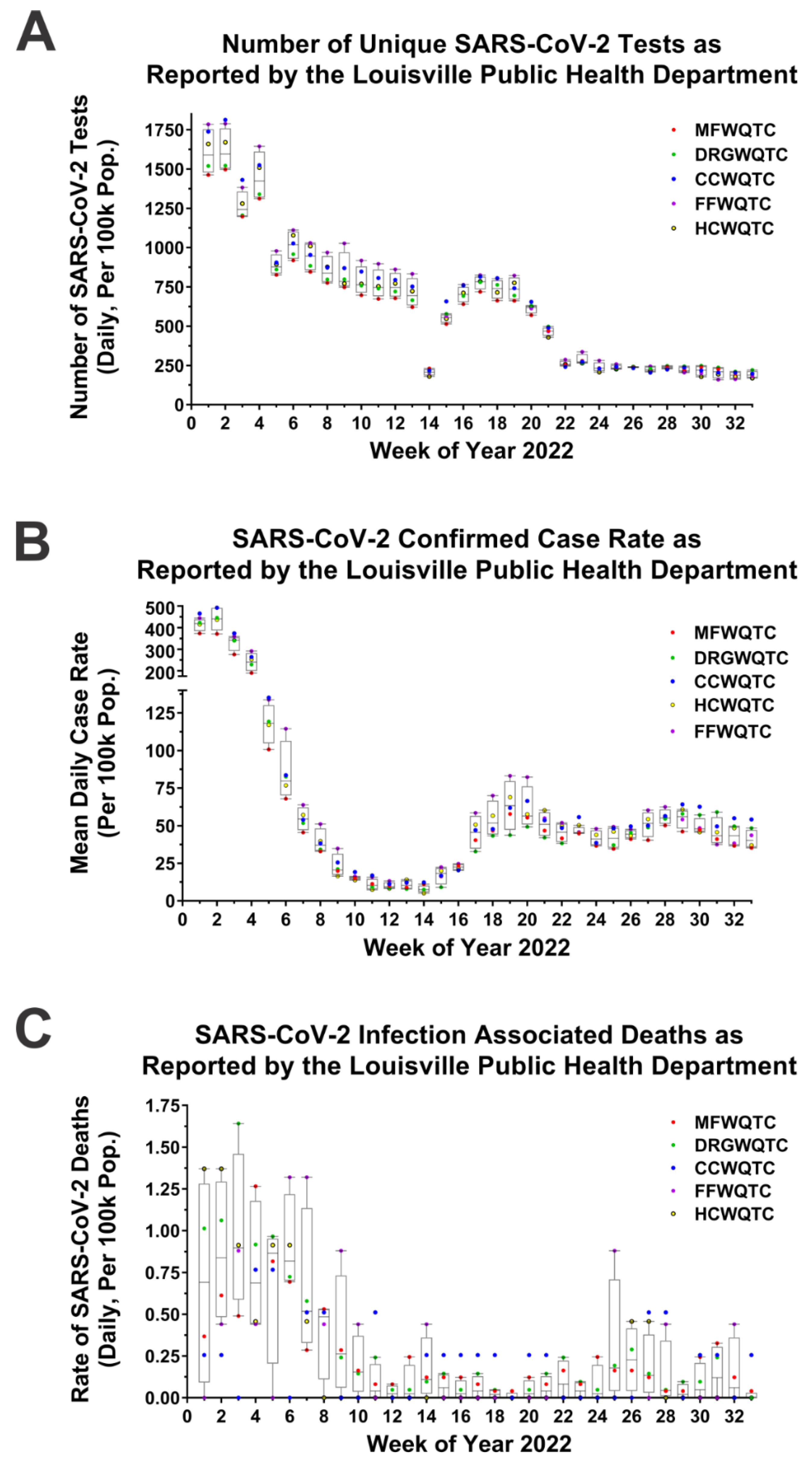
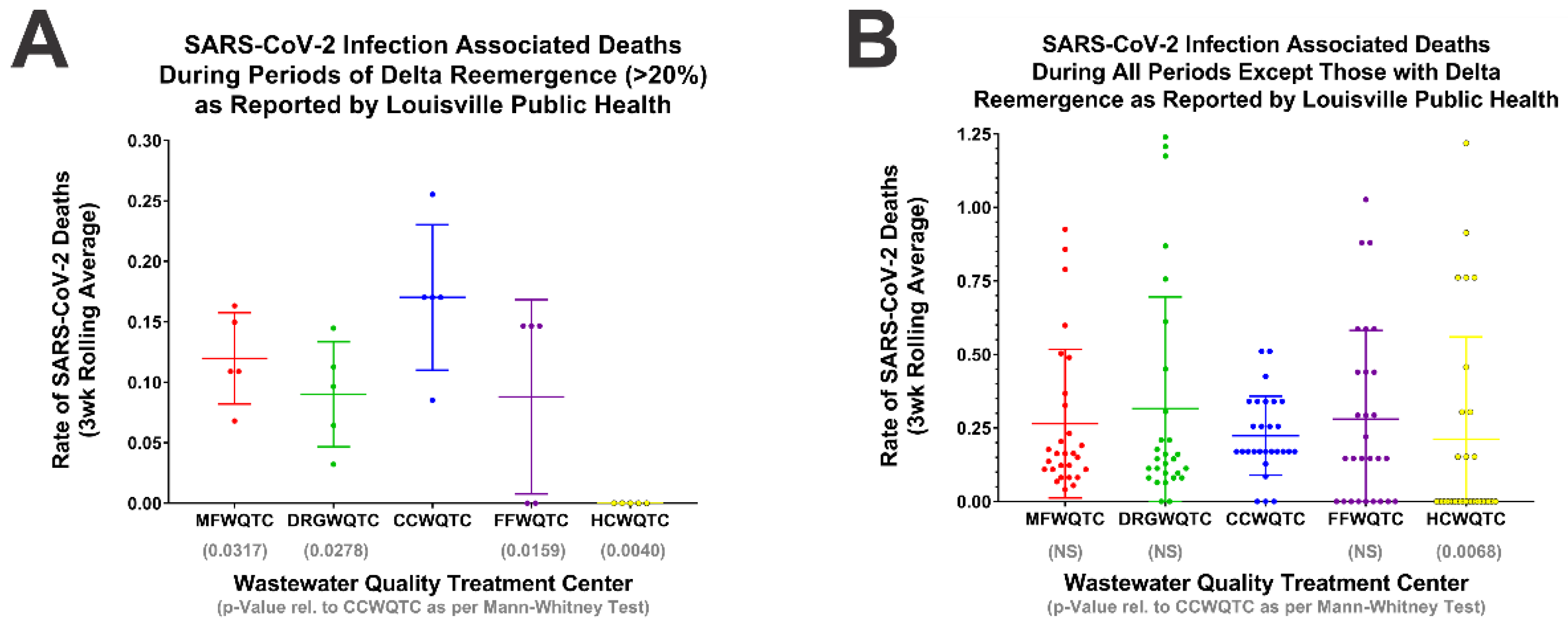
2.5. Statistical Analysis of the Impact of Delta Reemergence on SARS-CoV-2 Associated Deaths
3. Results
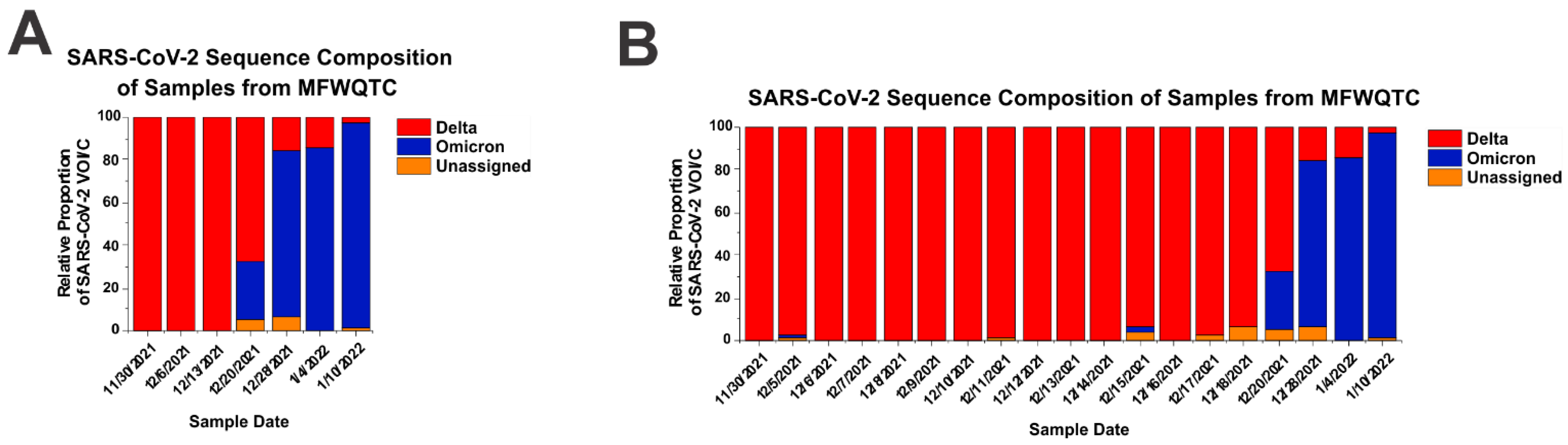
3.1. The Power and Importance of Wastewater Monitoring in the Detection of Variant Emergence
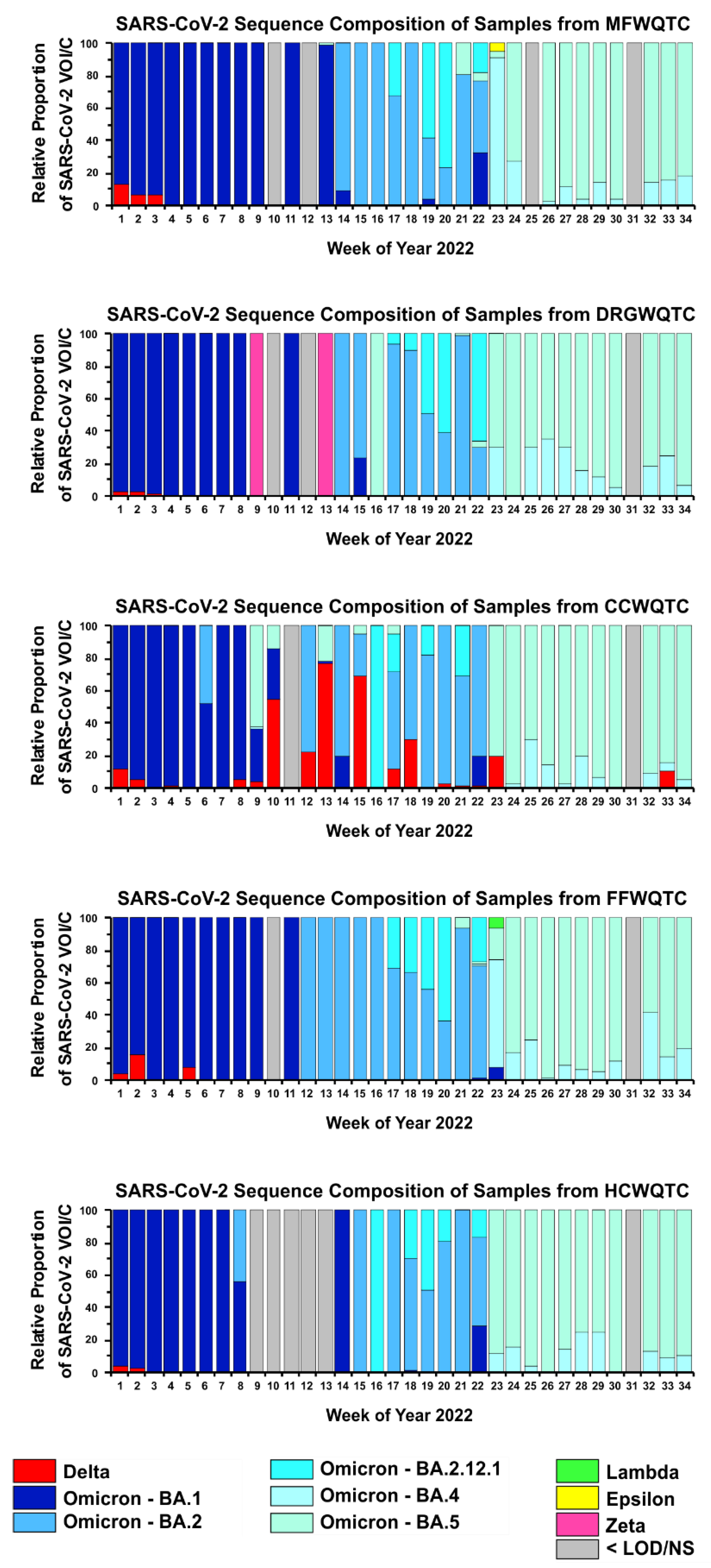
3.2. Unexplained Periodic Reemergence of the Delta Strain in Wastewatersheds of Louisville KY
4. Discussion
4.1. Weekly Monitoring of Wastewater Is Sufficient and Cost-Effective
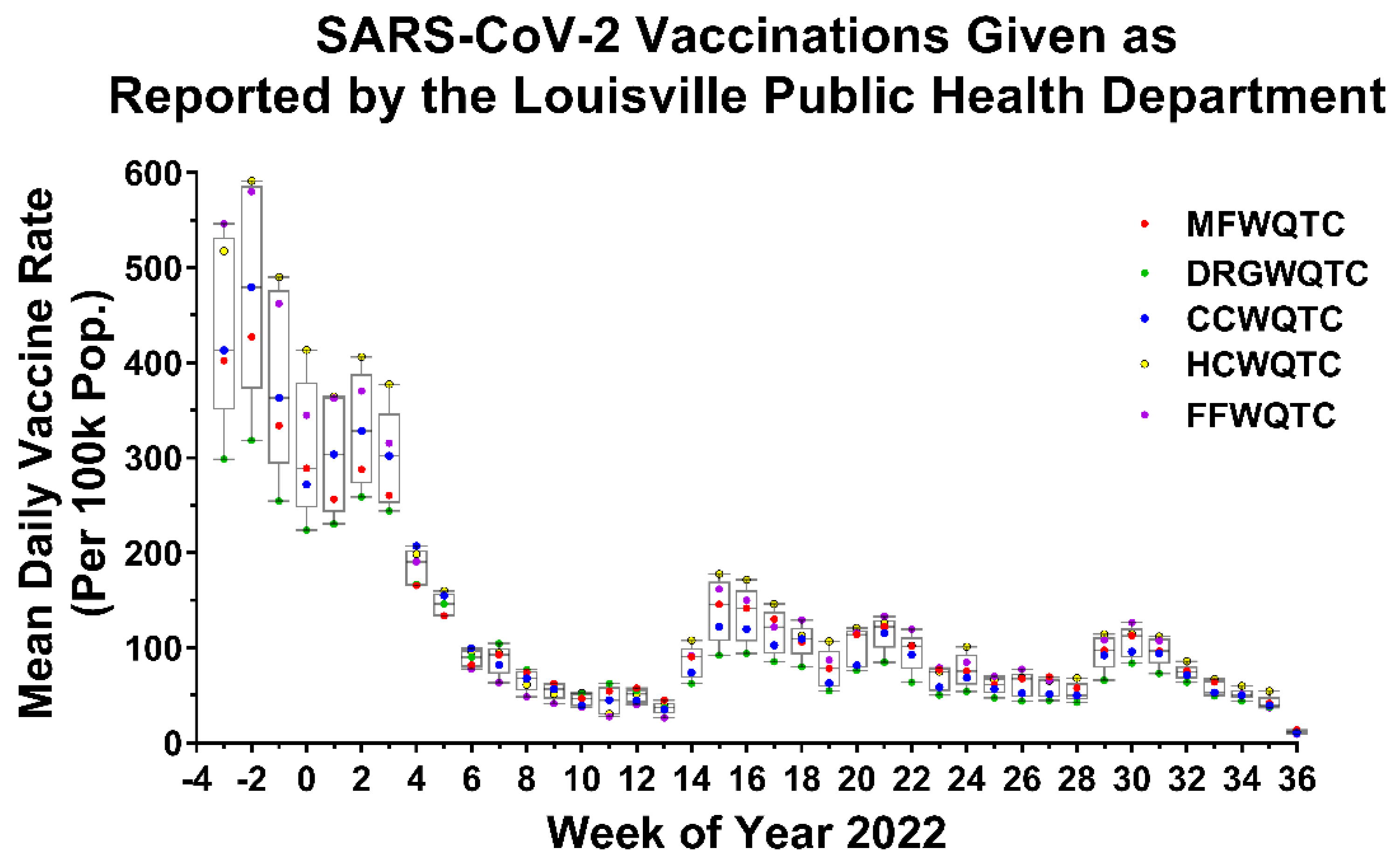
4.2. Low Vaccine Uptake Does Not Correlate with Reemergence of the SARS-CoV-2 Delta Strain, and Reemergence Correlates with Increased Negative Outcomes to Public Health
4.3. The Detection of SARS-CoV-2 Reemergence Events Underscores the Importance of Personalized Medicine in Infectious Disease
4.4. Forward Looking to the Next Pandemic
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Fontenele, R.S.; Kraberger, S.; Hadfield, J.; Driver, E.M.; Bowes, D.; Holland, L.A.; Faleye, T.O.C.; Adhikari, S.; Kumar, R.; Inchausti, R.; et al. High-throughput sequencing of SARS-CoV-2 in wastewater provides insights into circulating variants. Water Res. 2021, 205, 117710. [Google Scholar] [CrossRef] [PubMed]
- Pecson, B.M.; Darby, E.; Haas, C.N.; Amha, Y.M.; Bartolo, M.; Danielson, R.; Dearborn, Y.; Di Giovanni, G.; Ferguson, C.; Fevig, S.; et al. Consortium SA-C-I. Reproducibility and sensitivity of 36 methods to quantify the SARS-CoV-2 genetic signal in raw wastewater: Findings from an interlaboratory methods evaluation in the U.S. Environ.Sci. Water Res. Technol. 2021, 7, 504–520. [Google Scholar] [CrossRef] [PubMed]
- Wu, F.; Zhang, J.; Xiao, A.; Gu, X.; Lee, W.L.; Armas, F.; Kauffman, K.; Hanage, W.; Matus, M.; Ghaeli, N.; et al. SARS-CoV-2 Titers in Wastewater Are Higher than Expected from Clinically Confirmed Cases. Msystems 2020, 5, e00614-20. [Google Scholar] [CrossRef]
- Holm, R.H.; Mukherjee, A.; Rai, J.P.; Yeager, R.A.; Talley, D.; Rai, S.N.; Bhatnagar, A.; Smith, T. SARS-CoV-2 RNA abundance in wastewater as a function of distinct urban sewershed size. Environ.Sci. Water Res. Technol. 2022, 8, 807–819. [Google Scholar] [CrossRef]
- Smith, T.; Holm, R.H.; Keith, R.J.; Amraotkar, A.R.; Alvarado, C.R.; Banecki, K.; Choi, B.; Santisteban, I.C.; Bushau-Sprinkle, A.M.; Kitterman, K.T.; et al. Quantifying the relationship between sub-population wastewater samples and community-wide SARS-CoV-2 seroprevalence. Sci. Total Environ. 2022, 853, 158567. [Google Scholar] [CrossRef] [PubMed]
- Smith, T.; Holm, R.H.; Yeager, R.; Moore JBt Rouchka, E.C.; Sokoloski, K.J.; Elliott, E.M.; Talley, D.; Arora, V.; Moyer, S.; Bhatnagar, A. Combining Community Wastewater Genomic Surveillance with State Clinical Surveillance: A Framework for SARS-CoV-2 Public Health Practice. Food Environ. Virol. 2022. [Google Scholar] [CrossRef]
- Rouchka, E.C.; Chariker, J.H.; Saurabh, K.; Waigel, S.; Zacharias, W.; Zhang, M.; Talley, D.; Santisteban, I.; Puccio, M.; Moyer, S.; et al. The Rapid Assessment of Aggregated Wastewater Samples for Genomic Surveillance of SARS-CoV-2 on a City-Wide Scale. Pathogens 2021, 10, 1271. [Google Scholar] [CrossRef] [PubMed]
- Dhawan, M.; Saied, A.A.; Mitra, S.; Alhumaydhi, F.A.; Emran, T.B.; Wilairatana, P. Omicron variant (B.1.1.529) and its sublineages: What do we know so far amid the emergence of recombinant variants of SARS-CoV-2? Biomed. Pharm. 2022, 154, 113522. [Google Scholar] [CrossRef]
- Lyke, K.E.; Atmar, R.L.; Islas, C.D.; Posavad, C.M.; Szydlo, D.; Paul Chourdhury, R.; Deming, M.E.; Eaton, A.; Jackson, L.A.; Branche, A.R.; et al. Rapid decline in vaccine-boosted neutralizing antibodies against SARS-CoV-2 Omicron variant. Cell Rep. Med. 2022, 3, 100679. [Google Scholar] [CrossRef]
- Anichini, G.; Terrosi, C.; Gori Savellini, G.; Gandolfo, C.; Barbagli, F.; Carta, G.A.; Fabrizi, S.; Miceli, G.B.; Cusi, M.G. Antibody Response against Circulating Omicron Variants 8 Months after the Third Dose of mRNA Vaccine. Vaccines 2022, 10, 1512. [Google Scholar] [CrossRef]
- Amati, R.; Frei, A.; Kaufmann, M.; Sabatini, S.; Pellaton, C.; Fehr, J.; Albanese, E.; Puhan, M.A. Functional immunity against SARS-CoV-2 in the general population after a booster campaign and the Delta and Omicron waves, Switzerland, March 2022. Eurosurveillance 2022, 27, 2200561. [Google Scholar] [CrossRef] [PubMed]
- Khan, K.; Karim, F.; Cele, S.; Reedoy, K.; San, J.E.; Lustig, G.; Tegally, H.; Rosenberg, Y.; Bernstein, M.; Jule, Z.; et al. Omicron infection enhances Delta antibody immunity in vaccinated persons. Nature 2022, 607, 356–359. [Google Scholar] [CrossRef] [PubMed]
- La Rosa, G.; Mancini, P.; Bonanno Ferraro, G.; Veneri, C.; Iaconelli, M.; Lucentini, L.; Bonadonna, L.; Brusaferro, S.; Brandtner, D.; Fasanella, A.; et al. Rapid screening for SARS-CoV-2 variants of concern in clinical and environmental samples using nested RT-PCR assays targeting key mutations of the spike protein. Water Res. 2021, 197, 117104. [Google Scholar] [CrossRef] [PubMed]
- Bi, C.; Ramos-Mandujano, G.; Tian, Y.; Hala, S.; Xu, J.; Mfarrej, S.; Esteban, C.R.; Delicado, E.N.; Alofi, F.S.; Khogeer, A.; et al. Simultaneous detection and mutation surveillance of SARS-CoV-2 and multiple respiratory viruses by rapid field-deployable sequencing. Med 2021, 2, 689–700. [Google Scholar] [CrossRef]
- Crits-Christoph, A.; Kantor, R.S.; Olm, M.R.; Whitney, O.N.; Al-Shayeb, B.; Lou, Y.C.; Flamholz, A.; Kennedy, L.C.; Greenwald, H.; Hinkle, A. Genome sequencing of sewage detects regionally prevalent SARS-CoV-2 variants. MBio 2021, 12, e02703-20. [Google Scholar] [CrossRef]
- Martin, J.; Klapsa, D.; Wilton, T.; Zambon, M.; Bentley, E.; Bujaki, E.; Fritzsche, M.; Mate, R.; Majumdar, M. Tracking SARS-CoV-2 in sewage: Evidence of changes in virus variant predominance during COVID-19 pandemic. Viruses 2020, 12, 1144. [Google Scholar] [CrossRef]
- Nemudryi, A.; Nemudraia, A.; Wiegand, T.; Surya, K.; Buyukyoruk, M.; Cicha, C.; Vanderwood, K.K.; Wilkinson, R.; Wiedenheft, B. Temporal detection and phylogenetic assessment of SARS-CoV-2 in municipal wastewater. Cell Rep. Med. 2020, 1, 100098. [Google Scholar] [CrossRef]
- Challen, R.; Brooks-Pollock, E.; Read, J.M.; Dyson, L.; Tsaneva-Atanasova, K.; Danon, L. Risk of mortality in patients infected with SARS-CoV-2 variant of concern 202012/1: Matched cohort study. BMJ 2021, 372, n579. [Google Scholar] [CrossRef] [PubMed]
- Sigal, A.; Milo, R.; Jassat, W. Estimating disease severity of Omicron and Delta SARS-CoV-2 infections. Nat. Rev. Immunol. 2022, 22, 267–269. [Google Scholar] [CrossRef]
- Yaniv, K.; Ozer, E.; Shagan, M.; Paitan, Y.; Granek, R.; Kushmaro, A. Managing an evolving pandemic: Cryptic circulation of the Delta variant during the Omicron rise. Sci. Total Environ. 2022, 836, 155599. [Google Scholar] [CrossRef]
- Wolfe, M.; Hughes, B.; Duong, D.; Chan-Herur, V.; Wigginton, K.R.; White, B.J.; Boehm, A.B. Detection of SARS-CoV-2 Variants Mu, Beta, Gamma, Lambda, Delta, Alpha, and Omicron in Wastewater Settled Solids Using Mutation-Specific Assays Is Associated with Regional Detection of Variants in Clinical Samples. Appl. Environ. Microbiol. 2022, 88, e00045-22. [Google Scholar] [CrossRef] [PubMed]
- Lee, W.L.; Armas, F.; Guarneri, F.; Gu, X.; Formenti, N.; Wu, F.; Chandra, F.; Parisio, G.; Chen, H.; Xiao, A.; et al. Rapid displacement of SARS-CoV-2 variant Delta by Omicron revealed by allele-specific PCR in wastewater. Water Res. 2022, 221, 118809. [Google Scholar] [CrossRef]
- Takashita, E.; Kinoshita, N.; Yamayoshi, S.; Sakai-Tagawa, Y.; Fujisaki, S.; Ito, M.; Iwatsuki-Horimoto, K.; Chiba, S.; Halfmann, P.; Nagai, H.; et al. Efficacy of Antibodies and Antiviral Drugs against Covid-19 Omicron Variant. N. Engl. J. Med. 2022, 386, 995–998. [Google Scholar] [CrossRef]
- de Jonge, E.F.; Peterse, C.M.; Koelewijn, J.M.; van der Drift, A.R.; van der Beek, R.; Nagelkerke, E.; Lodder, W.J. The detection of monkeypox virus DNA in wastewater samples in the Netherlands. Sci. Total Environ. 2022, 852, 158265. [Google Scholar] [CrossRef] [PubMed]
- Zuckerman, N.S.; Bar-Or, I.; Sofer, D.; Bucris, E.; Morad, H.; Shulman, L.M.; Levi, N.; Weiss, L.; Aguvaev, I.; Cohen, Z.; et al. Emergence of genetically linked vaccine-originated poliovirus type 2 in the absence of oral polio vaccine, Jerusalem, April to July 2022. Eurosurveillance 2022, 27, 2200694. [Google Scholar] [CrossRef] [PubMed]
- Link-Gelles, R.; Lutterloh, E.; Schnabel Ruppert, P.; Backenson, P.B.; St George, K.; Rosenberg, E.S.; Anderson, B.J.; Fuschino, M.; Popowich, M.; Punjabi, C.; et al. Public Health Response to a Case of Paralytic Poliomyelitis in an Unvaccinated Person and Detection of Poliovirus in Wastewater—New York, June–August 2022. MMWR 2022, 71, 1065–1068. [Google Scholar]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Westcott, C.E.; Sokoloski, K.J.; Rouchka, E.C.; Chariker, J.H.; Holm, R.H.; Yeager, R.A.; Moore, J.B., IV; Elliott, E.M.; Talley, D.; Bhatnagar, A.; et al. The Detection of Periodic Reemergence Events of SARS-CoV-2 Delta Strain in Communities Dominated by Omicron. Pathogens 2022, 11, 1249. https://doi.org/10.3390/pathogens11111249
Westcott CE, Sokoloski KJ, Rouchka EC, Chariker JH, Holm RH, Yeager RA, Moore JB IV, Elliott EM, Talley D, Bhatnagar A, et al. The Detection of Periodic Reemergence Events of SARS-CoV-2 Delta Strain in Communities Dominated by Omicron. Pathogens. 2022; 11(11):1249. https://doi.org/10.3390/pathogens11111249
Chicago/Turabian StyleWestcott, Claire E., Kevin J. Sokoloski, Eric C. Rouchka, Julia H. Chariker, Rochelle H. Holm, Ray A. Yeager, Joseph B. Moore, IV, Erin M. Elliott, Daymond Talley, Aruni Bhatnagar, and et al. 2022. "The Detection of Periodic Reemergence Events of SARS-CoV-2 Delta Strain in Communities Dominated by Omicron" Pathogens 11, no. 11: 1249. https://doi.org/10.3390/pathogens11111249





