Trypanosoma brucei Acyl-Protein Thioesterase-like (TbAPT-L) Is a Lipase with Esterase Activity for Short and Medium-Chain Fatty Acids but Has No Depalmitoylation Activity
Abstract
:1. Introduction
2. Materials and Methods
2.1. Bioinformatics Analysis and Multiple Sequence Alignment (MSA)
2.2. Cell Culture
2.3. Alamar Blue Cell Viability Assay
2.4. Depalmitoylation Probe (DPP) Fluorescence Assays in Live Cells
2.5. DNA Construct Cloning and Transfections
2.6. Immunofluorescence Microscopy
2.7. In Vitro Growth Curves
2.8. qRT-PCR Detection of TbAPT-L Expression from cDNA
2.9. Recombinant Protein Expression and Purification
2.10. Western Blotting and Detection of TbAPT-L Protein from Whole Cell Lysates
2.11. Nitrophenyl Octanoate Assay
2.12. Fluorogenic Substrate Assays
3. Results
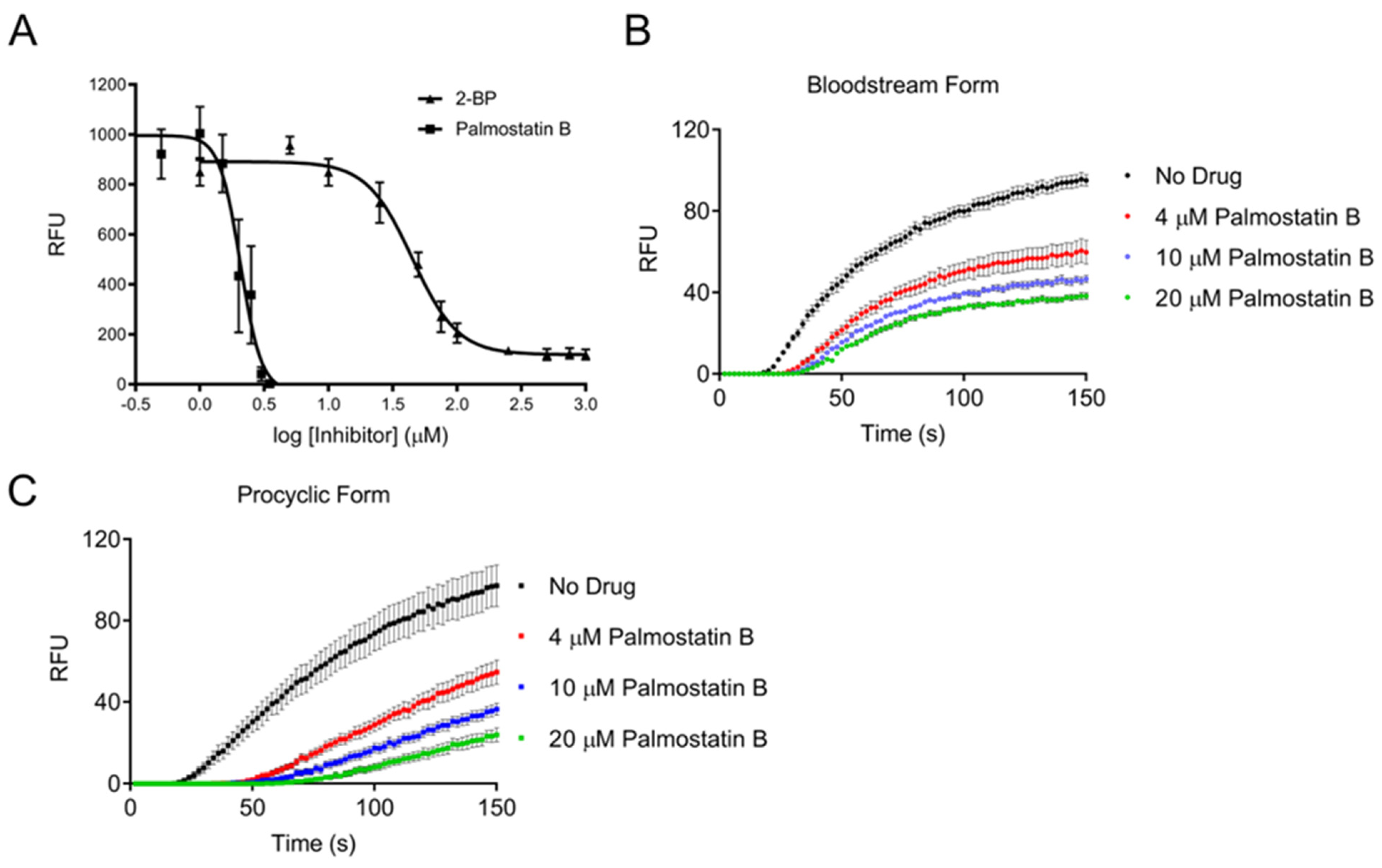
3.1. Trypanosoma brucei Cells Possess a Depalmitoylation Activity Sensitive to Palmostatin B
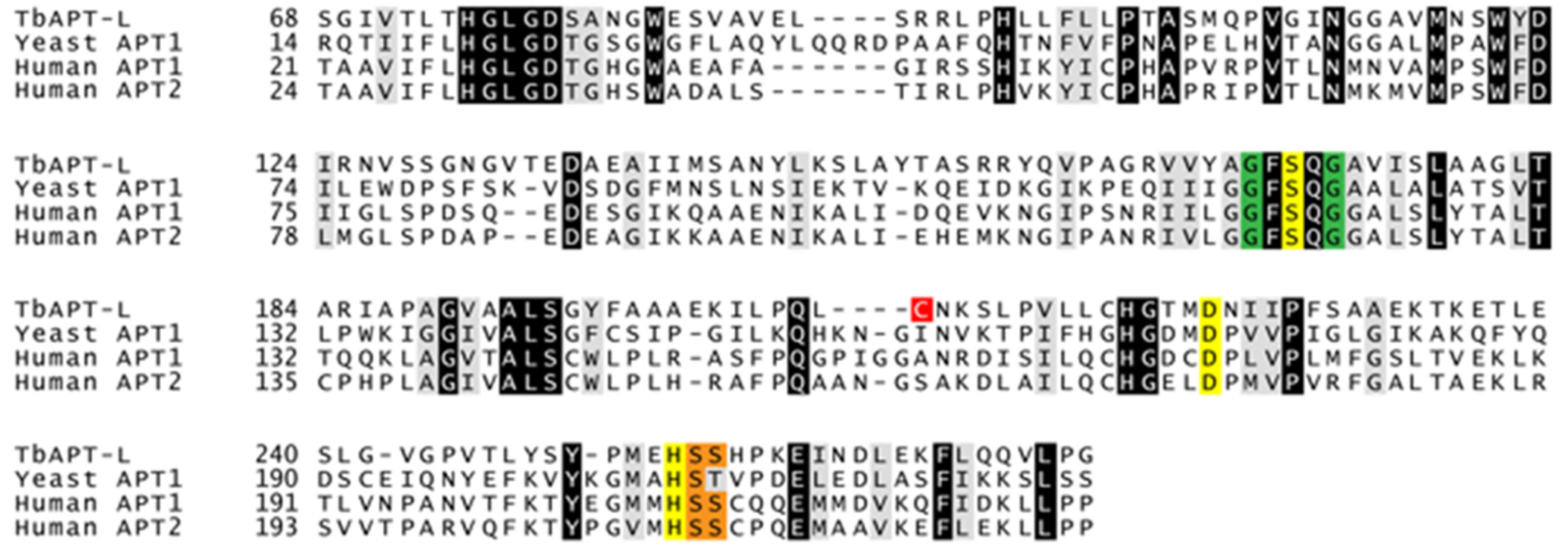
3.2. Identification of α/β Hydrolases and APT1 Homologue in Trypanosoma brucei
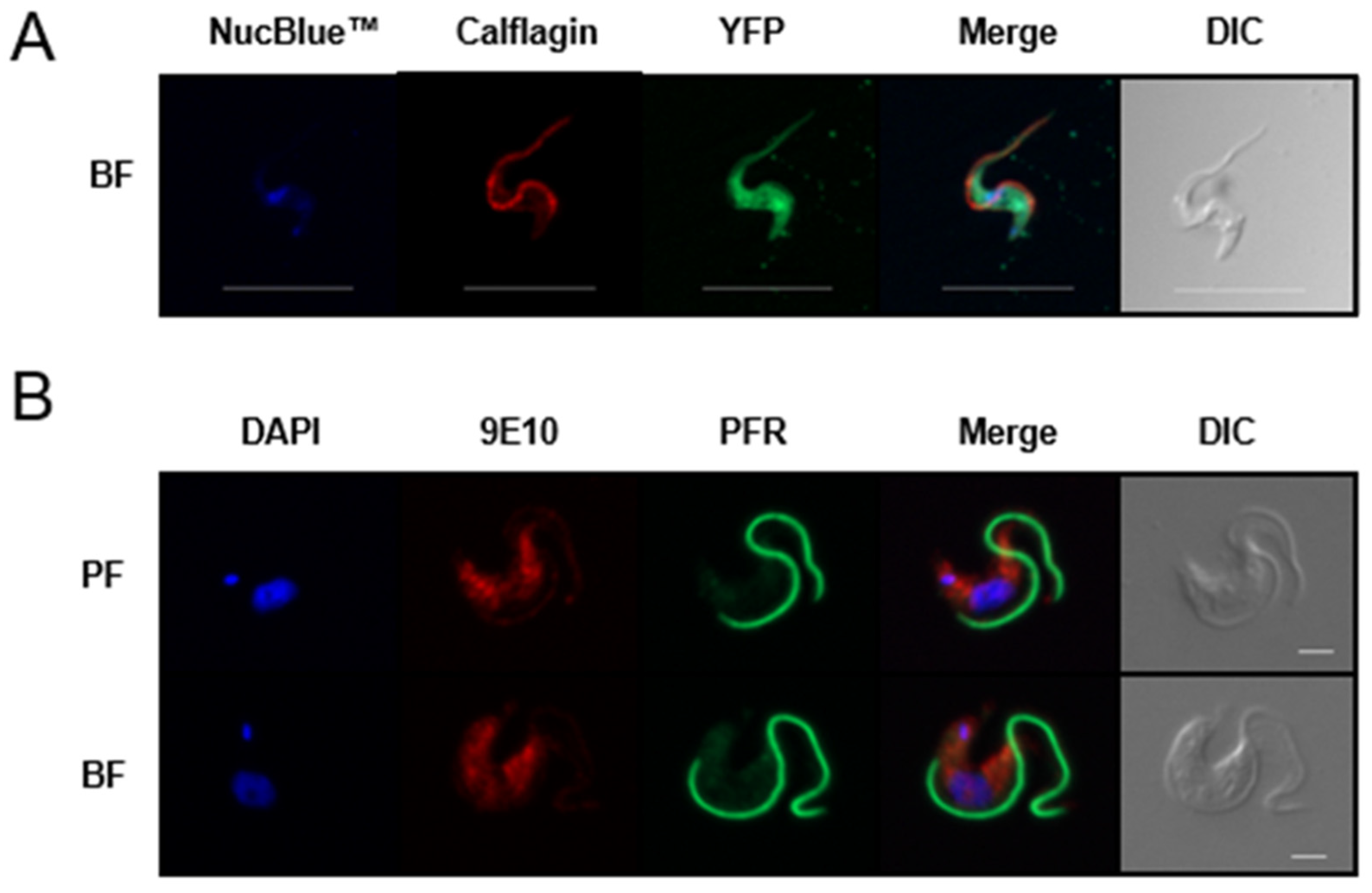
3.3. TbAPT-L Is a Soluble Enzyme with Cytoplasmic Localization
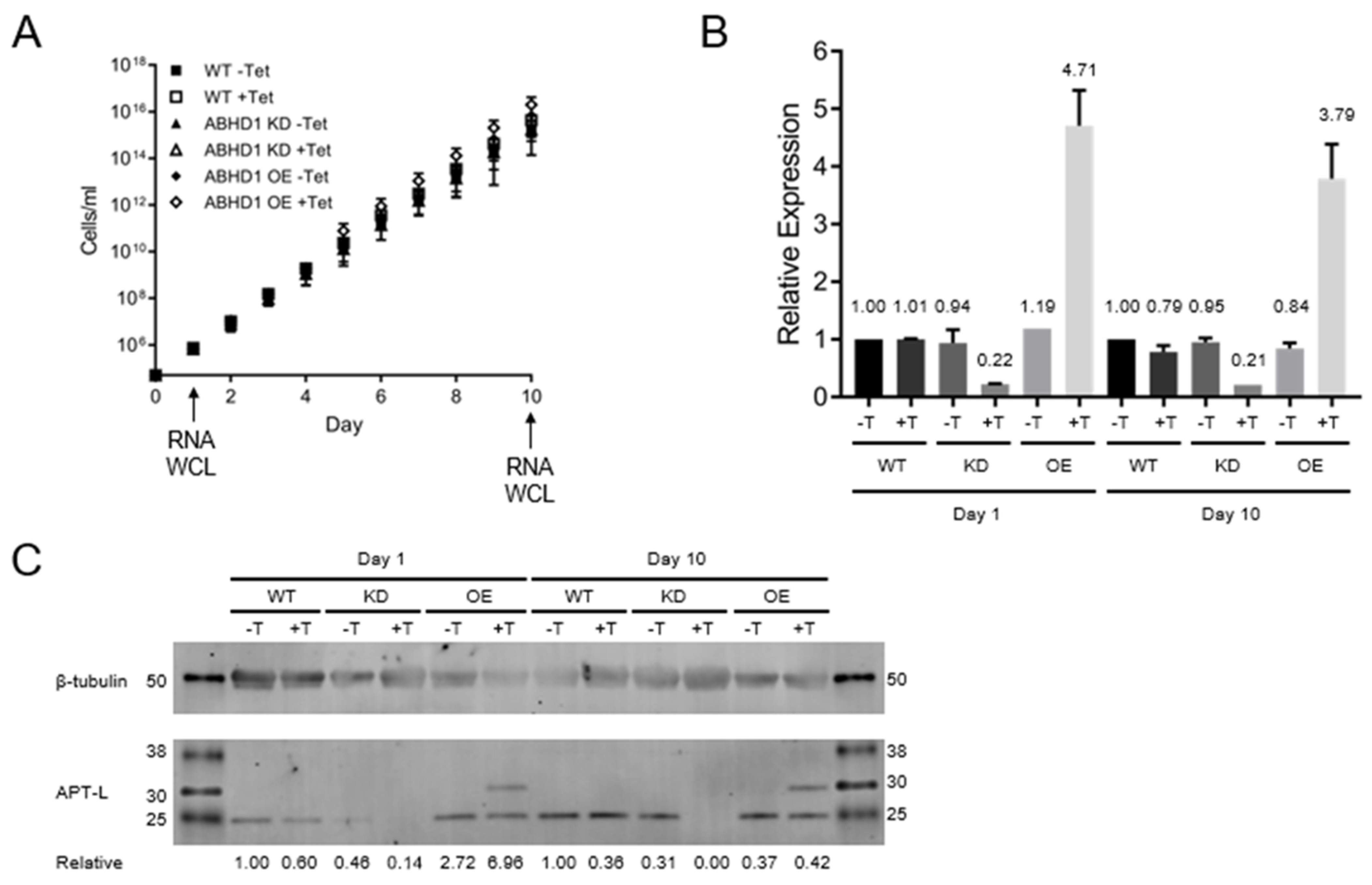
3.4. TbAPT-L Is Dispensable for In Vitro Growth in Bloodstream
3.5. Depalmitoylation Activity in Live Bloodstream form Cells Is Independent of TbAPT-L Expression
3.6. Esterase Activity in Bloodstream form Lysates Is Independent of TbAPT-L Expression
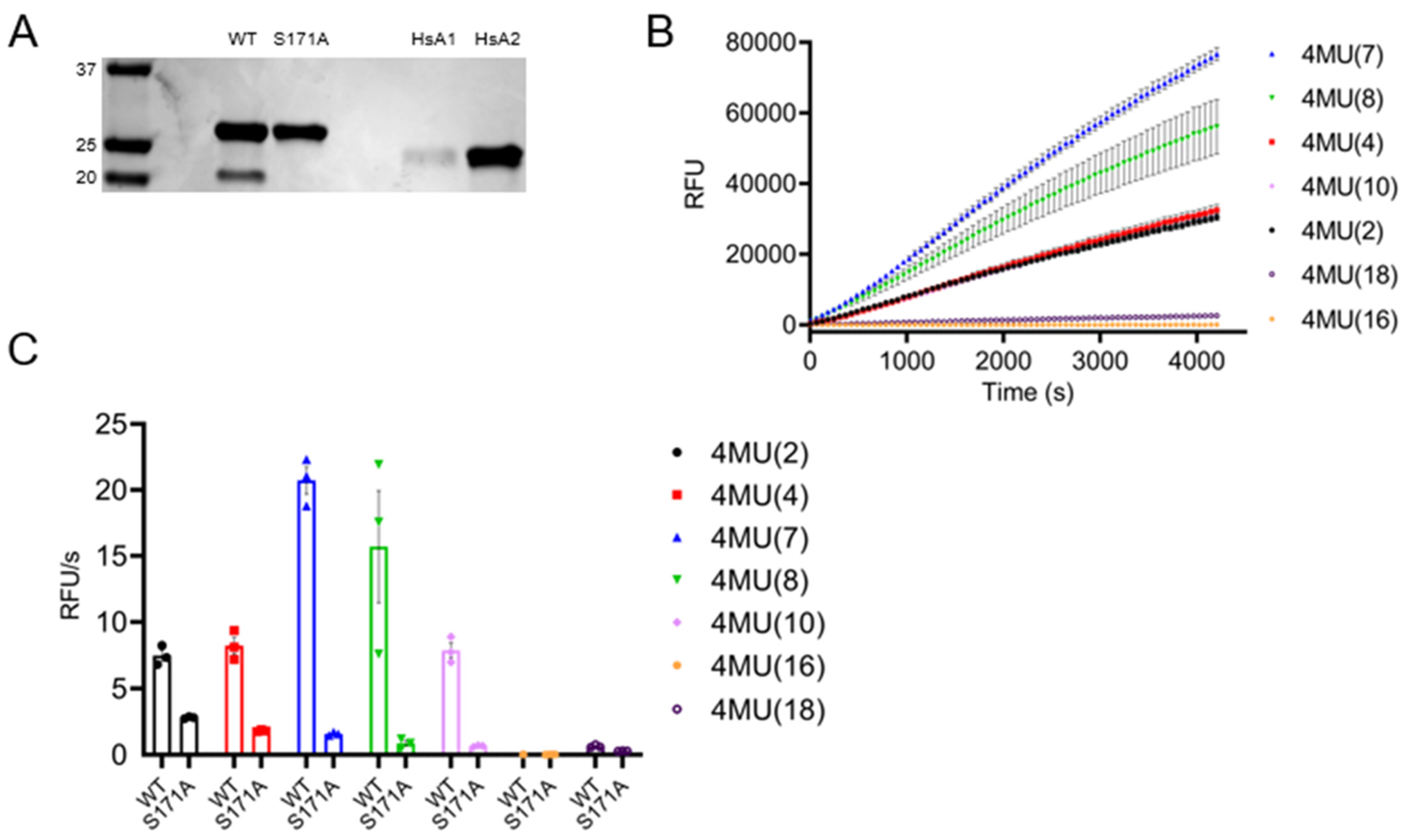
3.7. TbAPT-L Is a Bona Fide α/β Hydrolase with Esterase Activity for Synthetic Fluorogenic Short and Medium Acyl Chain Esters
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Chamberlain, L.H.; Shipston, M.J. The physiology of protein S-acylation. Physiol. Rev. 2015, 95, 341–376. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Muszbek, L.; Haramura, G.; Cluette-Brown, J.E.; Van Cott, E.M.; Laposata, M. The pool of fatty acids covalently bound to platelet proteins by thioester linkages can be altered by exogenously supplied fatty acids. Lipids 1999, 34 (Suppl. 1), S331–S337. [Google Scholar] [CrossRef] [PubMed]
- Greaves, J.; Munro, K.R.; Davidson, S.C.; Riviere, M.; Wojno, J.; Smith, T.K.; Tomkinson, N.C.O.; Chamberlain, L.H. Molecular basis of fatty acid selectivity in the zDHHC family of S-acyltransferases revealed by click chemistry. Proc. Natl. Acad. Sci. USA 2017, 114, E1365–E1374. [Google Scholar] [CrossRef] [Green Version]
- Zaballa, M.E.; van der Goot, F.G. The molecular era of protein S-acylation: Spotlight on structure, mechanisms, and dynamics. Crit. Rev. Biochem. Mol. Biol. 2018, 53, 420–451. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rana, M.S.; Kumar, P.; Lee, C.J.; Verardi, R.; Rajashankar, K.R.; Banerjee, A. Fatty acyl recognition and transfer by an integral membrane. Science 2018, 359, 6372. [Google Scholar] [CrossRef] [PubMed]
- Martin, B.R.; Cravatt, B.F. Large-scale profiling of protein palmitoylation in mammalian cells. Nat. Methods 2009, 6, 135–138. [Google Scholar] [CrossRef] [Green Version]
- Martin, B.R.; Wang, C.; Adibekian, A.; Tully, S.E.; Cravatt, B.F. Global profiling of dynamic protein palmitoylation. Nat. Methods 2011, 9, 84–89. [Google Scholar] [CrossRef] [Green Version]
- Wan, J.; Roth, A.F.; Bailey, A.O.; Davis, N.G. Palmitoylated proteins: Purification and identification. Nat. Protoc. 2007, 2, 1573–1584. [Google Scholar] [CrossRef]
- Yang, W.; Di Vizio, D.; Kirchner, M.; Steen, H.; Freeman, M.R. Proteome scale characterization of human S-acylated proteins in lipid raft-enriched and non-raft membranes. Mol. Cell. Proteom. 2010, 9, 54–70. [Google Scholar] [CrossRef] [Green Version]
- Roth, A.F.; Feng, Y.; Chen, L.; Davis, N.G. The yeast DHHC cysteine-rich domain protein Akr1p is a palmitoyl transferase. J. Cell. Biol. 2002, 159, 23–28. [Google Scholar] [CrossRef]
- Lobo, S.; Greentree, W.K.; Linder, M.E.; Deschenes, R.J. Identification of a Ras palmitoyltransferase in Saccharomyces cerevisiae. J. Biol. Chem. 2002, 277, 41268–41273. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ohno, Y.; Kihara, A.; Sano, T.; Igarashi, Y. Intracellular localization and tissue-specific distribution of human and yeast DHHC cysteine-rich domain-containing proteins. Biochim. Biophys. Acta Mol. Cell Biol. Lipids 2006, 1761, 474–483. [Google Scholar] [CrossRef] [PubMed]
- Mitchell, D.A.; Vasudevan, A.; Linder, M.E.; Deschenes, R.J. Protein palmitoylation by a family of DHHC protein S-acyltransferases. J. Lipid Res. 2006, 47, 1118–1127. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Won, S.J.; Cheung See Kit, M.; Martin, B.R. Protein depalmitoylases. Crit. Rev. Biochem. Mol. Biol. 2018, 53, 83–98. [Google Scholar] [CrossRef]
- Won, S.J.; Davda, D.; Labby, K.J.; Hwang, S.Y.; Pricer, R.; Majmudar, J.D.; Armacost, K.A.; Rodriguez, L.A.; Rodriguez, C.L.; Chong, F.S.; et al. Molecular Mechanism for Isoform-Selective Inhibition of Acyl Protein Thioesterases 1 and 2 (APT1 and APT2). ACS Chem. Biol. 2016, 11, 3374–3382. [Google Scholar] [CrossRef] [Green Version]
- Devedjiev, Y.; Dauter, Z.; Kuznetsov, S.R.; Jones, T.L.; Derewenda, Z.S. Crystal structure of the human acyl protein thioesterase I from a single X-ray data set to 1.5 A. Structure 2000, 8, 1137–1146. [Google Scholar] [CrossRef] [Green Version]
- Das, A.K.; Bellizzi, J.J., 3rd; Tandel, S.; Biehl, E.; Clardy, J.; Hofmann, S.L. Structural basis for the insensitivity of a serine enzyme (palmitoyl-protein thioesterase) to phenylmethylsulfonyl fluoride. J. Biol. Chem. 2000, 275, 23847–23851. [Google Scholar] [CrossRef] [Green Version]
- Lin, D.T.; Conibear, E. ABHD17 proteins are novel protein depalmitoylases that regulate N-Ras palmitate turnover and subcellular localization. eLife 2015, 4, e11306. [Google Scholar] [CrossRef]
- Camp, L.A.; Hofmann, S.L. Purification and properties of a palmitoyl-protein thioesterase that cleaves palmitate from H-Ras. J. Biol. Chem. 1993, 268, 22566–22574. [Google Scholar] [CrossRef]
- Toyoda, T.; Sugimoto, H.; Yamashita, S. Sequence, expression in Escherichia coli, and characterization of lysophospholipase, I.I. Biochim. Biophys. Acta 1999, 1437, 182–193. [Google Scholar] [CrossRef]
- Tomatis, V.M.; Trenchi, A.; Gomez, G.A.; Daniotti, J.L. Acyl-protein thioesterase 2 catalyzes the deacylation of peripheral membrane-associated GAP-43. PLoS ONE 2010, 5, e15045. [Google Scholar] [CrossRef] [Green Version]
- Amara, N.; Foe, I.T.; Onguka, O.; Garland, M.; Bogyo, M. Synthetic Fluorogenic Peptides Reveal Dynamic Substrate Specificity of Depalmitoylases. Cell. Chem. Biol. 2019, 26, 35–47. [Google Scholar] [CrossRef]
- Duncan, J.A.; Gilman, A.G. Characterization of Saccharomyces cerevisiae acyl-protein thioesterase 1, the enzyme responsible for G protein alpha subunit deacylation in vivo. J. Biol. Chem. 2002, 277, 31740–31752. [Google Scholar] [CrossRef] [Green Version]
- Bellizzi, J.J., 3rd; Widom, J.; Kemp, C.; Lu, J.Y.; Das, A.K.; Hofmann, S.L.; Clardy, J. The crystal structure of palmitoyl protein thioesterase 1 and the molecular basis of infantile neuronal ceroid lipofuscinosis. Proc. Natl. Acad. Sci. USA 2000, 97, 4573–4578. [Google Scholar] [CrossRef] [Green Version]
- Rocks, O.; Peyker, A.; Kahms, M.; Verveer, P.J.; Koerner, C.; Lumbierres, M.; Kuhlmann, J.; Waldmann, H.; Wittinghofer, A.; Bastiaens, P.I.H. An acylation cycle regulates localization and activity of palmitoylated Ras isoforms. Science 2005, 307, 1746–1752. [Google Scholar] [CrossRef] [PubMed]
- Rocks, O.; Gerauer, M.; Vartak, N.; Koch, S.; Huang, Z.P.; Pechlivanis, M.; Kuhlmann, J.; Brunsveld, L.; Chandra, A.; Ellinger, B.; et al. The palmitoylation machinery is a spatially organizing system for peripheral membrane proteins. Cell 2010, 141, 458–471. [Google Scholar] [CrossRef] [Green Version]
- Vartak, N.; Papke, B.; Grecco, H.E.; Rossmannek, L.; Waldmann, H.; Hedberg, C.; Bastiaens, P.I.H. The autodepalmitoylating activity of APT maintains the spatial organization of palmitoylated membrane proteins. Biophys. J. 2014, 106, 93–105. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Emmer, B.T.; Nakayasu, E.S.; Souther, C.; Choi, H.; Sobreira, T.J.; Epting, C.L.; Nesvizhskii, A.I.; Almeida, I.C.; Engman, D.M. Global analysis of protein palmitoylation in African trypanosomes. Eukaryot Cell 2011, 10, 455–463. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Aslett, M.; Aurrecoechea, C.; Berriman, M.; Brestelli, J.; Brunk, B.P.; Carrington, M.; Depledge, D.P.; Fischer, S.; Gajria, B.; Gao, X.; et al. TriTrypDB: A functional genomic resource for the Trypanosomatidae. Nucleic Acids Res. 2010, 38, D457–D462. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Coordinators, N.R. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2016, 44, D7–D19. [Google Scholar]
- Thompson, J.D.; Higgins, D.G.; Gibson, T.J. CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994, 22, 4673–4680. [Google Scholar] [CrossRef]
- Wirtz, E.; Leal, S.; Ochatt, C.; Cross, G.A. A tightly regulated inducible expression system for conditional gene knock-outs and dominant-negative genetics in Trypanosoma brucei. Mol. Biochem. Parasitol. 1999, 99, 89–101. [Google Scholar] [CrossRef]
- Brun, R.; Schoenenberger, M. Cultivation and in vitro cloning or procyclic culture forms of Trypanosoma brucei in a semi-defined medium. Acta Trop. 1979, 36, 289–292. [Google Scholar]
- Sykes, M.L.; Avery, V.M. Development of an Alamar Blue viability assay in 384-well format for high throughput whole cell screening of Trypanosoma brucei brucei bloodstream form strain 427. Am. J. Trop. Med. Hyg. 2009, 81, 665–674. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kathayat, R.S.; Cao, Y.; Elvira, P.D.; Sandoz, P.A.; Zaballa, M.E.; Springer, M.Z.; Drake, L.E.; Macleod, K.F.; van der Groot, F.G.; Dickinson, B.C. Active and dynamic mitochondrial S-depalmitoylation revealed by targeted fluorescent probes. Nat. Commun. 2018, 9, 334. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Redmond, S.; Vadivelu, J.; Field, M.C. RNAit: An automated web-based tool for the selection of RNAi targets in Trypanosoma brucei. Mol. Biochem. Parasitol. 2003, 128, 115–118. [Google Scholar] [CrossRef]
- Wang, Z.; Morris, J.C.; Drew, M.E.; Englund, P.T. Inhibition of Trypanosoma brucei gene expression by RNA interference using an integratable vector with opposing T7 promoters. J. Biol. Chem. 2000, 275, 40174–40179. [Google Scholar] [CrossRef] [Green Version]
- Burkard, G.; Fragoso, C.M.; Roditi, I. Highly efficient stable transformation of bloodstream forms of Trypanosoma brucei. Mol. Biochem. Parasitol. 2007, 153, 220–223. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Child, M.A.; Hall, C.I.; Beck, J.R.; Ofori, L.O.; Albrow, V.E.; Garland, M.; Bowyer, P.W.; Bradley, P.J.; Powers, J.C.; Boothroyd, J.C.; et al. Small-molecule inhibition of a depalmitoylase enhances Toxoplasma host-cell invasion. Nat. Chem. Biol. 2013, 9, 651–656. [Google Scholar] [CrossRef] [Green Version]
- Dekker, F.J.; Rocks, O.; Vartak, N.; Menninger, S.; Hedberg, C.; Balamurugan, R.; Wetzel, S.; Renner, S.; Gerauer, M.; Schölermann, B.; et al. Small-molecule inhibition of APT1 affects Ras localization and signaling. Nat. Chem. Biol. 2010, 6, 449–456. [Google Scholar] [CrossRef] [PubMed]
- Foe, I.T.; Onguka, O.; Amberg-Johnson, K.; Garner, R.M.; Amara, N.; Beatty, W.; Yeh, E.; Bogyo, M. The Toxoplasma gondii Active Serine Hydrolase 4 Regulates Parasite Division and Intravacuolar Parasite Architecture. mSphere 2018, 3, e00393-18. [Google Scholar] [CrossRef] [PubMed]
- Webb, Y.; Hermida-Matsumoto, L.; Resh, M.D. Inhibition of protein palmitoylation, raft localization, and T cell signaling by 2-bromopalmitate and polyunsaturated fatty acids. J. Biol. Chem. 2000, 275, 261–270. [Google Scholar] [CrossRef] [Green Version]
- Pedro, M.P.; Vilcaes, A.A.; Tomatis, V.M.; Oliveira, R.G.; Gomez, G.A.; Daniotti, J.L. 2-Bromopalmitate reduces protein deacylation by inhibition of acyl-protein thioesterase enzymatic activities. PLoS ONE 2013, 8, e75232. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Davda, D.; El Azzouny, M.A.; Tom, C.T.; Hernandez, J.L.; Majmudar, J.D.; Kennedy, R.T.; Martin, B.R. Profiling targets of the irreversible palmitoylation inhibitor 2-bromopalmitate. ACS Chem. Biol. 2013, 8, 1912–1917. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Savinainen, J.R.; Patel, J.Z.; Parkkari, T.; Navia-Paldanius, D.; Marjamaa, J.J.; Laitinen, T.; Nevalainen, T.; Laitinen, J.T. Biochemical and pharmacological characterization of the human lymphocyte antigen B-associated transcript 5 (BAT5/ABHD16A). PLoS ONE 2014, 9, e109869. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Azizi, S.A.; Kathayat, R.S.; Dickinson, B.C. Activity-Based Sensing of S-Depalmitoylases: Chemical Technologies and Biological Discovery. Acc. Chem. Res. 2019, 52, 3029–3038. [Google Scholar] [CrossRef]
- Kathayat, R.S.; Dickinson, B.C. Measuring S-Depalmitoylation Activity In Vitro and In Live Cells with Fluorescent Probes. Methods Mol. Biol. 2019, 2009, 99–109. [Google Scholar] [PubMed]
- Kathayat, R.S.; Elvira, P.D.; Dickinson, B.C. A fluorescent probe for cysteine depalmitoylation reveals dynamic APT signaling. Nat. Chem. Biol. 2017, 13, 150–152. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Duncan, J.A.; Gilman, A.G. A cytoplasmic acyl-protein thioesterase that removes palmitate from G protein alpha subunits and p21(RAS). J. Biol. Chem. 1998, 273, 15830–15837. [Google Scholar] [CrossRef] [Green Version]
- Holmquist, M. Alpha/Beta-hydrolase fold enzymes: Structures, functions and mechanisms. Curr. Protein Pept. Sci. 2000, 1, 209–235. [Google Scholar] [CrossRef]
- Ollis, D.L.; Cheah, E.; Cygler, M.; Dijkstra, B.; Frolow, F.; Franken, S.M.; Harel, M.; Remington, S.J.; Silman, I.; Schrag, J. The alpha/beta hydrolase fold. Protein Eng. 1992, 5, 197–211. [Google Scholar] [CrossRef]
- Kourist, R.; Jochens, H.; Bartsch, S.; Kuipers, R.; Padhi, S.K.; Gall, M.; Böttcher, D.; Joosten, H.-J.; Bornscheuer, U.T. The alpha/beta-hydrolase fold 3DM database (ABHDB) as a tool for protein engineering. Chembiochem. 2010, 11, 1635–1643. [Google Scholar] [CrossRef] [PubMed]
- Kemp, L.E.; Rusch, M.; Adibekian, A.; Bullen, H.E.; Graindorge, A.; Freymond, C.; Rottmann, M.; Braun-Breton, C.; Baumeister, S.; Profetye, A.T.; et al. Characterization of a serine hydrolase targeted by acyl-protein thioesterase inhibitors in Toxoplasma gondii. J. Biol. Chem. 2013, 288, 27002–27018. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yokoi, N.; Fukata, Y.; Sekiya, A.; Murakami, T.; Kobayashi, K.; Fukata, M. Identification of PSD-95 Depalmitoylating Enzymes. J. Neurosci. 2016, 36, 6431–6444. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Merritt, E.A.; Holmes, M.; Buckner, F.S.; Van Voorhis, W.C.; Quartly, E.; Phizicky, E.M.; Lauricella, A.; Luft, J.; DeTitta, G.; Neely, H.; et al. Structure of a Trypanosoma brucei alpha/beta-hydrolase fold protein with unknown function. Acta Cryst. Sect. F Struct. Biol. Cryst. Commun. 2008, 64, 474–478. [Google Scholar] [CrossRef] [Green Version]
- Sadeghi, R.S.; Kulej, K.; Kathayat, R.S.; Garcia, B.A.; Dickinson, B.C.; Brady, D.C.; Witze, E.S. Wnt5a signaling induced phosphorylation increases APT1 activity and promotes melanoma metastatic behavior. eLife 2018, 7, e34362. [Google Scholar] [CrossRef]
- Zhou, F.; Xue, Y.; Yao, X.; Xu, Y. CSS-Palm: Palmitoylation site prediction with a clustering and scoring strategy (CSS). Bioinformatics 2006, 22, 894–896. [Google Scholar] [CrossRef] [Green Version]
- Vasquez, J.J.; Hon, C.C.; Vanselow, J.T.; Schlosser, A.; Siegel, T.N. Comparative ribosome profiling reveals extensive translational complexity in different Trypanosoma brucei life cycle stages. Nucleic Acids Res. 2014, 42, 3623–3637. [Google Scholar] [CrossRef] [PubMed]
- Kong, E.; Peng, S.; Chandra, G.; Sarkar, C.; Zhang, Z.; Bagh, M.B.; Mukherjee, A.B. Dynamic palmitoylation links cytosol-membrane shuttling of acyl-protein thioesterase-1 and acyl-protein thioesterase-2 with that of proto-oncogene H-ras product and growth-associated protein-43. J. Biol. Chem. 2013, 288, 9112–9125. [Google Scholar] [CrossRef] [Green Version]
- Hirano, T.; Kishi, M.; Sugimoto, H.; Taguchi, R.; Obinata, H.; Ohshima, N.; Tatei, K.; Izumi, T. Thioesterase activity and subcellular localization of acylprotein thioesterase 1/lysophospholipase 1. Biochim. Biophys. Acta 2009, 1791, 797–805. [Google Scholar] [CrossRef] [PubMed]
- Brenndorfer, M.; Boshart, M. Selection of reference genes for mRNA quantification in Trypanosoma brucei. Mol. Biochem. Parasitol. 2010, 172, 52–55. [Google Scholar] [CrossRef]
- Burger, M.; Zimmermann, T.J.; Kondoh, Y.; Stege, P.; Watanabe, N.; Osada, H.; Waldmann, H.; Vetter, I.R. Crystal structure of the predicted phospholipase LYPLAL1 reveals unexpected functional plasticity despite close relationship to acyl protein thioesterases. J. Lipid Res. 2012, 53, 43–50. [Google Scholar] [CrossRef] [Green Version]
- Sugimoto, H.; Hayashi, H.; Yamashita, S. Purification, cDNA cloning, and regulation of lysophospholipase from rat liver. J. Biol. Chem. 1996, 271, 7705–7711. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Calero, G.; Gupta, P.; Nonato, M.C.; Tandel, S.; Biehl, E.R.; Hofmann, S.L.; Clardy, J. The crystal structure of palmitoyl protein thioesterase-2 (PPT2) reveals the basis for divergent substrate specificities of the two lysosomal thioesterases, PPT1 and PPT2. J. Biol. Chem. 2003, 278, 37957–37964. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Selvaraju, K.; Gowsalya, R.; Vijayakumar, R.; Nachiappan, V. MGL2/YMR210w encodes a monoacylglycerol lipase in Saccharomyces cerevisiae. FEBS Lett. 2016, 590, 1174–1186. [Google Scholar] [CrossRef] [Green Version]
- Kumar, N.M.; Thunuguntla, V.B.S.C.; Chandra Sekhar, B.; Bondili, J.S. Saccharomyces cerevisiae lipid droplet associated enzyme Ypr147cp shows both TAG lipase and ester hydrolase activities. J. Gen. Appl. Microbiol. 2018, 64, 76–83. [Google Scholar]
- Onguka, O.; Babin, B.M.; Lakemeyer, M.; Foe, I.T.; Amara, N.; Terrell, S.M.; Lum, K.M.; Cieplak, P.; Niphakis, M.J.; Long, J.Z.; et al. Toxoplasma gondii serine hydrolases regulate parasite lipid mobilization during growth and replication within the host. Cell Chem. Biol. 2021, 28, 1501–1513.e5. [Google Scholar] [CrossRef]
- Sadler, N.C.; Wright, A.T. Activity-based protein profiling of microbes. Curr. Opin. Chem. Biol. 2015, 24, 139–144. [Google Scholar] [CrossRef] [Green Version]
- Lord, C.C.; Thomas, G.; Brown, J.M. Mammalian alpha beta hydrolase domain (ABHD) proteins: Lipid metabolizing enzymes at the interface of cell signaling and energy metabolism. Biochim. Biophys. Acta 2013, 1831, 792–802. [Google Scholar] [CrossRef] [Green Version]
- Cao, Y.; Qiu, T.; Kathayat, R.S.; Azizi, S.A.; Thorne, A.K.; Ahn, D.; Fukata, Y.; Fukata, M.; Rice, P.A.; Dickinson, B.C. ABHD10 is an S-depalmitoylase affecting redox homeostasis through peroxiredoxin-5. Nat. Chem. Biol. 2019, 15, 1232–1240. [Google Scholar] [CrossRef]


| # | Primer Name | Purpose | Sequence 5′-3′ |
|---|---|---|---|
| 1 | Tb927.8.6390 RNAi Fwd | RNAi knockdown of TbAPT-L (XhoI) | CTCGAGAATGGTTGGGAGAGTGTTGC |
| 2 | Tb927.8.6390 RNAi Rev | RNAi knockdown of TbAPT-L (HindIII) | AAGCTTGATGATGTTGTCCATCGTGC |
| 3 | Tb427.8.6390 qPCR_F | qRT-PCR TbAPT-L | TTCAGTGCGGCTGAGAAAA |
| 4 | Tb427.8.6390 qPCR_R2 | qRT-PCR TbAPT-L | TTCTTTGGGATGAGAGGAGTG |
| 5 | TERT Fwd | qRT-PCR TERT | GAGCGTGTGACTTCCGAAGG |
| 6 | TERT Rev | qRT-PCR TERT | AGGAACTGTCACGGAGTTTGC |
| 7 | Tb427.8.6390 pLEW79 Fwd | TET-inducible over-expression of TbAPT-L-2×myc (HindIII) | AAGCTTATGTTTGGCACGCCGGTTG |
| 8 | Tb427.8.6390 pLEW79 Rev | TET-inducible over-expression of TbAPT-L-2×myc (XbaI) | TCTAGACGATTTCGATGAAGGTCCGGG |
| 9 | YFP-TbAPT-L pLEW100v5 F | TET-inducible over-expression YFP-TbAPT-L (XhoI) | GTCCTCGAGGGATGTTTGGCACGCCGGTTG |
| 10 | YFP-TbAPT-L pLEW100v5 R | TET-inducible over-expression YFP-TbAPT-L (BamHI) (STOP) | GTCGGATCCTTACGATTTCGATGAAGGTCCGG |
| 11 | TbAPT-L pET28a Fwd | IPTG-inducible TbAPT-L over-expression in E. coli (NdeI) | CATATGATGTTTGGCACGCCGGTTG |
| 12 | TbAPT-L pET28a Rev | IPTG-inducible TbAPT-L over-expression in E. coli (XhoI) (STOP) | CTCGAGTTACGATTTCGATGAAGGTCCGG |
| 13 | TbAPT-L S171A Fwd | Site-directed mutagenesis TbAPT-L S171A | GTGTATGCCGGATTCGCGCAAGGTGCTGTTATTTCGCTC |
| 14 | TbAPT-L S171A Rev | Site-directed mutagenesis TbAPT-L S171A | GAAATAACAGCACCTTGCGCGAATCCGGCATACACCACG |
| 15 | TbAPT-L seq Fwd | Sequencing TbAPT-L locus | GTATTAATGGAGGAACAG |
| 16 | TbAPT-L seq Rev | Sequencing TbAPT-L locus | GATTTCGATGAAGGTCCG |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Brown, R.W.B.; Sharma, A.I.; Villanueva, M.R.; Li, X.; Onguka, O.; Zilbermintz, L.; Nguyen, H.; Falk, B.A.; Olson, C.L.; Taylor, J.M.; et al. Trypanosoma brucei Acyl-Protein Thioesterase-like (TbAPT-L) Is a Lipase with Esterase Activity for Short and Medium-Chain Fatty Acids but Has No Depalmitoylation Activity. Pathogens 2022, 11, 1245. https://doi.org/10.3390/pathogens11111245
Brown RWB, Sharma AI, Villanueva MR, Li X, Onguka O, Zilbermintz L, Nguyen H, Falk BA, Olson CL, Taylor JM, et al. Trypanosoma brucei Acyl-Protein Thioesterase-like (TbAPT-L) Is a Lipase with Esterase Activity for Short and Medium-Chain Fatty Acids but Has No Depalmitoylation Activity. Pathogens. 2022; 11(11):1245. https://doi.org/10.3390/pathogens11111245
Chicago/Turabian StyleBrown, Robert W. B., Aabha I. Sharma, Miguel Rey Villanueva, Xiaomo Li, Ouma Onguka, Leeor Zilbermintz, Helen Nguyen, Ben A. Falk, Cheryl L. Olson, Joann M. Taylor, and et al. 2022. "Trypanosoma brucei Acyl-Protein Thioesterase-like (TbAPT-L) Is a Lipase with Esterase Activity for Short and Medium-Chain Fatty Acids but Has No Depalmitoylation Activity" Pathogens 11, no. 11: 1245. https://doi.org/10.3390/pathogens11111245






