RTA1 Is Involved in Resistance to 7-Aminocholesterol and Secretion of Fungal Proteins in Cryptococcus neoformans
Abstract
:1. Introduction
2. Materials and Methods
2.1. Strain Construction
2.2. Resistance to 7-Aminocholesterol
2.3. Capsule Induction
2.4. Capsule Shedding
2.5. Melanin Production
2.6. Urease Production
2.7. Laccase Production
2.8. Phospholipase B Secretion
2.9. Intracellular 7-Aminocholesterol Visualization
2.10. Homology Modeling
2.11. Phylogenetic Analysis
2.12. Promoter Analysis
2.13. Real-Time Quantitative PCR
2.14. Transmission Electron Microscopy (TEM)
2.15. Crystal Structure of 7-Aminocholesterol
2.16. Statistics
3. Results
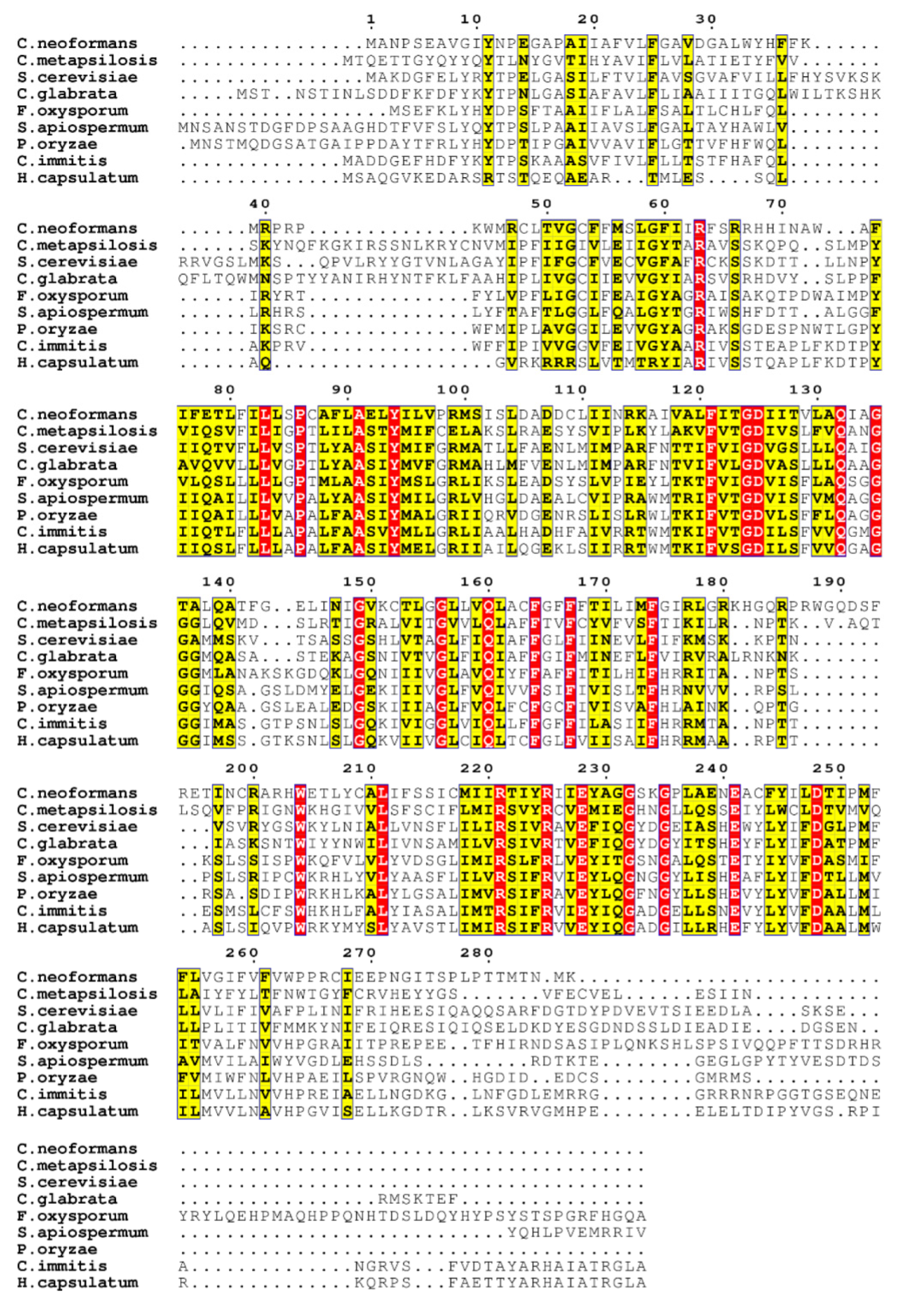
3.1. Sequence Homology
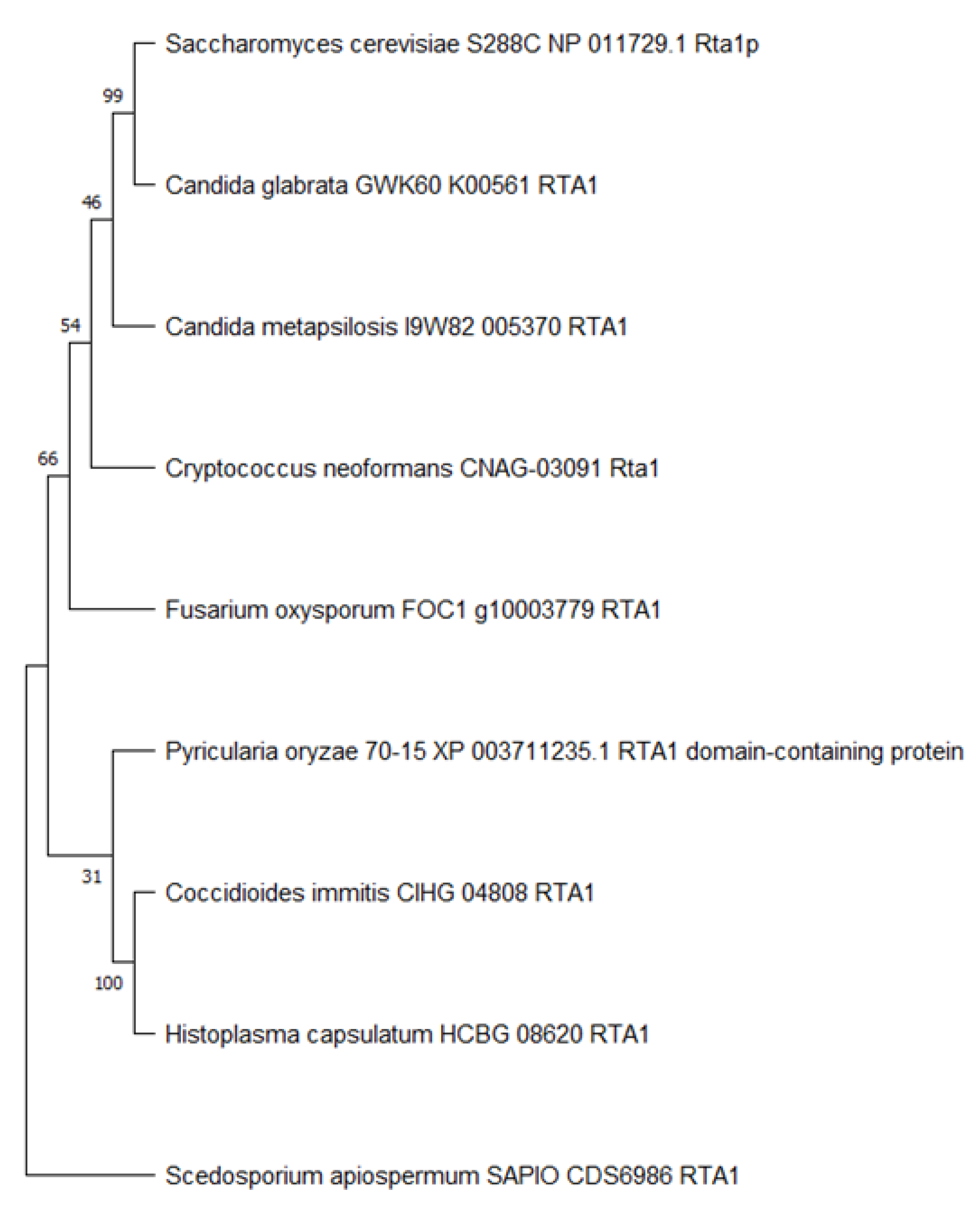
3.2. Rta1 Phylogeny
3.3. RTA1 Is Involved in Resistance to 7-Aminocholesterol
3.4. Intracellular 7-Aminocholesterol Visualization
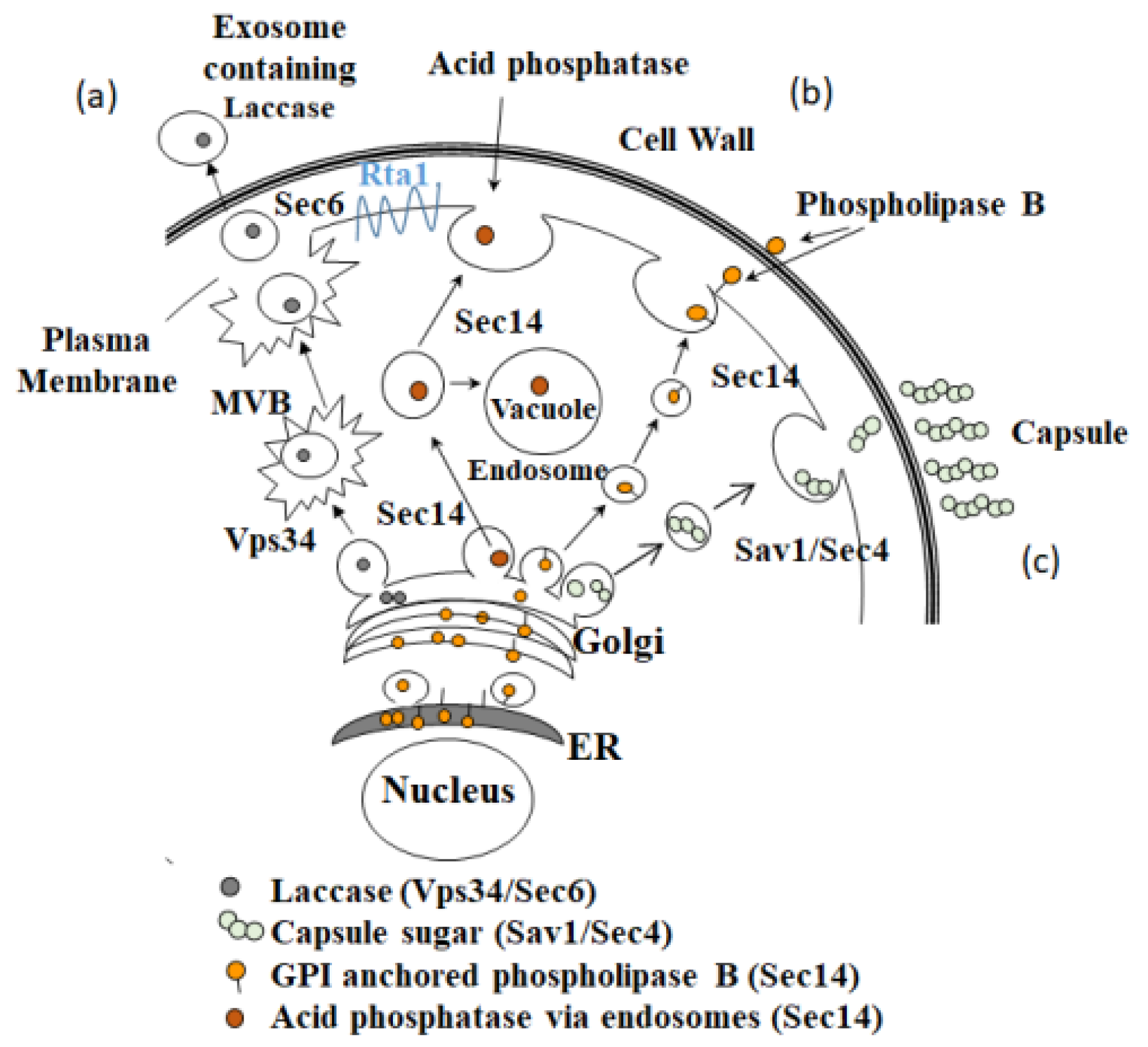
3.5. Secretion Phenotype of the H99, rta1Δ, and rta1Δ+RTA1 Strains
3.6. Urease Secretion in Different Secretion Mutants
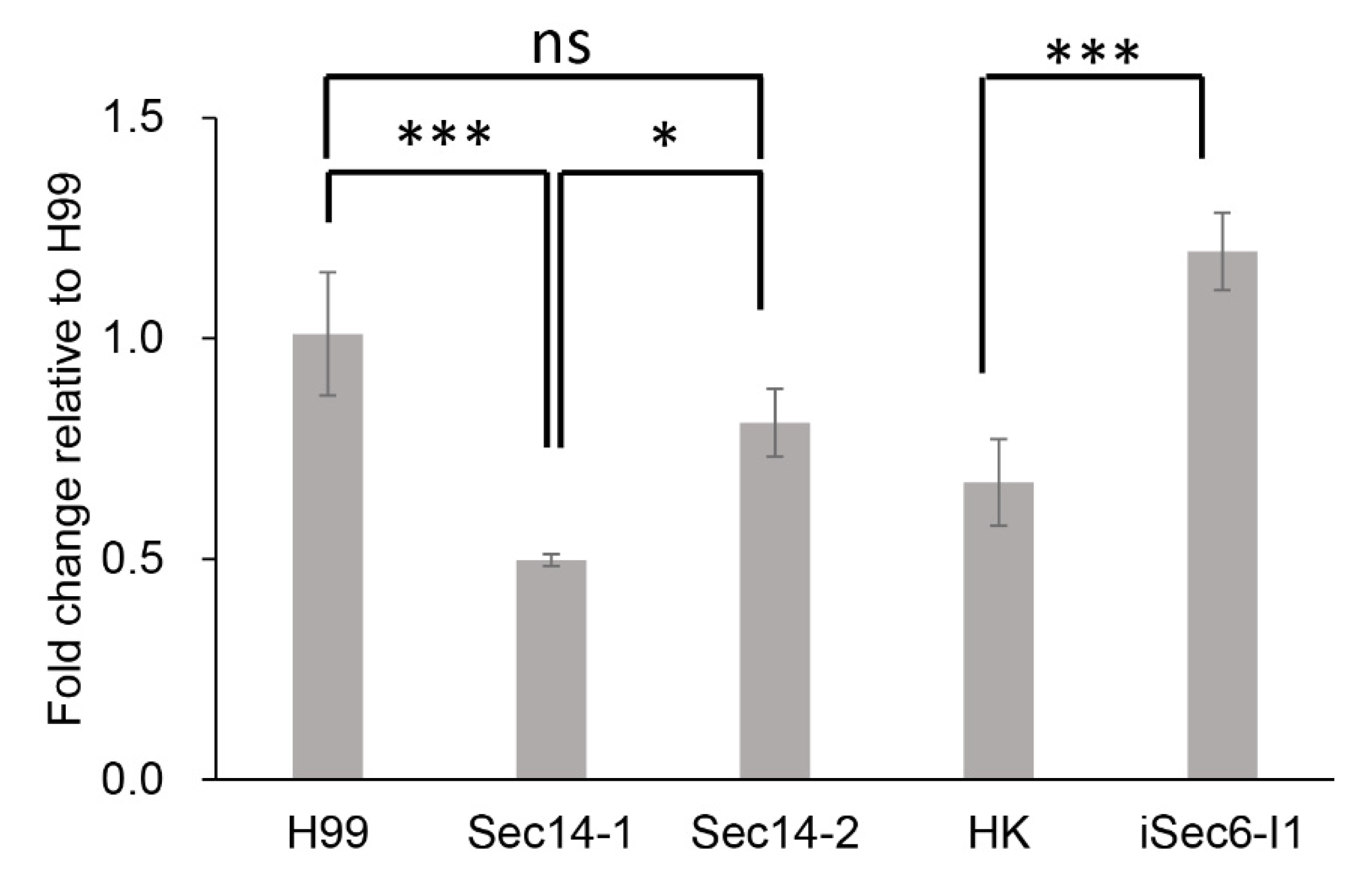
3.7. qPCR of RTA1 Expression in Different Secretion Mutants
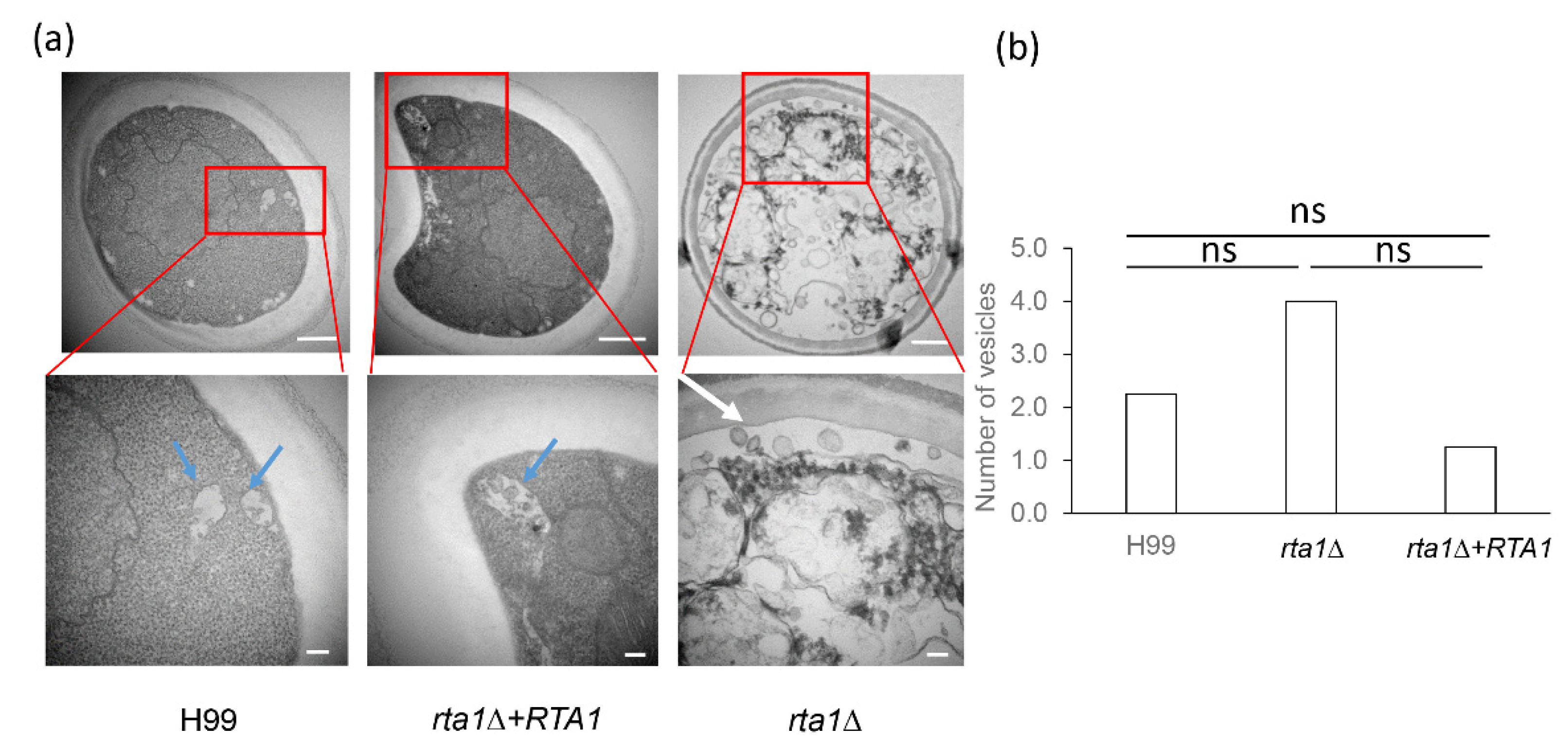
3.8. TEM of the H99, rta1Δ, and rta1Δ+RTA1 Strains
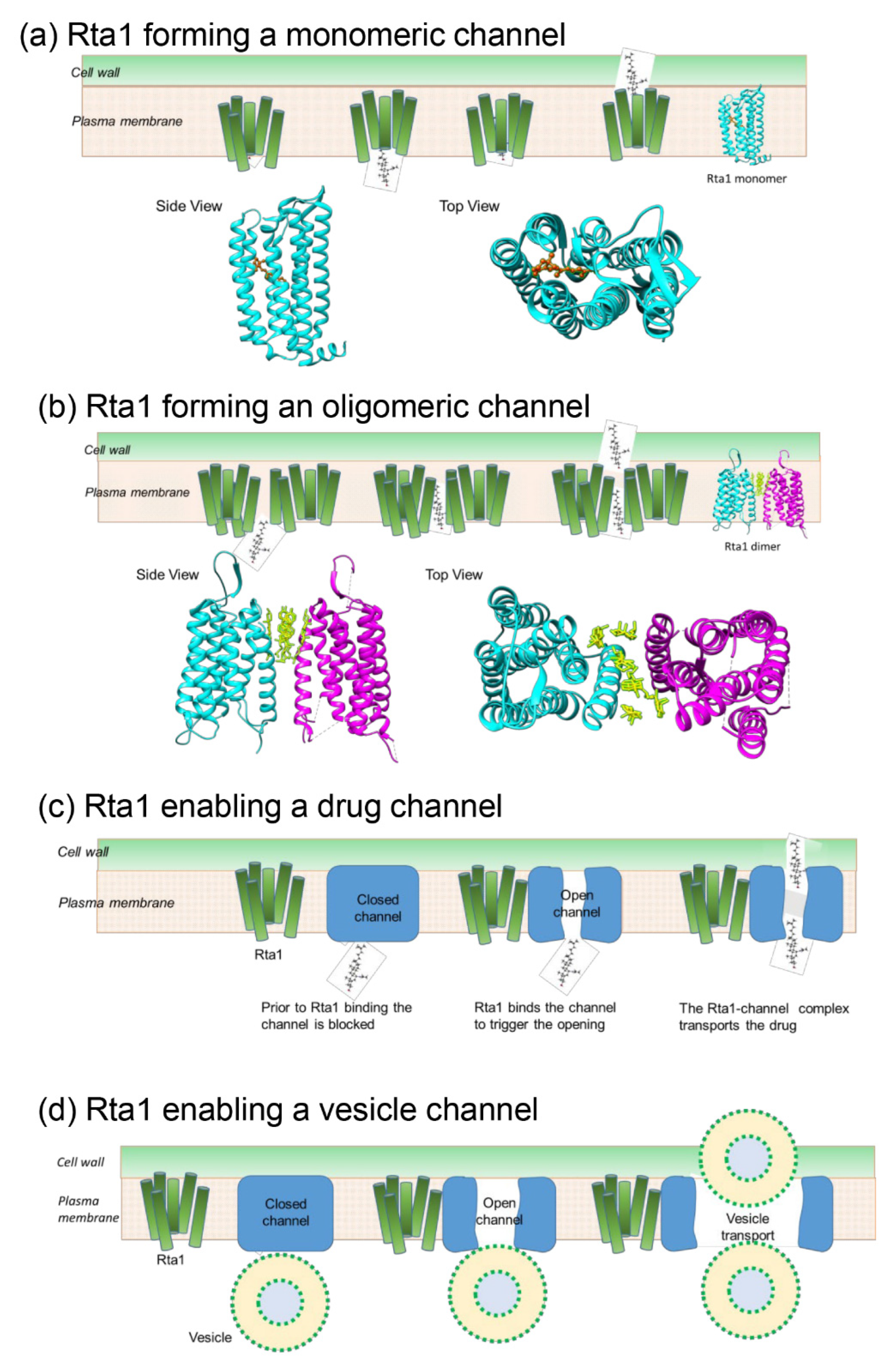
3.9. Potential Model of Rta1 Strucure in the Membrane
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Heitman, J.; Kozel, T.R.; Kwon-Chung, J.; Perfect, J.R.; Casadevall, A. Cryptococcus: From Human Pathogen to Model Yeast; ASM Press: Washington, DC, USA, 2011. [Google Scholar]
- Rajasingham, R.; Smith, R.M.; Park, B.J.; Jarvis, J.N.; Govender, N.P.; Chiller, T.M.; Denning, D.W.; Loyse, A.; Boulware, D.R. Global burden of disease of HIV-associated cryptococcal meningitis: An updated analysis. Lancet Infect. Dis. 2017, 17, 873–881. [Google Scholar] [CrossRef] [Green Version]
- McClelland, E.E.; Casadevall, A.; Eisenman, H.C. Pathogenesis of Cryptococcus neoformans. In New Insights in Medical Mycology; Kavanagh, K., Ed.; Springer: Dordrecht, The Netherlands, 2007; Volume 1, pp. 131–157. [Google Scholar]
- Soustre, I.; Letourneux, Y.; Karst, F. Characterization of the Saccharomyces cerevisiae RTA1 gene involved in 7-aminocholesterol resistance. Curr. Genet. 1996, 30, 121–125. [Google Scholar] [CrossRef] [PubMed]
- Alhanout, K.; Djouhri, L.; Vidal, N.; Brunel, J.M.; Piarroux, R.; Ranque, S. In Vitro activity of aminosterols against yeasts involved in blood stream infections. Med. Mycol. 2011, 49, 121–125. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Elkihel, L.; Soustre, I.; Karst, F.; Letourneux, Y. Amino- and aminomethylcholesterol derivatives with fungicidal activity. FEMS Microbiol. Lett. 1994, 120, 163–167. [Google Scholar] [CrossRef]
- Lorenz, R.T.; Parks, L.W. Cloning, sequencing, and disruption of the gene encoding sterol C-14 reductase in Saccharomyces cerevisiae. DNA Cell Biol. 1992, 11, 685–692. [Google Scholar] [CrossRef]
- Ashman, W.H.; Barbuch, R.J.; Ulbright, C.E.; Jarrett, H.W.; Bard, M. Cloning and disruption of the yeast C-8 sterol isomerase gene. Lipids 1991, 26, 628–632. [Google Scholar] [CrossRef]
- Weete, J.D. Structure and function of sterols in fungi. Adv. Lipid Res. 1989, 23, 115–167. [Google Scholar] [CrossRef]
- TerBush, D.R.; Novick, P. Sec6, Sec8, and Sec15 are components of a multisubunit complex which localizes to small bud tips in Saccharomyces cerevisiae. J. Cell Biol. 1995, 130, 299–312. [Google Scholar] [CrossRef] [Green Version]
- Panepinto, J.; Komperda, K.; Frases, S.; Park, Y.D.; Djordjevic, J.T.; Casadevall, A.; Williamson, P.R. Sec6-dependent sorting of fungal extracellular exosomes and laccase of Cryptococcus neoformans. Mol. Microbiol. 2009, 71, 1165–1176. [Google Scholar] [CrossRef]
- Rodrigues, M.L.; Nimrichter, L.; Oliveira, D.L.; Frases, S.; Miranda, K.; Zaragoza, O.; Alvarez, M.; Nakouzi, A.; Feldmesser, M.; Casadevall, A. Vesicular polysaccharide export in Cryptococcus neoformans is a eukaryotic solution to the problem of fungal trans-cell wall transport. Eukaryot. Cell 2007, 6, 48–59. [Google Scholar] [CrossRef]
- Yoneda, A.; Doering, T.L. A eukaryotic capsular polysaccharide is synthesized intracellularly and secreted via exocytosis. Mol. Biol. Cell 2006, 17, 5131–5140. [Google Scholar] [CrossRef] [PubMed]
- Salminen, A.; Novick, P.J. A ras-like protein is required for a post-Golgi event in yeast secretion. Cell 1987, 49, 527–538. [Google Scholar] [CrossRef]
- Curwin, A.J.; Fairn, G.D.; McMaster, C.R. Phospholipid transfer protein Sec14 is required for trafficking from endosomes and regulates distinct trans-Golgi export pathways. J. Biol. Chem. 2009, 284, 7364–7375. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chayakulkeeree, M.; Johnston, S.A.; Oei, J.B.; Lev, S.; Williamson, P.R.; Wilson, C.F.; Zuo, X.; Leal, A.L.; Vainstein, M.H.; Meyer, W.; et al. SEC14 is a specific requirement for secretion of phospholipase B1 and pathogenicity of Cryptococcus neoformans. Mol. Microbiol. 2011, 80, 1088–1101. [Google Scholar] [CrossRef] [Green Version]
- Lev, S.; Crossett, B.; Cha, S.Y.; Desmarini, D.; Li, C.; Chayakulkeeree, M.; Wilson, C.F.; Williamson, P.R.; Sorrell, T.C.; Djordjevic, J.T. Identification of Aph1, a phosphate-regulated, secreted, and vacuolar acid phosphatase in Cryptococcus neoformans. mBio 2014, 5, e01649-14. [Google Scholar] [CrossRef] [Green Version]
- Schekman, R. Lasker Basic Medical Research Award. SEC mutants and the secretory apparatus. Nat. Med. 2002, 8, 1055–1058. [Google Scholar] [CrossRef]
- Rodrigues, M.L.; Nakayasu, E.S.; Oliveira, D.L.; Nimrichter, L.; Nosanchuk, J.D.; Almeida, I.C.; Casadevall, A. Extracellular vesicles produced by Cryptococcus neoformans contain protein components associated with virulence. Eukaryot. Cell 2008, 7, 58–67. [Google Scholar] [CrossRef] [Green Version]
- Charlier, C.; Chretien, F.; Baudrimont, M.; Mordelet, E.; Lortholary, O.; Dromer, F. Capsule structure changes associated with Cryptococcus neoformans crossing of the blood-brain barrier. Am. J. Pathol. 2005, 166, 421–432. [Google Scholar] [CrossRef] [Green Version]
- Granger, D.L.; Perfect, J.R.; Durack, D.T. Virulence of Cryptococcus neoformans. Regulation of capsule synthesis by carbon dioxide. J. Clin. Investig. 1985, 76, 508–516. [Google Scholar] [CrossRef] [Green Version]
- Casadevall, A.; Mukherjee, J.; Scharff, M.D. Monoclonal antibody based ELISAs for cryptococcal polysaccharide. J. Immunol. Methods 1992, 154, 27–35. [Google Scholar] [CrossRef]
- Eisenman, H.C.; Chow, S.K.; Tse, K.K.; McClelland, E.E.; Casadevall, A. The effect of L-DOPA on Cryptococcus neoformans growth and gene expression. Virulence 2011, 2, 329–336. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kwon-Chung, K.J.; Wickes, B.L.; Booth, J.L.; Vishniac, H.S.; Bennett, J.E. Urease inhibition by EDTA in the two varieties of Cryptococcus neoformans. Infect. Immun. 1987, 55, 1751–1754. [Google Scholar] [CrossRef] [Green Version]
- Zhang, S.; Varma, A.; Williamson, P.R. The yeast Cryptococcus neoformans uses ‘mammalian’ enhancer sites in the regulation of the virulence gene, CNLAC1. Gene 1999, 227, 231–240. [Google Scholar] [CrossRef]
- Casadevall, A.; Perfect, J.R. Cryptococcus neoformans; ASM Press: Washington, DC, USA, 1998. [Google Scholar]
- Zimmermann, L.; Stephens, A.; Nam, S.Z.; Rau, D.; Kubler, J.; Lozajic, M.; Gabler, F.; Soding, J.; Lupas, A.N.; Alva, V. A completely reimplemented MPI bioinformatics toolkit with a new HHpred server at its core. J. Mol. Biol. 2018, 430, 2237–2243. [Google Scholar] [CrossRef]
- Buchan, D.W.; Minneci, F.; Nugent, T.C.; Bryson, K.; Jones, D.T. Scalable web services for the PSIPRED protein analysis workbench. Nucleic Acids Res. 2013, 41, W349–W357. [Google Scholar] [CrossRef] [Green Version]
- Mol, A.R.; Castro, M.S.; Fontes, W. NetWheels: A Web Application to Create High Quality Peptide Helical Wheel and Net Projections; Cold Spring Harbor Laboratory: Cold Spring Harbor, NY, USA, 2018. [Google Scholar]
- Rost, B.; Yachdav, G.; Liu, J. The PredictProtein server. Nucleic Acids Res. 2004, 32, W321–W326. [Google Scholar] [CrossRef] [Green Version]
- Consonni, S.V.; Maurice, M.M.; Bos, J.L. DEP domains: Structurally similar but functionally different. Nat. Rev. Mol. Cell Biol. 2014, 15, 357–362. [Google Scholar] [CrossRef]
- Petrey, D.; Fischer, M.; Honig, B. Structural relationships among proteins with different global topologies and their implications for function annotation strategies. Proc. Natl. Acad. Sci. USA 2009, 106, 17377–17382. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Valencia, A.; Kjeldgaard, M.; Pai, E.F.; Sander, C. GTPase domains of ras p21 oncogene protein and elongation factor Tu: Analysis of three-dimensional structures, sequence families, and functional sites. Proc. Natl. Acad. Sci. USA 1991, 88, 5443–5447. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Manente, M.; Ghislain, M. The lipid-translocating exporter family and membrane phospholipid homeostasis in yeast. FEMS Yeast Res. 2009, 9, 673–687. [Google Scholar] [CrossRef] [PubMed]
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Couch, G.S.; Greenblatt, D.M.; Meng, E.C.; Ferrin, T.E. UCSF Chimera--a visualization system for exploratory research and analysis. J. Comput. Chem. 2004, 25, 1605–1612. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, K.; Kwon, S.K.; Jun, S.H.; Cha, J.S.; Kim, H.; Lee, W.; Kim, J.F.; Cho, H.S. Crystal structure and functional characterization of a light-driven chloride pump having an NTQ motif. Nat. Commun. 2016, 7, 12677. [Google Scholar] [CrossRef] [PubMed]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11: Molecular Evolutionary Genetics Analysis Version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef] [PubMed]
- Bailey, T.L.; Elkan, C. Fitting a mixture model by expectation maximization to discover motifs in biopolymers. Proc. Int. Conf. Intell. Syst. Mol. Biol. 1994, 2, 28–36. [Google Scholar] [PubMed]
- Buske, F.A.; Boden, M.; Bauer, D.C.; Bailey, T.L. Assigning roles to DNA regulatory motifs using comparative genomics. Bioinformatics 2010, 26, 860–866. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Monteiro, P.T.; Oliveira, J.; Pais, P.; Antunes, M.; Palma, M.; Cavalheiro, M.; Galocha, M.; Godinho, C.P.; Martins, L.C.; Bourbon, N.; et al. YEASTRACT+: A portal for cross-species comparative genomics of transcription regulation in yeasts. Nucleic Acids Res. 2020, 48, D642–D649. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2 (-delta delta C(T)) method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- El Kihel, L.; Choucair, B.; Dhernomez, M.; Letourneux, Y. Stereoselective synthesis of 7α- and 7β-aminocholesterol as Δ8-Δ7 sterol isomerase inhibitors, with fungicidal activities towards resistant strains. Eur. J. Org. Chem. 2002, 23, 4075–4078. [Google Scholar] [CrossRef]
- Prabu, M.M.; Nagendra, H.G.; Suresh, S.; Vijayan, M. X-ray studies on crystalline complexes involving amino acids and peptides. XXXI. Effect of chirality on ionization state, stoichiometry and aggregation in the complexes of oxalic acid with L- and DL-histidine. J. Biomol. Struct. Dyn. 1996, 14, 387–392. [Google Scholar] [CrossRef]
- Sheldrick, G.M. A short history of SHELX. Acta Crystallogr. A 2008, 64, 112–122. [Google Scholar] [CrossRef]
- Farrugia, L.J. ORTEP-3 for Windows—A version of ORTEPIII with a Graphical user Interface (GUI). J. Appl. Cryst. 1997, 30, 565. [Google Scholar] [CrossRef]
- Madeira, F.; Pearce, M.; Tivey, A.R.N.; Basutkar, P.; Lee, J.; Edbali, O.; Madhusoodanan, N.; Kolesnikov, A.; Lopez, R. Search and sequence analysis tools services from EMBL-EBI in 2022. Nucleic Acids Res. 2022, 50, W276–W279. [Google Scholar] [CrossRef] [PubMed]
- Rice, P.; Longden, I.; Bleasby, A. EMBOSS: The European Molecular Biology Open Software Suite. Trends Genet. 2000, 16, 276–277. [Google Scholar] [CrossRef]
- Stothard, P. The sequence manipulation suite: JavaScript programs for analyzing and formatting protein and DNA sequences. Biotechniques 2000, 28, 1102–1104. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Robert, X.; Gouet, P. Deciphering key features in protein structures with the new ENDscript server. Nucleic Acids Res. 2014, 42, W320–W324. [Google Scholar] [CrossRef] [Green Version]
- Kolaczkowska, A.; Manente, M.; Kolaczkowski, M.; Laba, J.; Ghislain, M.; Wawrzycka, D. The regulatory inputs controlling pleiotropic drug resistance and hypoxic response in yeast converge at the promoter of the aminocholesterol resistance gene RTA1. FEMS Yeast Res. 2012, 12, 279–292. [Google Scholar] [CrossRef] [Green Version]
- Kolaczkowska, A.; Dylag, M.; Kolaczkowski, M. Differential expression of the Candida glabrata CgRTA1 and CgRSB1 genes in response to various stress conditions. Biochem. Biophys. Res. Commun. 2013, 432, 169–174. [Google Scholar] [CrossRef]
- Rodrigues, M.L.; Djordjevic, J.T. Unravelling secretion in Cryptococcus neoformans: More than one way to skin a cat. Mycopathologia 2012, 173, 407–418. [Google Scholar] [CrossRef]
- Skrzypek, M.S.; Nagiec, M.M.; Lester, R.L.; Dickson, R.C. Inhibition of amino acid transport by sphingoid long chain bases in Saccharomyces cerevisiae. J. Biol. Chem. 1998, 273, 2829–2834. [Google Scholar] [CrossRef] [Green Version]
- Kmetzsch, L.; Joffe, L.S.; Staats, C.C.; de Oliveira, D.L.; Fonseca, F.L.; Cordero, R.J.; Casadevall, A.; Nimrichter, L.; Schrank, A.; Vainstein, M.H.; et al. Role for Golgi reassembly and stacking protein (GRASP) in polysaccharide secretion and fungal virulence. Mol. Microbiol. 2011, 81, 206–218. [Google Scholar] [CrossRef]
- Djordjevic, J.T.; Lev, S. Fungal secretion: The next-gen target of antifungal agents? Cell Chem. Biol. 2018, 25, 233–235. [Google Scholar] [CrossRef] [PubMed]



| PDB ID | TC # | Description |
|---|---|---|
| 4OR2 | 9.A.14.7.1 | Human class C G protein-coupled metabotropic glutamate receptor 1 in complex with a negative allosteric modulator |
| 3RKO | 3.D.1.1.1 | Crystal structure of the membrane domain of respiratory complex I from E. coli at 3.0 angstrom resolution |
| 3AG3 | 3.D.4.7.1 | Bovine heart cytochrome C oxidase in the nitric oxide-bound fully reduced state at 100 K |
| 5G28 | 3.E.1.6.14 | The crystal structure of light-driven chloride pump ClR at pH 6.0 |
| 4HYJ | 3.E.1.6.5 | Crystal structure of Exiguobacterium sibiricum rhodopsin |
| 3EML | 9.A.14.3.8 | The 2.6 A crystal structure of a human A2A adenosine receptor bound to ZM241385 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Smith-Peavler, E.S.; Patel, R.; Onumajuru, A.M.; Bowring, B.G.; Miller, J.L.; Brunel, J.M.; Djordjevic, J.T.; Prabu, M.M.; McClelland, E.E. RTA1 Is Involved in Resistance to 7-Aminocholesterol and Secretion of Fungal Proteins in Cryptococcus neoformans. Pathogens 2022, 11, 1239. https://doi.org/10.3390/pathogens11111239
Smith-Peavler ES, Patel R, Onumajuru AM, Bowring BG, Miller JL, Brunel JM, Djordjevic JT, Prabu MM, McClelland EE. RTA1 Is Involved in Resistance to 7-Aminocholesterol and Secretion of Fungal Proteins in Cryptococcus neoformans. Pathogens. 2022; 11(11):1239. https://doi.org/10.3390/pathogens11111239
Chicago/Turabian StyleSmith-Peavler, Emily S., Ronakkumar Patel, Adejumoke Mary Onumajuru, Bethany G. Bowring, Joyce L. Miller, Jean Michel Brunel, Julianne T. Djordjevic, Moses M. Prabu, and Erin E. McClelland. 2022. "RTA1 Is Involved in Resistance to 7-Aminocholesterol and Secretion of Fungal Proteins in Cryptococcus neoformans" Pathogens 11, no. 11: 1239. https://doi.org/10.3390/pathogens11111239
APA StyleSmith-Peavler, E. S., Patel, R., Onumajuru, A. M., Bowring, B. G., Miller, J. L., Brunel, J. M., Djordjevic, J. T., Prabu, M. M., & McClelland, E. E. (2022). RTA1 Is Involved in Resistance to 7-Aminocholesterol and Secretion of Fungal Proteins in Cryptococcus neoformans. Pathogens, 11(11), 1239. https://doi.org/10.3390/pathogens11111239








