Responses to Ecopollutants and Pathogenization Risks of Saprotrophic Rhodococcus Species
Abstract
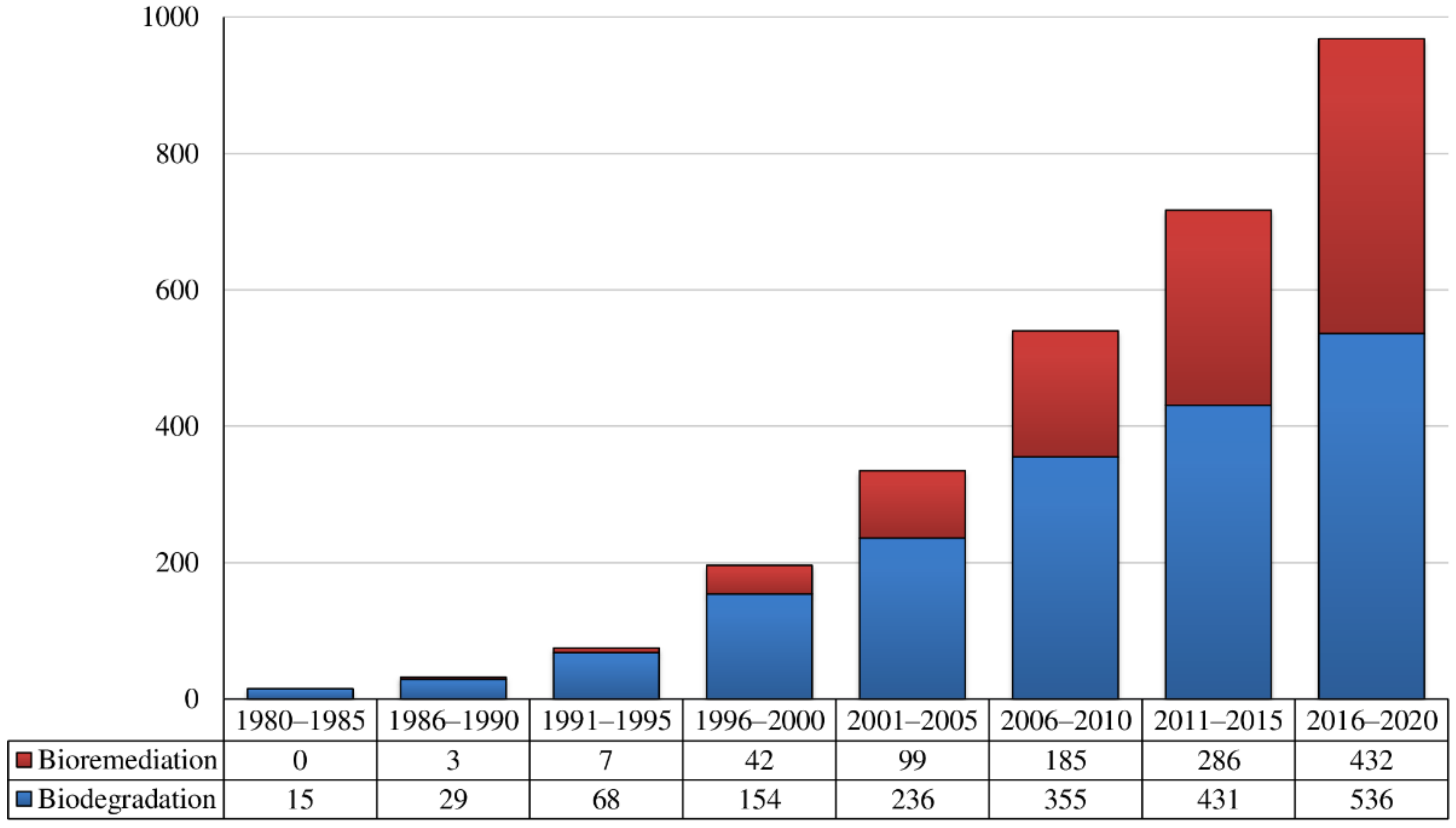
1. Introduction
2. Ubiquity of Rhodococcus and Existence of Pathogenic Species
2.1. Free-Living Rhodococci
2.2. Phytopatogenic Rhodococci
2.3. Animal and Human Rhodococcus Pathogens
3. Adaptive Cell Modifications of Rhodococci Exposed to Hydrocarbons and Other Environmental Pollutants
3.1. Adhesion, Cellular Autoaggregation, and Colonization as Survival Strategies
3.2. Changes in the Morphometric Parameters of Cells
3.3. Change in the Zeta Potential of Cells
4. Conclusions
Until recently, the image of an “enemy” (less often, a “companion”) dominated in the human–microbe relationships, now it becomes obvious that it is necessary to establish “peaceful coexistence” with this huge world.[155]
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Goodfellow, M. Phylum XXVI. Actinobacteria phyl. nov. In Bergey’s Manual of Systematic Bacteriology, 2nd ed.; Goodfellow, M., Kämpfer, P., Busse, H.-J., Trujillo, M.E., Suzuki, K., Ludwig, W., Whitman, W.B., Eds.; Springer: New York, NY, USA, 2012; Volume 5, pp. 33–34. [Google Scholar]
- Jones, A.L.; Goodfellow, M. Rhodococcus (Zopf 1891) emend. Goodfellow, Alderson and Chun 1998a. In Bergey’s Manual of Systematics of Archaea and Bacteria, Digital ed.; Trujillo, M.E., Dedysh, S., DeVos, P., Hedlund, B., Kämpfer, P., Rainey, F.A., Whitman, W.B., Eds.; John Wiley & Sons: New York, NY, USA, 2015; pp. 1–50. [Google Scholar]
- Gupta, R.S. Commentary: Genome-based taxonomic classification of the phylum Actinobacteria. Front. Microbiol. 2019, 10, 206. [Google Scholar] [CrossRef]
- Busch, H.; Hagedoorn, P.-L.; Hanefeld, U. Rhodococcus as a versatile biocatalyst in organic synthesis. Int. J. Mol. Sci. 2019, 20, 4787. [Google Scholar] [CrossRef]
- Ivshina, I.B.; Tyumina, E.A.; Kuzmina, M.V.; Vikhareva, E.V. Features of diclofenac biodegradation by Rhodococcus ruber IEGM 346. Sci. Rep. 2019, 9, 9159. [Google Scholar] [CrossRef]
- Garrido-Sanz, D.; Redondo-Nieto, M.; Martín, M.; Rivilla, R. Comparative genomics of the Rhodococcus genus shows wide distribution of biodegradation traits. Microorganisms 2020, 8, 774. [Google Scholar] [CrossRef]
- Alvarez, H.M.; Silva, R.A.; Cesari, A.C.; Zamit, A.L.; Peressutti, S.R.; Reichelt, R.; Keller, U.; Malkus, U.; Rasch, C.; Maskow, T.; et al. Physiolosgical and morphological responses of the soil bacterium Rhodococcus opacus strain PD630 to water stress. FEMS Microbiol. Ecol. 2004, 50, 75–86. [Google Scholar] [CrossRef] [PubMed]
- LeBlanc, J.C.; Gonçalves, E.R.; Mohn, W.W. Global response to desiccation stress in the soil actinomycete Rhodococcus jostii RHA1. Appl. Environ. Microbiol. 2008, 74, 2627–2636. [Google Scholar] [CrossRef]
- Fanget, N.V.; Foley, S. Starvation/stationary-phase survival of Rhodococcus 1454 erythropolis SQ1: A physiological and genetic analysis. Arch. Microbiol. 2011, 193, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Corno, G.; Coci, M.; Giardina, M.; Plechuk, S.; Campanile, F.; Stefani, S. Antibiotics promote aggregation within aquatic bacterial communities. Front. Microbiol. 2014, 5, 297. [Google Scholar] [CrossRef]
- Su, X.; Sun, F.; Wang, Y.; Hashmi, M.Z.; Guo, L.; Ding, L.; Shen, C. Identification, characterization and molecular analysis of the viable but nonculturable Rhodococcus biphenylivorans. Sci. Rep. 2015, 5, 18590. [Google Scholar] [CrossRef] [PubMed]
- Röttig, A.; Hauschild, P.; Madkour, M.H.; Al-Ansari, A.M.; Almakishah, N.H.; Steinbüchel, A. Analysis and optimization of triacylglycerol synthesis in novel oleaginous Rhodococcus and Streptomyces strains isolated from desert soil. J. Biotechnol. 2016, 225, 48–56. [Google Scholar] [CrossRef]
- Zhang, C.; Yang, L.; Ding, Y.; Wang, Y.; Lan, L.; Ma, Q.; Chi, X.; Wei, P.; Zhao, Y.; Steinbüchel, A.; et al. Bacterial lipid droplets bind to DNA via an intermediary protein that enhances survival under stress. Nat. Commun. 2017, 8, 15979. [Google Scholar] [CrossRef] [PubMed]
- Raymond-Bouchard, I.; Tremblay, J.; Altshuler, I.; Greer, C.W.; Whyte, L.G. Comparative transcriptomics of cold growth and adaptive features of a eury- and steno-psychrophile. Front. Microbiol. 2018, 9, 1565. [Google Scholar] [CrossRef] [PubMed]
- Firrincieli, A.; Presentato, A.; Favoino, G.; Marabottini, R.; Allevato, E.; Stazi, S.-R.; Mugnozza, G.-S.; Harfouche, A.; Petruccioli, M.; Turner, R.J.; et al. Identification of resistance genes and response to arsenic in Rhodococcus aetherivorans BCP1. Front. Microbiol. 2019, 110, 888. [Google Scholar] [CrossRef]
- Cappelletti, M.; Presentato, A.; Piacenza, E.; Firrincieli, A.; Turner, R.J.; Zannon, D. Biotechnology of Rhodococcus for the production of valuable compounds. Appl. Microbiol. Biotechnol. 2020, 104, 8567–8594. [Google Scholar] [CrossRef] [PubMed]
- Hu, X.; Li, D.; Qiao, Y.; Song, Q.; Guan, Z.; Qiu, K.; Cao, J.; Huang, L. Salt tolerance mechanism of a hydrocarbon-degrading strain: Salt tolerance mediated by accumulated betaine in cells. J. Hazard. Mater. 2020, 392, 122326. [Google Scholar] [CrossRef]
- Sundararaghavan, A.; Mukherjee, A.; Suraishkumar, G.K. Investigating the potential use of an oleaginous bacterium, Rhodococcus opacus PD630, for nano-TiO2 remediation. Environ. Sci. Pollut. Res. 2020, 27, 27394–27406. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Chen, Y.; Zhou, H.; Li, X.; Tan, Z. Adaptation mechanisms of Rhodococcus sp. CNS16 under different temperature gradients: Physiological and transcriptome. Chemosphere 2020, 238, 124571. [Google Scholar] [CrossRef]
- Pátek, M.; Grulich, M.; Nešvera, J. Stress response in Rhodococcus strains. Biotechnol. Adv. 2021, 107698. [Google Scholar] [CrossRef]
- Goodfellow, M.; Jones, A.L.; Maldonado, L.A.; Salanitro, J. Rhodococcus aetherivorans sp. nov., a new species that contains methyl t-butyl ether-degrading actinomycetes. Syst. Appl. Microbiol. 2004, 27, 61–65. [Google Scholar] [CrossRef]
- de Carvalho, C.C.C.R.; Costa, S.S.; Fernandes, P.; Couto, I.; Viveiros, M. Membrane transport systems and the biodegradation potential and pathogenicity of genus Rhodococcus. Front. Physiol. 2014, 5, 133. [Google Scholar] [CrossRef]
- Ivshina, I.B.; Kuyukina, M.S.; Krivoruchko, A.V. Hydrocarbon-oxidizing bacteria and their potential in eco-biotechnology and bioremediation. In Microbial Resources: From Functional Existence in Nature to Industrial Applications; Kurtböke, I., Ed.; Elsevier: New York, NY, USA, 2017; pp. 121–148. [Google Scholar]
- Cappelletti, M.; Fedi, S.; Zannoni, D. Degradation of alkanes in Rhodococcus. In Biology of Rhodococcus. Microbiology Monographs, 2nd ed.; Alvarez, H.M., Ed.; Springer Nature: Cham, Switzerland, 2019; Volume 16, pp. 137–171. [Google Scholar]
- Acosta-González, A.; Martirani-von Abercron, S.-M.; Rosselló-Móra, R.; Wittich, R.M.; Marqués, S. The effect of oil spills on the bacterial diversity and catabolic function in coastal sediments: A case study on the Prestige oil spill. Environ. Sci. Pollut. Res. 2016, 22, 15200–15214. [Google Scholar] [CrossRef]
- Kuyukina, M.S.; Ivshina, I.B. Bioremediation of contaminated environments using Rhodococcus. In Biology of Rhodococcus. Microbiology Monographs, 2nd ed.; Alvarez, H.M., Ed.; Springer Nature: Cham, Switzerland, 2019; Volume 16, pp. 231–270. [Google Scholar]
- Voronina, E.; Balandina, A.; Frolova, N.; Dubrovina, S.; Loginova, T. Effect of benzo(a)pyrene on the number of soil microorganisms of the genus Pseudomonas and Rhodococcus. In IOP Conference Series: Earth and Environmental Science; IOP Publishing: Bristol, UK, 2021; Volume 723, p. 042010. [Google Scholar]
- Bej, A.K. Cold-tolerant alkane-degrading Rhodococcus species from Antarctica. Pol. Biol. 2000, 23, 100–105. [Google Scholar] [CrossRef]
- de Carvalho, C.C.C.R. Adaptation of Rhodococcus erythropolis cells for growth and bioremediation under extreme conditions. Res. Microbiol. 2012, 163, 125–136. [Google Scholar] [CrossRef]
- Goordial, J.; Raymond-Bouchard, I.; Zolotarov, Y.; de Bethencourt, L.; Ronholm, J.; Shapiro, N.; Woyke, T.; Stromvik, M.; Greer, C.W.; Bakermans, C.; et al. Cold adaptive traits revealed by comparative genomic analysis of the eurypsychrophile Rhodococcus sp. JG3 isolated from high elevation McMurdo Dry Valley permafrost, Antarctica. FEMS Microbiol. Ecol. 2016, 92, fiv154. [Google Scholar] [PubMed]
- Ivshina, I.B. Current situation and challenges of specialized microbial resource centres in Russia. Microbiology 2012, 81, 509–516. [Google Scholar] [CrossRef]
- Catalogue of Strains of Regional Specialised Collection of Alkanotrophic Microorganisms. Available online: http://www.iegmcol/strains/index.html (accessed on 17 May 2021).
- Cuello, O.H.; Caorlin, M.J.; Reviglio, V.E.; Carvajal, L.; Juarez, C.P.; de Guerra, E.P.; Luna, J.D. Rhodococcus globerulus keratitis after laser in situ keratomileusis. J. Cat. Refr. Surg. 2002, 28, 2235–2237. [Google Scholar] [CrossRef]
- Jones, A.L. Rhodococcus gordoniae sp. nov., an actinomycete isolated from clinical material and phenol-contaminated soil. Inter. J. Syst. Evol. Microbiol. 2004, 54, 407–411. [Google Scholar] [CrossRef]
- Letek, M.; Gonzalez, P.; MacArthur, I.; Rodriguez, H.; Freeman, T.C.; Valero-Rello, A.; Blonco, M.; Buckley, T.; Cherevach, I.; Fahey, R.; et al. The genome of a pathogenic Rhodococcus: Cooptive virulence underpinned by key gene acquisitions. PLoS Genet. 2010, 6, e1001145. [Google Scholar] [CrossRef]
- Ng, S.; King, C.S.; Hang, J.; Clifford, R.; Lesho, E.P.; Kuschner, R.A.; Cox, E.D.; Stelsel, R.; Mody, R. Severe cavitary pneumonia caused by a non-egui Rhodococcus species in an immunocompetent patient. Respir. Care 2013, 58, 47–50. [Google Scholar]
- Anastasi, E.; MacArthur, I.; Scortti, M.; Alvarez, S.; Giguère, S.; Vázquez-Boland, J.A. Pangenome and phylogenomic analysis of the pathogenic actinobacterium Rhodococcus equi. GBE 2016, 8, 3140–3148. [Google Scholar]
- Rahdar, H.A.; Mahmoudi, S.; Bahador, A.; Ghiasvand, F.; Heravi, F.S.; Feizabadi, M.M. Molecular identification and antibiotic resistance pattern of actinomycetes isolates among immunocompromised patients in Iran, emerging of new infections. Sci. Rep. 2021, 11, 10745. [Google Scholar] [CrossRef]
- Luz, A.P.; Pellizari, V.H.; Whyte, L.G.; Greer, C.W. A survey of indigenous microbial hydrocarbon degradation genes in soils from Antarctica and Brazil. Can. J. Microbiol. 2004, 50, 323–333. [Google Scholar] [CrossRef] [PubMed]
- Sheng, H.M.; Gao, H.S.; Xue, L.G.; Ding, S.; Song, C.L.; Feng, H.Y.; An, L.Z. Analysis of the composition and characteristics of culturable endophytic bacteria within subnival plants of the Tianshan Mountains, northwestern China. Curr. Microbiol. 2011, 62, 923–932. [Google Scholar] [CrossRef] [PubMed]
- Hassanshahian, M.; Ahmadinejad, M.; Tebyanian, H.; Kariminik, A. Isolation and characterization of alkane degrading bacteria from petroleum reservoir waste water in Iran (Kerman and Tehran provenances). Mar. Pollut. Bullet. 2013, 73, 300–305. [Google Scholar] [CrossRef] [PubMed]
- Konishi, M.; Nishi, S.; Fukuoka, T.; Kitamoto, D.; Watsuji, T.-O.; Nagano, Y.; Yabuki, A.; Nakagawa, S.; Hatada, Y.; Horiuchi, J.-I. Deep-sea Rhodococcus sp. BS-15, lacking the phytopathogenic fas genes, produces a novel glucotriose lipid biosurfactant. Mar. Biotechnol. 2014, 16, 484–493. [Google Scholar] [CrossRef]
- Mikolasch, A.; Omirbekova, A.; Schumann, P.; Reinhard, A.; Sheikhany, H.; Berzhanova, R.; Mukasheva, T.; Schauer, F. Enrichment of aliphatic, alicyclic and aromatic acids by oil-degrading bacteria isolated from the rhizosphere of plants growing in oil-contaminated soil from Kazakhstan. Appl. Microbiol. Biotechnol. 2015, 99, 4071–4084. [Google Scholar] [CrossRef]
- Viggor, S.; Jõesaar, M.; Vedler, E.; Kiiker, R.; Pärnpuu, L.; Heinaru, A. Occurrence of diverse alkane hydroxylase alkB genes in indigenous oil-degrading bacteria of Baltic Sea surface water. Mar. Pollut. Bull. 2015, 101, 507–516. [Google Scholar] [CrossRef] [PubMed]
- Sinha, R.K.; Krishnan, K.P.; Hatha, A.A.; Rahiman, M.; Thresyamma, D.D.; Kerkar, S. Diversity of retrievable heterotrophic bacteria in Kongsfjorden, an Arctic fjord. Braz. J. Microbiol. 2017, 48, 51–61. [Google Scholar] [CrossRef]
- Auta, H.S.; Emenike, C.U.; Jayanthi, B.; Fauziah, S.H. Growth kinetics and biodeterioration of polypropylene microplastics by Bacillus sp. and Rhodococcus sp. isolated from mangrove sediment. Mar. Pollut. Bull. 2018, 127, 15–21. [Google Scholar] [CrossRef]
- Habib, S.; Ahmad, S.A.; Johari, W.L.W.; Shukor, M.Y.A.; Alias, S.A.; Khalil, K.A.; Yasid, N.A. Evaluation of conventional and response surface level optimisation of n-dodecane (n-C12) mineralisation by psychrotolerant strains isolated from pristine soil at Southern Victoria Island, Antarctica. Microb. Cell Fact. 2018, 17, 44. [Google Scholar] [CrossRef]
- Yuste, L.; Corbella, M.E.; Turiégano, M.J.; Karlson, U.; Puyet, A.; Rojo, F. Characterization of bacterial strains able to grow on high molecular mass residues from crude oil processing. FEMS Microbiol. Ecol. 2000, 32, 69–75. [Google Scholar] [CrossRef] [PubMed]
- Hamamura, N.; Olson, S.H.; Ward, D.M.; Inskeep, W.P. Microbial population dynamics associated with crude-oil biodegradation in diverse soils. Appl. Environ. Microbiol. 2006, 72, 6316–6324. [Google Scholar] [CrossRef] [PubMed]
- de Oliveira, V.M.; Sette, L.D.; Simioni, K.C.M.; Santos Neto, E.V.D. Bacterial diversity characterization in petroleum samples from Brazilian reservoirs. Braz. J. Microbiol. 2008, 39, 445–452. [Google Scholar] [CrossRef][Green Version]
- Li, J.; Zhao, G.-Z.; Long, L.-J.; Wang, F.-Z.; Tian, X.-P.; Zhang, S.; Li, W.-J. Rhodococcus nanhaiensis sp. nov., an actinobacterium isolated from marine sediment. Int. J. Syst. Evol. Microbiol. 2012, 62, 2517–2521. [Google Scholar] [CrossRef] [PubMed]
- Urbano, S.B.; Albarracín, V.H.; Ordoñez, O.F.; Farías, M.E.; Alvarez, H.M. Lipid storage in high-altitude Andean Lakes extremophiles and its mobilization under stress conditions in Rhodococcus sp. A5, a UV-resistant actinobacterium. Extremophiles 2013, 17, 217–227. [Google Scholar] [CrossRef]
- Dastager, S.G.; Mawlankar, R.; Tang, S.-K.; Krishnamurthi, S.; Ramana, V.V.; Joseph, N.; Shouche, Y.S. Rhodococcus enclensis sp. nov., a novel member of the genus Rhodococcus. Int. J. Syst. Evol. Microbiol. 2014, 64, 2693–2699. [Google Scholar] [CrossRef]
- Ivshina, I.B.; Kuyukina, M.S.; Krivoruchko, A.V.; Barbe, V.; Fischer, C. Draft genome sequence of propane- and butane-oxidizing actinobacterium Rhodococcus ruber IEGM 231. Genome Announc. 2014, 2, e01297-14. [Google Scholar] [CrossRef]
- Hwang, C.Y.; Lee, I.; Cho, Y.; Lee, Y.M.; Baek, K.; Jung, Y.J.; Yang, Y.Y.; Lee, T.; Rhee, T.S.; Lee, H.K. Rhodococcus aerolatus sp. nov., isolated from subarctic rainwater. Int. J. Syst. Evol. Microbiol. 2015, 65, 465–471. [Google Scholar] [CrossRef]
- Aggarwal, R.K.; Dawar, C.; Phanindranath, R.; Mutnuri, L.; Dayal, A.M. Draft genome sequence of a versatile hydrocarbon-degrading bacterium, Rhodococcus pyridinivorans strain KG-16, collected from oil fields in India. Genome Announc. 2016, 4, e01704-15. [Google Scholar] [CrossRef]
- Táncsics, A.; Máthe, I.; Benedek, T.; Toth, E.M.; Atasayar, E.; Sproer, C.; Márialigeti, K.; Felfoldi, T.; Kriszt, B. Rhodococcus sovatensis sp. nov., an actinomycete isolated from the hypersaline and heliothermal Lake Ursu. Int. J. Syst. Evol. Microbiol. 2017, 67, 190–196. [Google Scholar] [CrossRef]
- Ramaprasad, E.V.V.; Mahidhara, G.; Sasikala, C.; Ramana, C.V. Rhodococcus electrodiphilus sp. nov., a marine electro active actinobacterium isolated from coral reef. Int. J. Syst. Evol. Microbiol. 2018, 68, 2644–2649. [Google Scholar] [CrossRef]
- Campbell, C.A.; Beiderbeck, V.O.; Warder, F.G. Influence of simulated fall and sping conditions on the soil system. III. Effect of method of simulating spring temperatures of ammonification, nitrification and microbial populations. Soil Sci. Soc. Am. Proc. 1973, 37, 382–386. [Google Scholar] [CrossRef]
- Anandan, R.; Dhanasekaran, D.; Gopinath, P.M. An introduction to actinobacteria. In Actinobacteria–Basics and Biotechnological Applications; Dhanasekaran, D., Jiang, Y., Eds.; IntechOpen: London, UK, 2016; pp. 3–37. [Google Scholar]
- Zhao, G.-Z.; Li, J.; Zhu, W.-Y.; Tian, S.-Z.; Zhao, L.-X.; Yang, L.-L.; Xu, L.-H.; Li, W.-J. Rhodococcus artemisiae sp. nov., an endophytic actinobacterium isolated from the pharmaceutical plant Artemisia annua L. Int. J. Syst. Evol. Microbiol. 2012, 62, 900–905. [Google Scholar] [CrossRef]
- Kämpfer, P.; Wellner, S.; Lohse, K.; Lodders, N.; Martin, K. Rhodococcus cerastii sp. nov. and Rhodococcus trifolii sp. nov., two novel species isolated from leaf surfaces. Int. J. Syst. Evol. Microbiol. 2013, 63, 1024–1029. [Google Scholar] [CrossRef] [PubMed]
- Ma, J.; Zhang, L.; Wang, G.; Zhang, S.; Zhang, X.; Wang, Y.; Shi, C.; Si, L.; Zhao, H.; Liu, F.; et al. Rhodococcus gannanensis sp. nov., a novel endophytic actinobacterium isolated from root of sunflower (Helianthus annuus L.). Antonie van Leeuwenhoek 2017, 110, 1113–1120. [Google Scholar] [CrossRef]
- Silva, L.J.; Souza, D.T.; Genuario, D.B.; Hoyos, H.A.V.; Santos, S.N.; Rosa, L.H.; Zucchi, T.D.; Melo, I.S. Rhodococcus psychrotolerans sp. nov., isolated from rhizosphere of Deschampsia antarctica. Antonie van Leeuwenhoek 2018, 111, 629–636. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Cao, P.; Jiang, M.; Hou, Y.; Du, C.; Xiang, W.; Zhao, J.; Wang, X. Rhodococcus oryzae sp. nov., a novel actinobacterium isolated from rhizosphere soil of rice (Oryza sativa L.). Int. J. Syst. Evol. Microbiol. 2020, 70, 3300–3308. [Google Scholar] [CrossRef] [PubMed]
- Hamedi, J.; Mohammadipanah, F. Biotechnological application and taxonomical distribution of plant growth promoting actinobacteria. J. Ind. Microbiol. Biotechnol. 2015, 42, 157–171. [Google Scholar] [CrossRef]
- Stes, E.; Francis, I.; Pertry, I.; Dolzblasz, A.; Depuydt, S.; Vereecke, D. The leafy gall syndrome induced by Rhodococcus fascians. FEMS Microbiol. Lett. 2013, 342, 187–194. [Google Scholar] [CrossRef]
- Creason, A.L.; Vandeputte, O.M.; Savory, E.A.; Davis, E.W.; Putnam, M.L.; Hu, E.; Swader-Hines, D.; Mol, A.; Baucher, M.; Prinsen, E.; et al. Analysis of genome sequences from plant pathogenic Rhodococcus reveals genetic novelties in virulence loci. PLoS ONE 2014, 9, e101996. [Google Scholar] [CrossRef]
- Goethals, K.; Vereecke, D.; Jaziri, M.; Van, M.M.; Holsters, M. Leafy gall formation by Rhodococcus fascians. Ann. Rev. Phytopathol. 2001, 39, 27–52. [Google Scholar] [CrossRef] [PubMed]
- Depuydt, S.; Putnam, M.; Holsters, M.; Vereecke, D. Rhodococcus fascians, an emerging threat for ornamental crops. In Floriculture, Ornamental, and Plant Biotechnology: Advances and Topical Issues, 1st ed.; Teixeira da Silva, J.A., Ed.; Global Science Books: Ikenobe, Japan, 2008; Volume 5, pp. 480–489. [Google Scholar]
- Stamler, R.A.; Kilcrease, J.; Kallsen, C.; Fichtner, E.J.; Cooke, P.; Heerema, R.J.; Randall, J.J. First report of Rhodococcus isolates causing Pistachio Bushy Top Syndrome on ‘UCB-1’ rootstock in California and Arizona. Plant Dis. 2015, 99, 1468–1476. [Google Scholar] [CrossRef]
- Vereecke, D.; Zhang, Y.; Francis, I.M.; Lambert, P.Q.; Venneman, J.; Stamler, R.A.; Kilcrease, J.; Randall, J.J. Functional genomics insights into the pathogenicity, habitat fitness, and mechanisms modifying plant development of Rhodococcus sp. PBTS1 and PBTS2. Front. Microbiol. 2020, 11, 14. [Google Scholar] [CrossRef]
- Vereecke, D.; Fichtner, E.J.; Lambert, P.Q.; Cooke, P.; Kilcrease, J.; Stamler, R.A.; Zhang, Y.; Francis, I.M.; Randall, J.J. Colonization and survival capacities underlying the multifaceted life of Rhodococcus sp. PBTS1 and PBTS2. Plant Pathol. 2021, 70, 567–583. [Google Scholar] [CrossRef]
- Park, J.M.; Koo, J.; Kang, S.W.; Jo, S.H.; Park, J.M. Detection of Rhodococcus fascians, the causative agent of lily fasciation in South Korea. Pathogens 2021, 10, 241. [Google Scholar] [CrossRef] [PubMed]
- Vereecke, D.; Cornelis, K.; Temmerman, W.; Jaziri, M.; Van Montagu, M.; Holsters, M.; Goerhals, K. Chromosomal locus that affects the pathogenicity of Rhodococcus fascians. J. Bacteriol. 2002, 184, 1112–1120. [Google Scholar] [CrossRef] [PubMed]
- Savory, E.A.; Fuller, S.L.; Weisberg, A.J.; Thomas, W.J.; Gordon, M.I.; Stevens, D.M.; Creason, A.L.; Belcher, M.S.; Serdani, M.; Wiseman, M.S.; et al. Evolutionary transitions between beneficial and phytopathogenic Rhodococcus challenge disease management. eLife 2017, 6, e30925. [Google Scholar] [CrossRef]
- von Bargen, K.; Haas, A. Molecular and infection biology of the horse pathogen Rhodococcus equi. FEMS Microbiol. Rev. 2009, 33, 870–891. [Google Scholar] [CrossRef] [PubMed]
- Bell, K.S.; Philp, J.C.; Aw, D.W.; Christofi, N. The genus Rhodococcus. J. Appl. Microbiol. 1998, 85, 195–210. [Google Scholar] [CrossRef]
- Vázquez-Boland, J.A.; Wim, G.; Meijer, W.G. The pathogenic actinobacterium Rhodococcus equi: What’s in a name? Mol. Microbiol. 2019, 112, 1–15. [Google Scholar] [CrossRef]
- Takai, S.; Sawada, N.; Nakayama, Y.; Ishizuka, S.; Nakagawa, R.; Kawashima, G.; Sangkanjanavanich, N.; Sasaki, Y.; Kakuda, T.; Suzuk, Y. Reinvestigation of the virulence of Rhodococcus equi isolates from patients with and without AIDS. Lett. Appl. Microbiol. 2020, 71, 679–683. [Google Scholar] [CrossRef]
- Scotton, P.G.; Tonon, E.; Giobbia, M.; Gallucci, M.; Rigoli, R.; Vaglia, A. Rhodococcus equi nosocomial meningitis cured by levofloxacin and shunt removal. Clin. Infect. Dis. 2000, 30, 223–224. [Google Scholar] [CrossRef][Green Version]
- Matsushita, H.; Hanayama, N.; Hobo, K.; Kuba, K.; Takazawa, A. Infectious endocarditis caused by Rhodococcus equi. Ann. Thorac. Surg. 2010, 89, 957–959. [Google Scholar] [CrossRef] [PubMed]
- Kedlaya, I.; Ing, M.B.; Wong, S.S. Rhodococcus equi infections in immunocompetent hosts: Case report and review. Clin. Infect. Dis. 2001, 32, 39–46. [Google Scholar] [CrossRef] [PubMed]
- Gurtler, V.; Mayall, B.C.; Seviour, R. Can whole genome analysis refine the taxonomy of the genus Rhodococcus? FEMS Microbiol. Rev. 2004, 28, 377–403. [Google Scholar] [CrossRef]
- Muscatello, G.; Leadon, D.P.; Klay, M.; Ocampo-Sosa, A.; Lewis, D.A.; Fogarty, U.; Buckley, T.; Gilkerson, J.R.; Meijer, W.G.; Vazquez-Boland, J.A. Rhodococcus equi infection in foals: The science of ‘rattles’. Equine Vet. J. 2007, 39, 470–478. [Google Scholar] [CrossRef] [PubMed]
- Vázquez-Boland, J.A.; Prescott, J.F.; Meijer, W.G.; Leadon, D.P.; Hines, S.A. Havermeyer workshop report: Rhodococcus equi comes of age. Equine Vet. J. 2009, 41, 93–95. [Google Scholar] [CrossRef] [PubMed]
- MacArthur, I.; Anastasi, E.; Alvarez, S.; Scortti, M.; Vázquez-Boland, J.A. Comparative genomics of Rhodococcus equi virulence plasmids indicates host-driven evolution of the vap pathogenicity island. GBE 2017, 9, 1241–1247. [Google Scholar] [CrossRef] [PubMed]
- Sangal, V.; Goodfellow, M.; Jones, A.L.; Seviour, R.J.; Sutcliffe, I.C. Refined systematics of the genus Rhodococcus based on whole genome analyses. In Biology of Rhodococcus. Microbiology Monographs, 2nd ed.; Alvarez, H.M., Ed.; Springer Nature: Cham, Switzerland, 2019; Volume 16, pp. 1–21. [Google Scholar]
- Álvarez-Narváez, S.; Huber, L.; Giguère, S.; Hart, K.A.; Berghaus, R.D.; Sanchez, S.; Cohen, N.D. Epidemiology and molecular basis of multidrug resistance in Rhodococcus equi. Microbiol. Mol. Biol. Rev. 2021, 85, e00011–e00021. [Google Scholar] [CrossRef] [PubMed]
- Willingham-Lane, J.M.; Coulson, G.B.; Hondalus, M.K. Identification of a VapA virulence factor functional homolog in Rhodococcus equi isolates housing the pVAPB plasmid. PLoS ONE 2018, 13, e0204475. [Google Scholar] [CrossRef]
- Heller, M.C.; Jackson, K.A.; Watson, J.L. Identification of immunologically relevant genes in mare and foal dendritic cells responding to infection by Rhodococcus equi. Vet. Immunol. Immunopathol. 2010, 136, 144–150. [Google Scholar] [CrossRef]
- Mourenza, Á.; Bravo-Santano, N.; Pradal, I.; Gil, J.A.; Mateos, L.M.; Letek, M. Mycoredoxins are required for redox homeostasis and intracellular survival in the actinobacterial pathogen Rhodococcus equi. Antioxidants 2019, 8, 558. [Google Scholar] [CrossRef]
- von Bargen, K.; Scraba, M.; Krämer, I.; Ketterer, M.; Nehls, C.; Krokowski, S.; Repnik, U.; Wittlich, M.; Maaser, A.; Zapka, P.; et al. Virulence-associated protein A from Rhodococcus equi is an intercompartmental pH-neutralising virulence factor. Cell. Microbiol. 2019, 21, e12958. [Google Scholar] [CrossRef]
- Valero-Rello, A.; Hapeshi, A.; Anastasi, E.; Alvarez, S.; Scortti, M.; Meijer, W.G.; MacArthur, I.; Vázquez-Boland, J.A. An invertron-like linear plasmid mediates intracellular survival and virulence in bovine isolates of Rhodococcus equi. Infect. Immun. 2015, 83, 2725–2737. [Google Scholar] [CrossRef]
- Paterson, M.L.; Ranasinghe, D.; Blom, J.; Dover, L.G.; Sutcliffe, I.C.; Lopes, B.; Sangal, V. Genomic analysis of a novel Rhodococcus (Prescottella) equi isolate from a bovine host. Arch. Microbiol. 2019, 201, 1317–1321. [Google Scholar] [CrossRef]
- Ying, J.; Ye, J.; Xu, T.; Wang, Q.; Bao, Q.; Li, A. Comparative genomic analysis of Rhodococcus equi: An insight into genomic diversity and genome evolution. Int. J. Genom. 2019, 2019, 8987436. [Google Scholar] [CrossRef]
- Álvarez-Narváez, S.; Giguère, S.; Berghaus, L.J.; Dailey, C.; Vazquez-Boland, J.A. Horizontal spread of Rhodococcus equi macrolide resistance plasmid pRErm46 across environmental actinobacteria. Appl. Environ. Microbiol. 2020, 86, e00108-20. [Google Scholar] [CrossRef]
- Baba, H.; Nada, T.; Ohkusu, K.; Ezaki, T.; Hasagawa, Y.; Paterson, D.L. First case of bloodstream infection caused by Rhodococcus erythropolis. J. Clin. Microbiol. 2009, 47, 2667–2669. [Google Scholar] [CrossRef] [PubMed]
- Khairy, H.; Meinert, C.; Wubbeler, J.H.; Poehlein, A.; Daniel, R.; Voigt, B.; Riedel, K.; Steinbüchel, A. Genome and proteome analysis of Rhodococcus erythropolis MI2: Elucidation of the 4,4′-dithiodibutyric acid catabolism. PLoS ONE 2016, 11, e0167539. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Sazykin, I.; Makarenko, M.; Khmelevtsova, L.; Seliverstova, E.; Rakin, A.; Sazykina, M. Cyclohexane, naphthalene, and diesel fuel increase oxidative stress, CYP153, sodA, and recA gene expression in Rhodococcus erythropolis. Microbiologyopen 2019, 8, e00855. [Google Scholar]
- Zheng, Y.T.; Toyofuku, M.; Nomura, N.; Shigeto, S. Correlation of carotenoid accumulation with aggregation and biofilm development in Rhodococcus sp. SD-74. Anal. Chem. 2013, 85, 7295–7301. [Google Scholar] [CrossRef]
- Weathers, T.S.; Higgins, C.P.; Sharp, J.O. Enhanced biofilm production by a toluene-degrading Rhodococcus observed after exposure to perfluoroalkyl acids. Envir. Sci. Technol. 2015, 49, 5458–5466. [Google Scholar] [CrossRef]
- Kuyukina, M.S.; Ivshina, I.B. Production of trehalolipid biosurfactants by Rhodococcus. In Biology of Rhodococcus. Microbiology Monographs, 2nd ed.; Alvarez, H.M., Ed.; Springer Nature: Cham, Switzerland, 2019; Volume 16, pp. 271–298. [Google Scholar]
- Franzetti, A.; Gandolfi, I.; Bestetti, G.; Smyth, T.J.P.; Banat, I.M. Production and applications of trehalose lipid biosurfactants. Eur. J. Lipid Sci. Technol. 2010, 112, 617–627. [Google Scholar] [CrossRef]
- Pacwa-Płociniczak, M.; Płaza, G.A.; Piotrowska-Seget, Z.; Cameotra, S.S. Characterization of hydrocarbon-degrading and biosurfactant-producing Pseudomonas sp. P-1 strain as a potential tool for bioremediation of petroleum-contaminated soil. Int. J. Mol. Sci. 2011, 12, 633–654. [Google Scholar] [CrossRef] [PubMed]
- Kuyukina, M.S.; Ivshina, I.B.; Korshunova, I.O.; Rubtsova, E.V. Assessment of bacterial resistance to organic solvents using a combined confocal laser scanning and atomic force microscopy (CLSM/AFM). J. Microbiol. Methods. 2014, 107, 23–29. [Google Scholar] [CrossRef] [PubMed]
- Serebrennikova, M.K.; Kuyukina, M.S.; Krivoruchko, A.V.; Ivshina, I.B. Adaptation of coimmobilized Rhodococcus cells to oil hydrocarbons in a column bioreactor. Appl. Biochem. Microbiol. 2014, 50, 265–272. [Google Scholar] [CrossRef]
- Korshunova, I.O.; Pistsova, O.N.; Kuyukina, M.S.; Ivshina, I.B. The effect of organic solvents on the viability and morphofunctional properties of Rhodococcus. Appl. Biochem. Microbiol. 2016, 52, 43–50. [Google Scholar] [CrossRef]
- Tarasova, E.V.; Grishko, V.V.; Ivshina, I.B. Cell adaptations of Rhodococcus rhodochrous IEGM 66 to betulin biotransformation. Proc. Biochem. 2017, 52, 1–9. [Google Scholar] [CrossRef]
- Cheremnykh, K.M.; Luchnikova, N.A.; Grishko, V.V.; Ivshina, I.B. Bioconversion of ecotoxic dehydroabietic acid using Rhodococcus actinobacteria. J. Hazard. Mater. 2018, 346, 103–112. [Google Scholar] [CrossRef] [PubMed]
- Bos, R.; van der Mei, H.C.; Busscher, H.J. Physico-chemistry of initial microbial adhesive interactions—Its mechanisms and methods for study. FEMS Microbiol. Rev. 1999, 23, 179–230. [Google Scholar] [CrossRef]
- Matz, C.; Kjelleberg, S. Off the hook—How bacteria survive protozoan grazing. Trends Microbiol. 2005, 13, 909–915. [Google Scholar] [CrossRef] [PubMed]
- Lekfeldt, J.D.S.; Rønn, R. A common soil fagellate (Cercomonas sp.) grows slowly when feeding on the bacterium Rhodococcus fascians in isolation, but does not discriminate against it in a mixed culture with Sphingopyxis witfariensis. FEMS Microbiol. Ecol. 2008, 65, 113–124. [Google Scholar] [CrossRef][Green Version]
- Corno, G.; Villiger, J.; Pernthaler, J. Coaggregation in a microbial predator–prey system affects competition and trophic transfer efficiency. Ecology. 2013, 94, 870–881. [Google Scholar] [CrossRef]
- Rubtsova, E.V.; Kuyukina, M.S.; Ivshina, I.B. Effect of cultivation conditions on the adhesive activity of rhodococci towards n-hexadecane. Appl. Biochem. Microbiol. 2012, 48, 452–459. [Google Scholar] [CrossRef]
- Denich, T.J.; Beaudette, L.A.; Lee, H.; Trevors, J.T. Effect of selected environmental and physico-chemical factors on bacterial cytoplasmic membranes. J. Microbiol. Methods. 2003, 52, 149–182. [Google Scholar] [CrossRef]
- de Carvalho, C.C.C.R. Adaptation of Rhodococcus to organic solvents. In Biology of Rhodococcus. Microbiology Monographs, 2nd ed.; Alvarez, H.M., Ed.; Springer Nature: Cham, Switzerland, 2019; Volume 16, pp. 103–135. [Google Scholar]
- de Carvalho, C.C.C.R.; Fischer, M.A.; Kirsten, S.; Wurz, B.; Wick, L.Y.; Heipieper, H.J. Adaptive response of Rhodococcus opacus PWD4 to salt and phenolic stress on the level of mycolic acids. AMB Express 2016, 6, 66. [Google Scholar] [CrossRef]
- Henson, W.R.; Hsu, F.F.; Dantas, G.; Moon, T.S.; Foston, M. Lipid metabolism of 1480 phenol-tolerant Rhodococcus opacus strains for lignin bioconversion. Biotechnol. Biofuels 2018, 11, 339. [Google Scholar] [CrossRef] [PubMed]
- Su, X.; Guo, L.; Ding, L.; Qu, K.; Shen, C. Induction of viable but nonculturable state in Rhodococcus and transcriptome analysis using RNA-seq. PLoS ONE 2016, 11, e0147593. [Google Scholar] [CrossRef] [PubMed]
- Alvarez, H.; Steinbüchel, A. Biology of triacylglycerol accumulation by Rhodococcus. In Biology of Rhodococcus. Microbiology Monographs, 2nd ed.; Alvarez, H.M., Ed.; Springer Nature: Cham, Switzerland, 2019; Volume 16, pp. 299–332. [Google Scholar]
- Kuyukina, M.S.; Ivshina, I.B.; Rychkova, M.I.; Chumakov, O.B. Effect of cell lipid composition on the formation of nonspecific antibiotic resistance in alkanotrophic rhodococci. Microbiology 2000, 69, 51–57. [Google Scholar] [CrossRef]
- Sokolovská, I.; Rozenberg, R.; Riez, C.; Rouxhet, P.G.; Agathos, S.N.; Wattiau, P. Carbon source-induced modifications in the mycolic acid content and cell wall permeability of Rhodococcus erythropolis El. Appl. Environ. Microbiol. 2003, 69, 7019–7027. [Google Scholar] [CrossRef]
- Barbey, C.; Chane, A.; Burini, J.F.; Maillot, O.; Merieau, A.; Gallique, M.; Beury-Cirou, A.; Konto-Ghiorghi, Y.; Feuilloley, M.; Gobert, V.; et al. A rhodococcal transcriptional regulatory mechanism detects the common lactone ring of AHL quorum-sensing signals and triggers the quorum-quenching response. Front. Microbiol. 2018, 9, 2800. [Google Scholar] [CrossRef]
- Müller, C.; Birmes, F.S.; Rückert, C.; Kalinowski, J.; Fetzner, S. Rhodococcus erythropolis BG43 genes mediating Pseudomonas aeruginosa quinolone signal degradation and virulence factor attenuation. Appl. Environ. Microbiol. 2015, 81, 7720–7729. [Google Scholar] [CrossRef] [PubMed]
- Iwabuchi, N.; Sunairi, M.; Anzai, H.; Morisaki, H.; Nakajima, M. Relationships among colony morphotypes, cell-surface properties and bacterial adhesion to substrata in Rhodococcus. Colloid. Surface. B 2003, 30, 51–60. [Google Scholar] [CrossRef]
- Iwabuchi, N.; Sharma, P.K.; Sunairi, M.; Kishi, E.; Sugita, K.; van der Mei, H.C.; Nakajima, M.; Busscher, H.J. Role of interfacial tensions in the translocation of Rhodococcus erythropolis during growth in a two phase culture. Environ. Sci. Technol. 2009, 43, 8290–8294. [Google Scholar] [CrossRef]
- Laczi, K.; Kis, Á.; Horváth, B.; Maróti, G.; Hegedüs, B.; Perei, K.; Rákhely, G. Metabolic responses of Rhodococcus erythropolis PR4 grown on diesel oil and various hydrocarbons. Appl. Microbiol. Biotechnol. 2015, 99, 9745–9759. [Google Scholar] [CrossRef]
- Pen, Y.; Zhang, Z.J.; Morales-García, A.L.; Mears, M.; Tarmey, D.S.; Edyvean, R.G.; Banwart, S.A.; Geoghegan, M. Effect of extracellular polymeric substances on the mechanical properties of Rhodococcus. Biochim. Biophys. Acta 2015, 1848, 518–526. [Google Scholar] [CrossRef]
- Ivshina, I.B.; Kuyukina, M.S. Isolation of propane-oxidizing rhodococci on selective media with antibiotics. Microbiology 1997, 66, 413–418. [Google Scholar]
- Marris, D.P.; Hagr, A. Biofilm: Why the sudden interest? J. Otolaryngol. 2005, 34, 56–59. [Google Scholar]
- Ivshina, I.B.; Kuyukina, M.S.; Philp, J.C.; Christofi, N. Oil desorption from mineral and organic materials using biosurfactant complexes produced by Rhodococcus species. World J. Microbiol. Biotechnol. 1998, 14, 711–717. [Google Scholar] [CrossRef]
- Kuyukina, M.S.; Ivshina, I.B.; Makarov, S.O.; Litvinenko, L.V.; Cunningham, C.J.; Philp, J.C. Effect of biosurfactants on crude oil desorption and mobilization in a soil system. Environ. Intern. 2005, 31, 155–161. [Google Scholar] [CrossRef] [PubMed]
- Ivshina, I.; Kostina, L.; Krivoruchko, A.; Kuyukina, M.; Peshkur, T.; Anderson, P.; Cunningham, C. Removal of polycyclic aromatic hydrocarbons in soil spiked with model mixtures of petroleum hydrocarbons and heterocycles using biosurfactants from Rhodococcus ruber IEGM 231. J. Hazard. Mater. 2016, 312, 8–17. [Google Scholar] [CrossRef]
- Ward, O.; Singh, A.; van Hamme, J. Accelerated biodegradation of petroleum hydrocarbon waste. J. Ind. Microbiol. Biotechnol. 2003, 30, 260–270. [Google Scholar] [CrossRef]
- Alvarez, H.M. Relationship between β-oxidation and the hydrocarbon-degrading profile and actinomycetes bacteria. Int. Biodeter. Biodegrad. 2003, 52, 35–42. [Google Scholar] [CrossRef]
- Ramos, J.L.; Duque, E.; Gallegos, M.-T.; Godoy, P.; Ramos-González, M.I.; Rojas, A.; Terán, W.; Segura, A. Mechanisms of solvent tolerance in gram-negative bacteria. Ann. Rev. Microbiol. 2002, 56, 743–768. [Google Scholar] [CrossRef] [PubMed]
- Belfiore, C.; Curia, M.V.; Farías, M.E. Characterization of Rhodococcus sp. A5wh isolated from a high altitude Andean lake to unravel the survival strategy under lithium stress. Rev. Argent. Microbiol. 2018, 50, 311–322. [Google Scholar]
- Allocati, N.; Masulli, M.; Di Ilio, C.; de Laurenzi, V. Die for the community: An overview of programmed cell death in bacteria. Cell Death Dis. 2015, 6, e1609. [Google Scholar] [CrossRef]
- Atrat, P.; Hösel, P.; Richter, W.; Meyer, H.W.; Hörhold, C. Interactions of Mycobacterium fortuitum with solid sterol substrate particles. J. Basic Microbiol. 1991, 31, 413–422. [Google Scholar] [CrossRef]
- Neumann, G.; Veeranagouda, Y.; Karegoudar, T.B.; Sahin, O.; Mäusezahl, I.; Kabelitz, N.; Kappelmeyer, U.; Heipieper, H.J. Cells of Pseudomonas putida and Enterobacter sp. adapt to toxic organic compounds by increasing their size. Extremophiles 2005, 9, 163–168. [Google Scholar] [CrossRef] [PubMed]
- Nithya, C.; Gnanalakshmi, B.; Pandian, S.K. Assessment and characterization of heavy metal resistance in Palk Bay sediment bacteria. Mar. Environ. Res. 2011, 71, 283–294. [Google Scholar] [CrossRef] [PubMed]
- Veeranagouda, Y.; Karegoudar, T.B.; Neumann, G.; Heipieper, H.J. Enterobacter sp. VKGH12 growing with n-butanol as the sole carbon source and cells to which the alcohol is added as pure toxin show considerable differences in their adaptive responses. FEMS Microbiol. Lett. 2006, 254, 48–54. [Google Scholar] [CrossRef] [PubMed]
- Shen, Y.; Zhao, S.; Zhao, X.; Sun, H.; Shao, M.; Xu, H. In vitro adsorption mechanism of acrylamide by lactic acid bacteria. LWT 2019, 100, 119–125. [Google Scholar] [CrossRef]
- Du, L.N.; Wang, B.; Li, G.; Wang, S.; Crowley, D.E.; Zhao, Y.H. Biosorption of the metal-complex dye Acid Black 172 by live and heat-treated biomass of Pseudomonas sp. strain DY1: Kinetics and sorption mechanisms. J. Hazard. Mater. 2012, 205–206, 47–54. [Google Scholar] [CrossRef]
- Uzoechi, S.C.; Abu-Lail, N.I. The effects of β-lactam antibiotics on surface modifications of multidrug-resistant Escherichia coli: A multiscale approach. Microsc. Microanal. 2019, 25, 135–150. [Google Scholar] [CrossRef] [PubMed]
- Jia, Y.; Yu, C.; Fan, J.; Fu, Y.; Ye, Z.; Guo, X.; Xu, Y.; Shen, C. Alterations in the cell wall of Rhodococcus biphenylivorans under norfloxacin stress. Front. Microbiol. 2020, 11, 554957. [Google Scholar] [CrossRef] [PubMed]
- Halder, S.; Yadav, K.K.; Sarkar, R.; Mukherjee, S.; Saha, P.; Haldar, S.; Karmakar, S.; Sen, T. Alteration of Zeta potential and membrane permeability in bacteria: A study with cationic agents. SpringerPlus 2015, 4, 672. [Google Scholar] [CrossRef] [PubMed]
- Wilson, W.W.; Wade, M.M.; Holman, S.C.; Champlin, F.R. Status of methods for assessing bacterial cell surface charge. J. Microbiol. Methods 2001, 43, 153–164. [Google Scholar] [CrossRef]
- Gibson, K.J.C.; Gilleron, M.; Constant, P.; Puzo, G.; Nigou, J.; Besra, G.S. Structural and functional features of Rhodococcus ruber lipoarabinomannan. Microbiology 2003, 149, 1437–1445. [Google Scholar] [CrossRef]
- Kaczorek, E.; Pacholak, A.; Zdarta, A.; Smułek, W. The impact of biosurfactants on microbial cell properties leading to hydrocarbon bioavailability increase. Colloids Interfaces 2018, 2, 35. [Google Scholar] [CrossRef]
- Arakha, M.; Saleem, M.; Mallick, B.C.; Jha, S. The effects of interfacial potential on antimicrobial propensity of ZnO nanoparticle. Sci. Rep. 2015, 5, 9578. [Google Scholar] [CrossRef]
- Strahl, H.; Hamoen, L.W. Membrane potential is important for bacterial cell division. Proc. Natl. Acad. Sci. USA 2010, 107, 12281–12286. [Google Scholar] [CrossRef]
- Bai, N.; Wang, S.; Abuduaini, R.; Zhang, M.; Zhu, X.; Zhao, Y. Rhamnolipid-aided biodegradation of carbendazim by Rhodococcus sp. D-1: Characteristics, products, and phytotoxicity. Sci. Total Environ. 2017, 590–591, 343–351. [Google Scholar] [CrossRef] [PubMed]
- Kalakoutskii, L.V. In the flow of time. In Proceedings of the IV International Conference “Microbial Diversity: Resource Potential”, Moscow, Russia, 23–25 November 2016; Ivshina, I.B., Kuyukina, M.S., Kamenskikh, T.N., Alfimova, L.A., Eds.; Institute of Ecology and Genetics of Microorganisms, Ural Branch, Perm State University, Russian Academy of Sciences: Moscow, Russia; Perm, Russia, 2016; pp. 121–122. [Google Scholar]

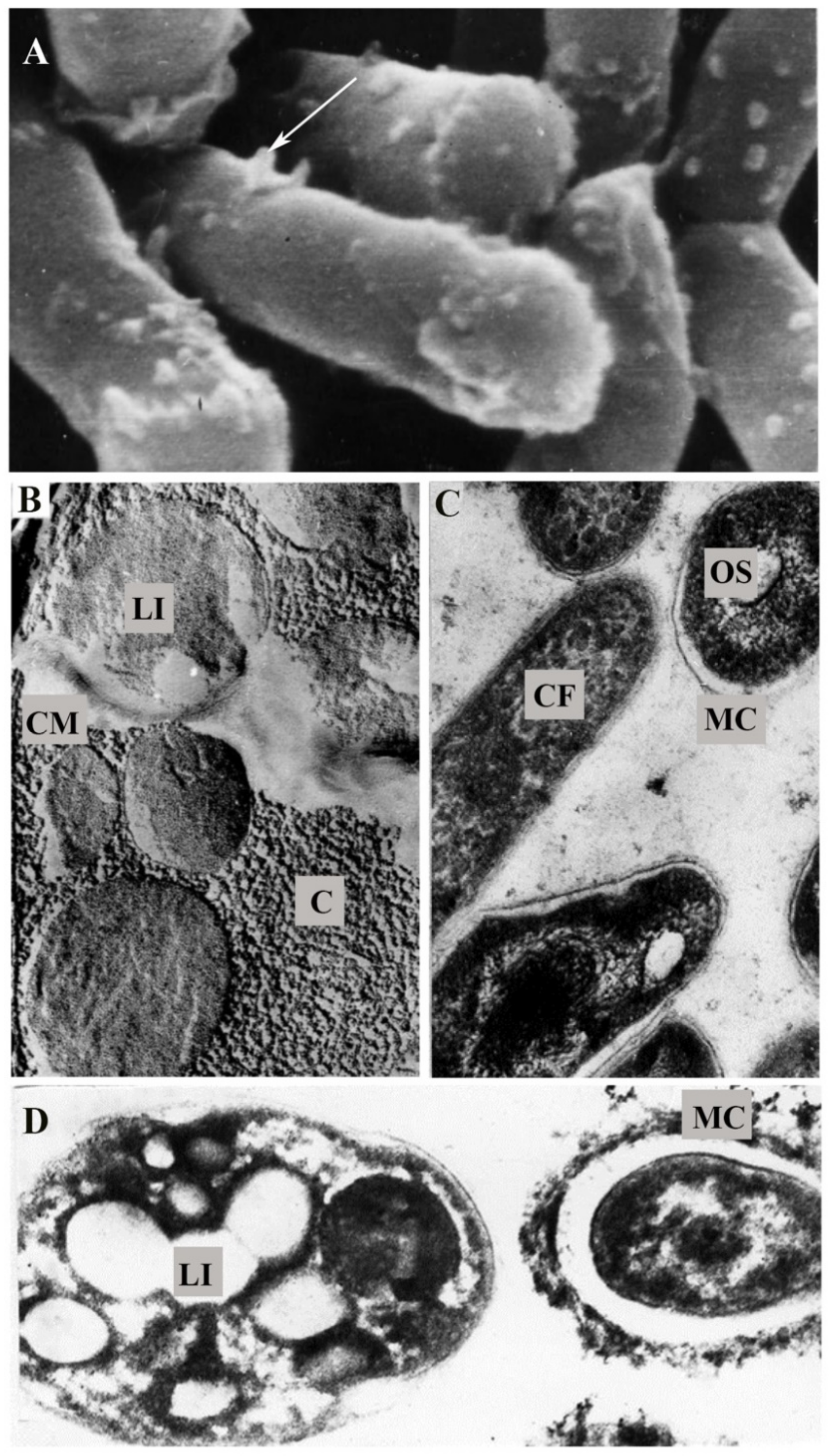


| Strain | Variant | Length, μm | Width, μm | Volume, V, μm3 | Area, S, μm2 | S/V, μm−1 | Roughness, nm |
|---|---|---|---|---|---|---|---|
| Diclofenac | |||||||
| R. ruber IEGM 346 | Control | 3.0 ± 0.02 | 0.9 ± 0.05 | 1.9 ± 0.03 | 5.5 ± 0.05 | 2.9 ± 0.02 | 197.8 ± 2.30 |
| 50 mg/L | 3.5 ± 0,13 | 1.1 ± 0.02 | 3.3 ± 0.05 | 7.9 ± 0.10 | 2.4 ± 0.08 | 216.1 ± 5.51 | |
| 50 μg/L | 2.2 ± 0.05 | 0.8 ± 0.01 | 1.0 ± 0.02 | 3.6 ±0.03 | 3.6 ± 0.02 | 249.6 ± 6.64 | |
| Betulin | |||||||
| R. rhodochrous IEGM 66 | Control | 2.1 ± 0.30 | 0.8 ± 0.10 | 1.1 ± 0.39 | 6.3 ± 0.45 | 5.7 ± 0.69 | 268.5 ± 12.72 |
| 500 mg/L | 1.9 ± 0.47 | 0.7 ± 0.11 | 0.9 ± 0.33 | 5.4 ± 0.39 | 6.0 ± 0.85 | 359.6 ± 9.13 | |
| 3000 mg/L | 1.8 ± 0.48 | 0.8 ± 0.12 | 0.9 ± 0.12 | 5.6 ± 1.07 | 6.2 ± 0.88 | 400.9 ± 7.92 | |
| Dehydroabietic acid | |||||||
| R. rhodochrous IEGM 107 | Control | 1.3 ± 0.28 | 1.1 ± 0.19 | 1.3 ± 0.16 | 4.3 ± 0.28 | 3.2 ± 0.11 | 206.5 ± 10.72 |
| 500 mg/L | 1.8 ± 0.26 | 1.2 ± 0.24 | 2.2 ± 0.13 | 6.1 ± 0.23 | 2.7 ± 0.09 | 365.9 ± 6.92 | |
| Strain | Variant | Zeta Potential |
|---|---|---|
| Dehydroabietic acid | ||
| R. rhodochrous IEGM 107 | Control | −26.6 ± 0.91 |
| 500 mg/L | −27.3 ± 1.11 | |
| R. erythropolis IEGM 267 | Control | −15.5 ± 1.42 |
| 500 mg/L | −19.8 ± 1.23 | |
| Diclofenac | ||
| R. ruber IEGM 346 | Control | −35.3 ± 2.33 |
| 50 mg/L | −31.3 ± 0.83 | |
| Betulin | ||
| R. rhodochrous IEGM 66 | Control | −34.8 ± 0.91 |
| 3000 mg/L | −35.2 ± 1.11 | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ivshina, I.B.; Kuyukina, M.S.; Krivoruchko, A.V.; Tyumina, E.A. Responses to Ecopollutants and Pathogenization Risks of Saprotrophic Rhodococcus Species. Pathogens 2021, 10, 974. https://doi.org/10.3390/pathogens10080974
Ivshina IB, Kuyukina MS, Krivoruchko AV, Tyumina EA. Responses to Ecopollutants and Pathogenization Risks of Saprotrophic Rhodococcus Species. Pathogens. 2021; 10(8):974. https://doi.org/10.3390/pathogens10080974
Chicago/Turabian StyleIvshina, Irina B., Maria S. Kuyukina, Anastasiia V. Krivoruchko, and Elena A. Tyumina. 2021. "Responses to Ecopollutants and Pathogenization Risks of Saprotrophic Rhodococcus Species" Pathogens 10, no. 8: 974. https://doi.org/10.3390/pathogens10080974
APA StyleIvshina, I. B., Kuyukina, M. S., Krivoruchko, A. V., & Tyumina, E. A. (2021). Responses to Ecopollutants and Pathogenization Risks of Saprotrophic Rhodococcus Species. Pathogens, 10(8), 974. https://doi.org/10.3390/pathogens10080974








