Evaluation of Biofilm Formation and Prevalence of Multidrug-Resistant Strains of Staphylococcus epidermidis Isolated from Neonates with Sepsis in Southern Poland
Abstract
1. Introduction
2. Results
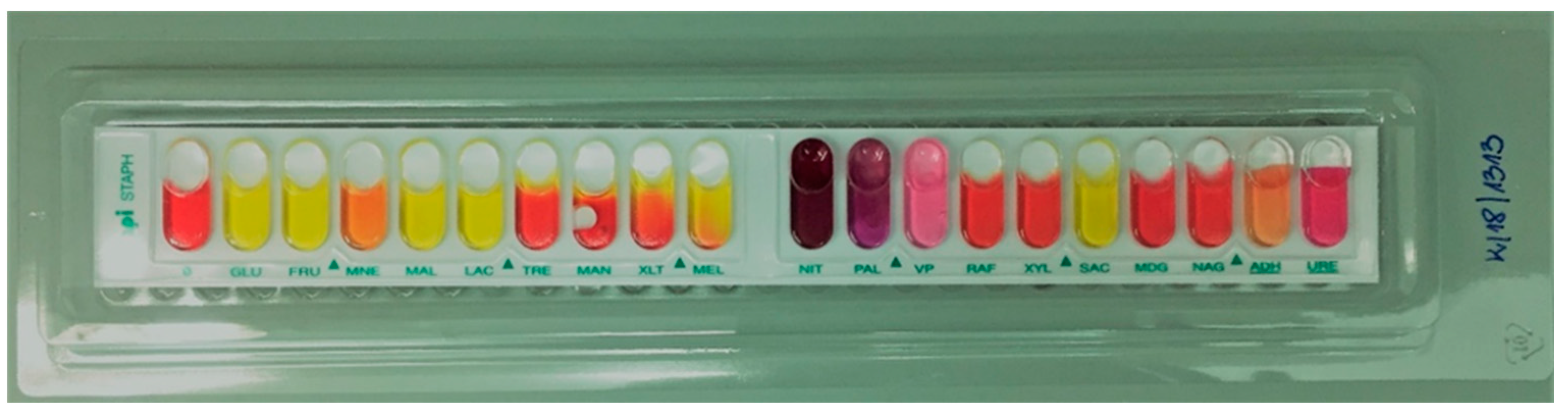
2.1. Species Identification of the Tested Strains
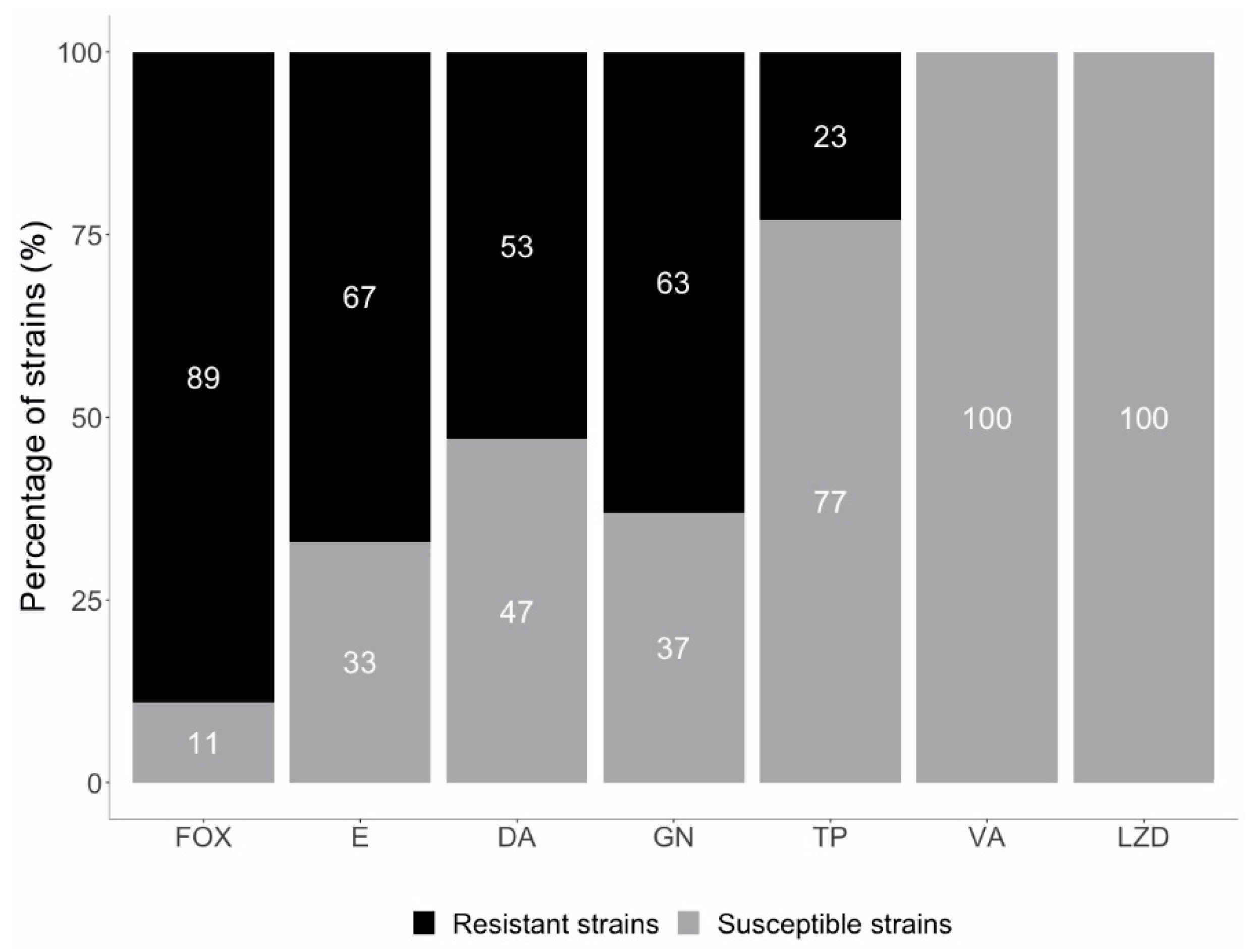
2.2. Resistance to Methicillin
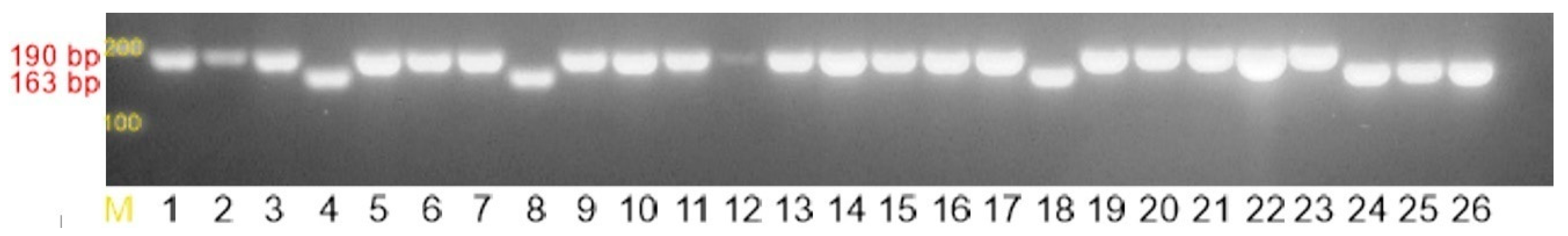
2.3. Macrolide, Lincosamide, and Streptogramin (MLS) Resistance Mechanisms
2.4. Biofilm-Forming Capacity
2.5. Application of Logistic Regression to Predict A Dichotomous Variable for Three Biofilm Incidence Study Models
3. Discussion
4. Materials and Methods
4.1. Phenotypic Methods
4.1.1. Bacterial Strains
4.1.2. Antimicrobial Susceptibility Testing
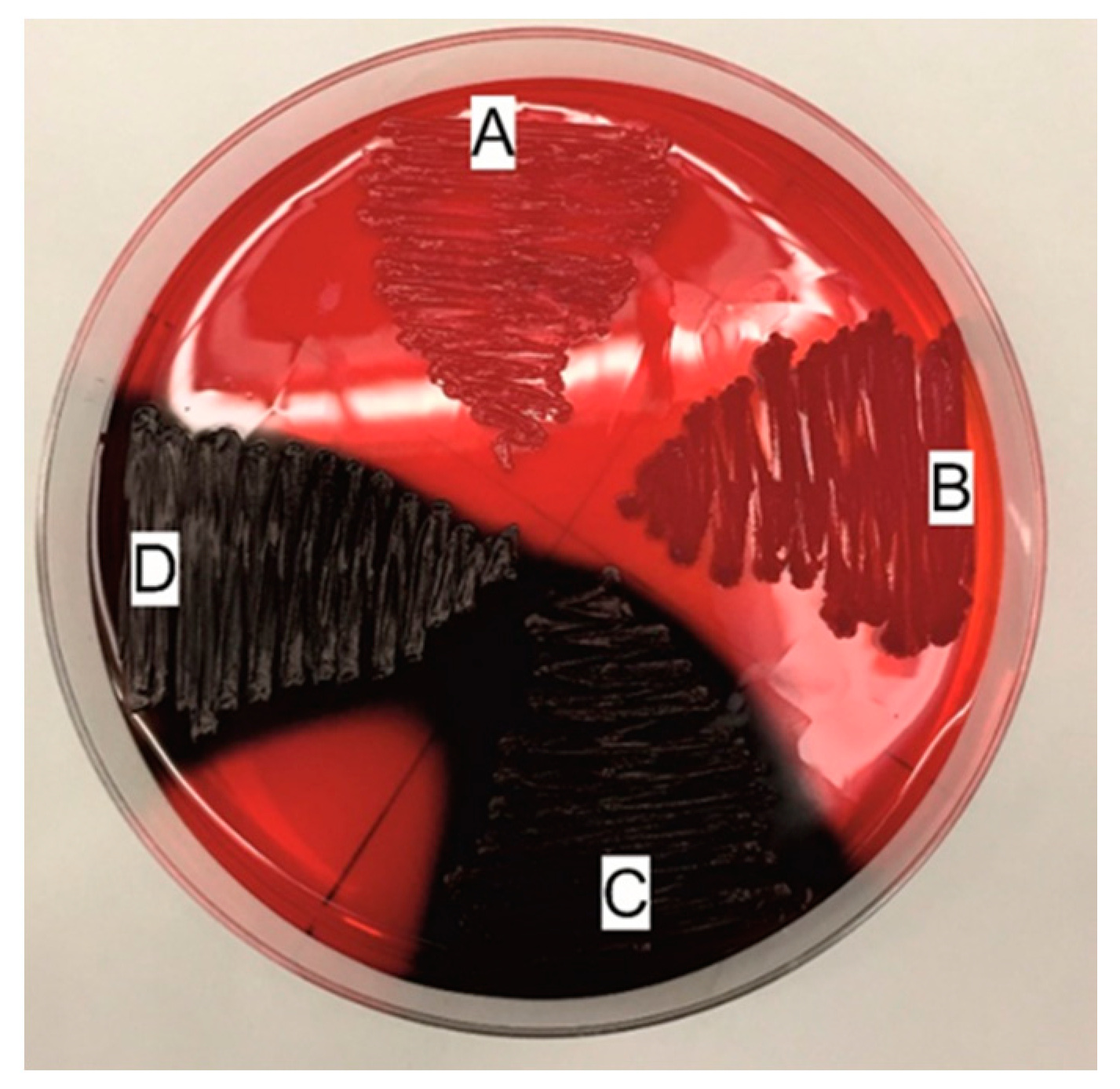
4.1.3. Biofilm Detection
Freeman Method
Christensen Method
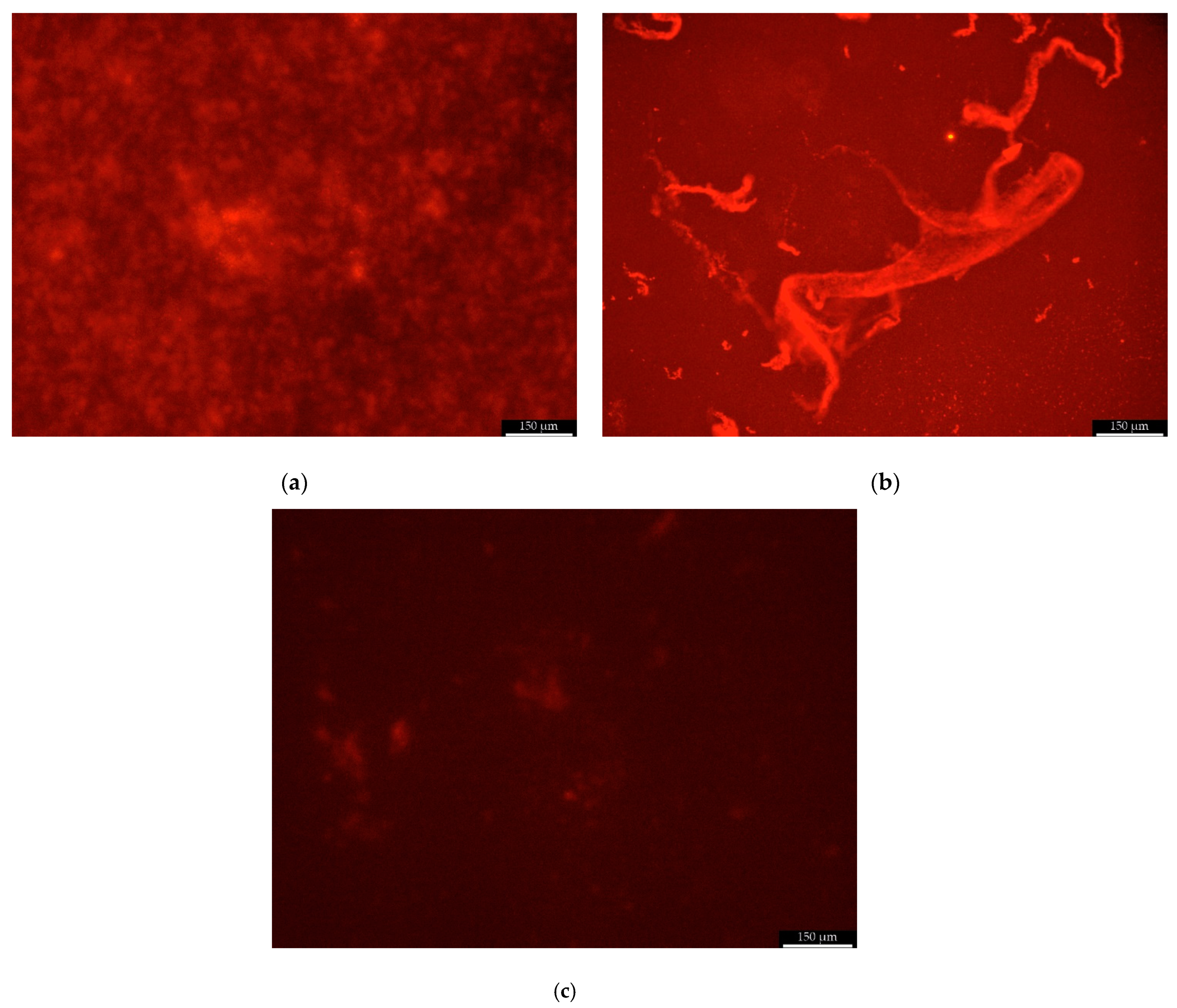
Scanning Fluorescent Microscope
4.2. Genetic Methods
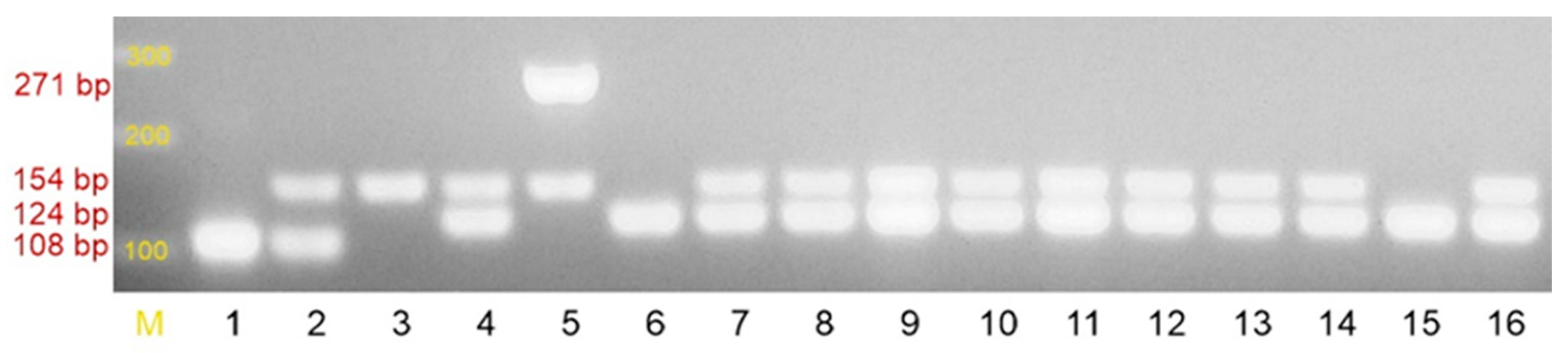
4.2.1. Identification of the Tested Strains to the Species Level and Detection of Selected Resistance Mechanisms
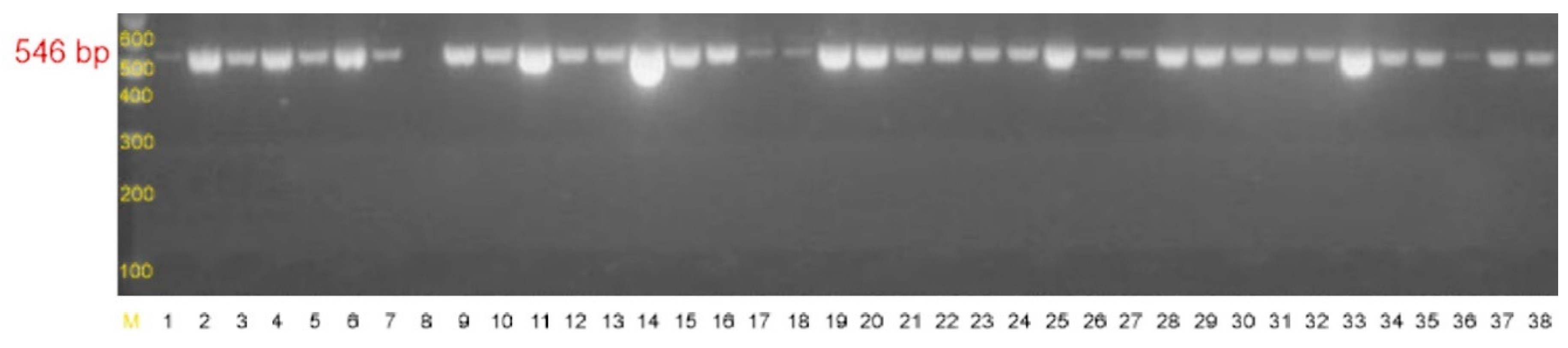
4.2.2. Detection of Genes Associated with Biofilm Formation
4.2.3. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Friedrich, A.W. Control of hospital acquired infections and antimicrobial resistance in Europe: the way to go. Wien. Med. Wochenschr. 2019, 169, 25–30. [Google Scholar] [CrossRef] [PubMed]
- Becker, K.; Both, A.; Weißelberg, S.; Heilmann, C.; Rohde, H. Emergence of coagulase-negative staphylococci. Expert Rev. Anti-Infective Ther. 2020, 18, 349–366. [Google Scholar] [CrossRef]
- Moles, L.; Gómez, M.; Moroder, E.; Bustos, G.; Melgar, A.; Del Campo, R.; Rodríguez, J. Staphylococcus epidermidis in feedings and feces of preterm neonates. PLoS ONE 2020, 15, e0227823. [Google Scholar] [CrossRef]
- Wang, L.; Du, K.-N.; Zhao, Y.-L.; Yu, Y.-J.; Sun, L.; Jiang, H.-B. Risk Factors of Nosocomial Infection for Infants in Neonatal Intensive Care Units: A Systematic Review and Meta-Analysis. Med. Sci. Monit. 2019, 25, 8213–8220. [Google Scholar] [CrossRef]
- Konstantinidi, A.; Sokou, R.; Panagiotounakou, P.; Lampridou, M.; Parastatidou, S.; Tsantila, K.; Gounari, E.; Gounaris, A.K. Umbilical Venous Catheters and Peripherally Inserted Central Catheters: Are They Equally Safe in VLBW Infants? A Non-Randomized Single Center Study. Medicina 2019, 55, 442. [Google Scholar] [CrossRef]
- Bhatta, D.R.; Subramanya, S.H.; Hamal, D.; Shrestha, R.; Gauchan, E.; Basnet, S.; Nayak, N.; Gokhale, S. Bacterial contamination of neonatal intensive care units: How safe are the neonates? Antimicrob. Resist. Infect. Control. 2021, 10, 1–6. [Google Scholar] [CrossRef]
- Freitas, F.T.D.M.; Viegas, A.P.B.; Romero, G.A.S. Neonatal healthcare-associated infections in Brazil: systematic review and meta-analysis. Arch. Public Health 2021, 79, 1–10. [Google Scholar] [CrossRef]
- Dong, Y.; Speer, C.P.; Glaser, K. Beyond sepsis: Staphylococcus epidermidis is an underestimated but significant contributor to neonatal morbidity. Virulence 2018, 9, 621–633. [Google Scholar] [CrossRef]
- França, A.; Gaio, V.; Lopes, N.; Melo, L. Virulence Factors in Coagulase-Negative Staphylococci. Pathogens 2021, 10, 170. [Google Scholar] [CrossRef] [PubMed]
- Yong, Y.Y.; Dykes, G.; Choo, W.S. Biofilm formation by staphylococci in health-related environments and recent reports on their control using natural compounds. Crit. Rev. Microbiol. 2019, 45, 201–222. [Google Scholar] [CrossRef] [PubMed]
- Kranjec, C.; Angeles, D.M.; Mårli, M.T.; Fernández, L.; García, P.; Kjos, M.; Diep, D. Staphylococcal Biofilms: Challenges and Novel Therapeutic Perspectives. Antibiotics 2021, 10, 131. [Google Scholar] [CrossRef]
- Yin, W.; Wang, Y.; Liu, L.; He, J. Biofilms: The Microbial “Protective Clothing” in Extreme Environments. Int. J. Mol. Sci. 2019, 20, 3423. [Google Scholar] [CrossRef]
- Shresthar, L.B.; Bhattarai, N.R.; Khanal, B. Comparative evaluation of methods for the detection of biofilm formation in coagulase-negative staphylococci and correlation with antibiogram. Infect. Drug Resist. 2018, 11, 607–613. [Google Scholar] [CrossRef] [PubMed]
- Chon, J.-W.; Lee, U.J.; Bensen, R.; West, S.; Paredes, A.; Lim, J.; Khan, S.; Hart, M.E.; Phillips, K.S.; Sung, K. Virulence Characteristics of mecA-Positive Multidrug-Resistant Clinical Coagulase-Negative Staphylococci. Microorganisms 2020, 8, 659. [Google Scholar] [CrossRef] [PubMed]
- Fyfe, C.; Grossman, T.H.; Kerstein, K.; Sutcliffe, J. Resistance to Macrolide Antibiotics in Public Health Pathogens. Cold Spring Harb. Perspect. Med. 2016, 6, a025395. [Google Scholar] [CrossRef] [PubMed]
- Teodoro, C.R.S.; Mattos, C.S.; Cavalcante, F.S.; Pereira, E.M.; Dos Santos, K.R.N. Characterization of MLSb resistance among Staphylococcus aureus and Staphylococcus epidermidis isolates carrying different SCCmec types. Microbiol. Immunol. 2012, 56, 647–650. [Google Scholar] [CrossRef]
- Khashei, R.; Malekzadegan, Y.; Ebrahim-Saraie, H.S.; Razavi, Z. Phenotypic and genotypic characterization of macrolide, lincosamide and streptogramin B resistance among clinical isolates of staphylococci in southwest of Iran. BMC Res. Notes 2018, 11, 711. [Google Scholar] [CrossRef]
- Michels, R.; Last, K.; Becker, S.; Papan, C. Update on Coagulase-Negative Staphylococci—What the Clinician Should Know. Microorganisms 2021, 9, 830. [Google Scholar] [CrossRef] [PubMed]
- Sánchez, A.; Benito, N.; Rivera, A.; García, L.; Miró, E.; Mur, I.; González, Y.; Gutiérrez, C.; Horcajada, J.; Espinal, P.; et al. Pathogenesis of Staphylococcus epidermidis in prosthetic joint infections: can identification of virulence genes differentiate between infecting and commensal strains? J. Hosp. Infect. 2020, 105, 561–568. [Google Scholar] [CrossRef]
- Pereira, E.M.; Schuenck, R.P.; Malvar, K.L.; Iório, N.L.; Matos, P.D.; Olendzki, A.N.; Oelemann, W.M.; Dos Santos, K.R. Staphylococcus aureus, Staphylococcus epidermidis and Staphylococcus haemolyticus: Methicillin-resistant isolates are detected directly in blood cultures by multiplex PCR. Microbiol. Res. 2010, 165, 243–249. [Google Scholar] [CrossRef]
- Chaieb, K.; Zmantar, T.; Chehab, O.; Bouchami, O.; Ben Hasen, A.; Mahdouani, K.; Bakhrouf, A. Antibiotic Resistance Genes Detected by Multiplex PCR Assay in Staphylococcus epidermidis Strains Isolated from Dialysis Fluid and Needles in a Dialysis Service. Jpn. J. Infect. Dis. 2007, 60, 183–187. [Google Scholar] [CrossRef]
- Freeman, D.J.; Falkiner, F.R.; Keane, C.T. New method for detecting slime production by coagulase negative staphylococci. J. Clin. Pathol. 1989, 42, 872–874. [Google Scholar] [CrossRef]
- Christensen, G.D.; A Simpson, W.; Younger, J.J.; Baddour, L.M.; Barrett, F.F.; Melton, D.M.; Beachey, E.H. Adherence of coagulase-negative staphylococci to plastic tissue culture plates: a quantitative model for the adherence of staphylococci to medical devices. J. Clin. Microbiol. 1985, 22, 996–1006. [Google Scholar] [CrossRef]
- Ferreira, A.A.; Tette, P.A.S.; Mendonça, R.C.S.; Soares, A.D.S.; De Carvalho, M.M. Detection of exopolysaccharide production and biofilm-related genes in Staphylococcus spp. isolated from a poultry processing plant. Food Sci. Technol. 2014, 34, 710–716. [Google Scholar] [CrossRef]
- Jakiel, G.; Wilińska, M.; Bińkowska, M.; Kowal, A.; Rumowska, S.; Ciebiera, M. Late preterm infants–impact of perinatal factors on neonatal results. A clinical study. Ann. Agric. Environ. Med. 2015, 22, 536–541. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Sadkowska-Todys, M.; Zieliński, A.; Czarkowski, M.P. Infectious diseases in Poland in 2013. Prz. Epidemiol. 2015, 69, 329–334. [Google Scholar]
- Turlej, A.; Hryniewicz, W.; Empel, J. Staphylococcal Cassette Chromosome mec (SCCmec) Classification and Typing Methods: an Overview. Pol. J. Microbiol. 2011, 60, 95–103. [Google Scholar] [CrossRef]
- Brzychczy-Wloch, M.; Borszewska-Kornacka, M.; Gulczyńska, E.; Wojkowska-Mach, J.; Sulik, M.; Grzebyk, M.; Luchter, M.; Heczko, P.B.; Bulanda, M. Prevalence of antibiotic resistance in multi-drug resistant coagulase-negative staphylococci isolated from invasive infection in very low birth weight neonates in two Polish NICUs. Ann. Clin. Microbiol. Antimicrob. 2013, 12, 41. [Google Scholar] [CrossRef]
- Al-Mulla, N.A.; Taj-Aldeen, S.J.; Elshafie, S.S.; Janahi, M.; Al-Nasser, A.A.; Chandra, P. Bacterial bloodstream infections and antimicrobial susceptibility pattern in pediatric hematology/oncology patients after anticancer chemotherapy. Infect. Drug Resist. 2014, 7, 289–299. [Google Scholar] [CrossRef] [PubMed]
- Najar-Peerayeh, S.; Moghadas, A.J.; Behmanesh, M. Antibiotic Susceptibility and mecA Frequency in Staphylococcus epidermidis, Isolated from Intensive Care Unit Patients. Jundishapur J. Microbiol. 2014, 7, e11188. [Google Scholar] [CrossRef] [PubMed]
- Hassoun, A.; Linden, P.K.; Friedman, B. Incidence, prevalence, and management of MRSA bacteremia across patient populations—a review of recent developments in MRSA management and treatment. Crit. Care 2017, 21, 1–10. [Google Scholar] [CrossRef]
- Szczuka, E.; Prawda-Zolotar, J.; Nowakiewicz, M.; Kaznowski, A. Wrażliwość na antybiotyki i zdolność wytwarzania śluzu przez szczepy gronkowców koagulazo-ujemnych. Med. Dosw. Mikrobiol. 2011, 63, 1. [Google Scholar]
- Wojtyczka, R.D.; Orlewska, K.; Kępa, M.; Idzik, D.; Dziedzic, A.; Mularz, T.; Krawczyk, M.; Miklasińska, M.; Wąsik, T.J. Biofilm Formation and Antimicrobial Susceptibility of Staphylococcus epidermidis Strains from a Hospital Environment. Int. J. Environ. Res. Public Health 2014, 11, 4619–4633. [Google Scholar] [CrossRef] [PubMed]
- Juda, M.; Chudzik-Rzad, B.; Malm, A. The prevalence of genotypes that determine resistance to macrolides, lincosamides, and streptogramins B compared with spiramycin susceptibility among erythromycin-resistant Staphylococcus epidermidis. Mem. Inst. Oswaldo Cruz 2016, 111, 155–160. [Google Scholar] [CrossRef] [PubMed]
- Szemraj, M.; Czekaj, T.; Kalisz, J.; Szewczyk, E.M. Differences in distribution of MLS antibiotics resistance genes in clinical isolates of staphylococci belonging to species: S. epidermidis, S. hominis, S. haemolyticus, S. simulans and S. warneri. BMC Microbiol. 2019, 19, 124. [Google Scholar] [CrossRef] [PubMed]
- Feßler, A.T.; Wang, Y.; Wu, C.; Schwarz, S. Mobile lincosamide resistance genes in staphylococci. Plasmid 2018, 99, 22–31. [Google Scholar] [CrossRef]
- Bialkowska-Hobrzanska, H.; Jaskot, D.; Hammerberg, O. Molecular characterization of the coagulase-negative staphylococcal surface flora of premature neonates. J. Gen. Microbiol. 1993, 139, 2939–2944. [Google Scholar] [CrossRef][Green Version]
- Available online: http://www.korld.edu.pl/spec_rekomendacje-eucast.php (accessed on 28 August 2020).
- Dzierżanowska, D. Antybiotykoterapia Praktyczna; Alfa Medica Press: Bielsko-Biała, Poland, 2018. [Google Scholar]
- Tevell, S.; Claesson, C.; Hellmark, B.; Söderquist, B.; Nilsdotter-Augustinsson, Å. Heterogeneous glycopeptide intermediate Staphylococcus epidermidis isolated from prosthetic joint infections. Eur. J. Clin. Microbiol. Infect. Dis. 2014, 33, 911–917. [Google Scholar] [CrossRef]
- Wu, H.; Moser, C.; Wang, H.-Z.; Høiby, N.; Song, Z.-J. Strategies for combating bacterial biofilm infections. Int. J. Oral Sci. 2015, 7, 1–7. [Google Scholar] [CrossRef]
- Zheng, Y.; He, L.; Asiamah, T.K.; Otto, M. Colonization of medical devices by staphylococci. Environ. Microbiol. 2018, 20, 3141–3153. [Google Scholar] [CrossRef]
- Grzebyk, M.; Brzychczy-Włoch, M.; Piotrowska, A.; Krzyściak, P.; Heczko, P.B.; Bulanda, M. Phenotypic evaluation of hydrophobi-city and the ability to produce biofilm in coagulase-negative staphylococci isolated from infected very-low-birthweight new-borns. Med. Dosw. Mikrobiol. 2013, 65, 149–159. [Google Scholar]
- Oliveira, A.; Cunha, M.D.L.R. Comparison of methods for the detection of biofilm production in coagulase-negative staphylococci. BMC Res. Notes 2010, 3, 260. [Google Scholar] [CrossRef] [PubMed]
- Cafiso, V.; Bertuccio, T.; Santagati, M.; Campanile, F.; Amicosante, G.; Perilli, M.; Selan, L.; Artini, M.; Nicoletti, G.; Stefani, S. Presence of the ica operon in clinical isolates of Staphylococcus epidermidis and its role in biofilm production. Clin. Microbiol. Infect. 2004, 10, 1081–1088. [Google Scholar] [CrossRef] [PubMed]
- Mathur, T.; Singhal, S.; Khan, S.; Upadhyay, D.J.; Fatma, T.; Rattan, A. Detection of biofilm formation among the clinical isolates of Staphylococci: An evaluation of three different screening methods. Indian J. Med Microbiol. 2006, 24, 25–29. [Google Scholar] [CrossRef]
- Līduma, I.; Tračevska, T.; Bērs, U.; Žileviča, A. Phenotypic and genetic analysis of biofilm formation by Staphylococcus epidermidis. Medicina 2012, 48, 305–309. [Google Scholar] [CrossRef]
- Fraiha, R.O.; Pereira, A.P.R.; Brito, E.D.C.A.; Borges, C.L.; Parente, A.F.A.; Perdomo, R.T.; Macedo, M.L.; Weber, S.S. Stress conditions in the host induce persister cells and influence biofilm formation by Staphylococcus epidermidis RP62A. Rev. Soc. Bras. Med. Trop. 2019, 52, e20180001. [Google Scholar] [CrossRef] [PubMed]
- Fey, P.D.; E Olson, M. Current concepts in biofilm formation of Staphylococcus epidermidis. Futur. Microbiol. 2010, 5, 917–933. [Google Scholar] [CrossRef]
- Büttner, H.; Emack, D.; Erohde, H. Structural basis of Staphylococcus epidermidis biofilm formation: mechanisms and molecular interactions. Front. Cell. Infect. Microbiol. 2015, 5, 14. [Google Scholar] [CrossRef]
- Gajewska, J.; Chajęcka-Wierzchowska, W. Biofilm Formation Ability and Presence of Adhesion Genes among Coagulase-Negative and Coagulase-Positive Staphylococci Isolates from Raw Cow’s Milk. Pathogens 2020, 9, 654. [Google Scholar] [CrossRef]
- Del Pozo, J.L. Biofilm-related disease. Expert Rev. Anti-Infective Ther. 2018, 16, 51–65. [Google Scholar] [CrossRef]
- Moryl, M. Extracellular matrix as a microbial virulence factor in the development of human diseases. Postęp. Hig. Med. Dośw. 2015, 69, 1485–1498. [Google Scholar]
- Pietruczuk-Padzik, A.; Stefańska, J.; Semczuk, K.; Dzierzanowska, D.; Tyski, S. Evaluation of biofilm formation by Staphylococcus aureus isolated from sputum of cystic fibrosis patients. Med. Dosw. Mikrobiol. 2010, 62, 1–8. [Google Scholar] [PubMed]
- Available online: https://www.eucast.org/fileadmin/src/media/PDFs/EUCAST_files/Disk_test_documents/2021_manuals/Manual_v_9.0_EUCAST_Disk_Test_2021.pdf (accessed on 15 June 2021).
- Stepanović, S.; Vukovic, D.; Dakić, I.; Savić, B.; Švabić-Vlahović, M. A modified microtiter-plate test for quantification of staphylococcal biofilm formation. J. Microbiol. Methods 2000, 40, 175–179. [Google Scholar] [CrossRef]
- Available online: https://www.thermofisher.com/document-connect/document-connect.html?url=https://assets.thermofisher.com/TFS-Assets/LSG/manuals/mp10318.pdf (accessed on 15 June 2021).
| Average OD Value | N (%) | Biofilm Formation |
|---|---|---|
| OD ≤ 0.09 | 0 (0%) | No |
| 0.09 < OD ≤ 0.18 | 11 (11%) | Weak |
| 0.18 < OD ≤ 0.36 | 3 (3%) | Moderate |
| 0.36 < OD | 86 (86%) | Strong |
| * ODc = 0.09 |
| Resistance Mechanisms | Biofilm Formation | ||||||
|---|---|---|---|---|---|---|---|
| Christensen Method N (%) | Freeman Method N (%) | icaADB Gene Cluster N (%) | |||||
| Strong | Moderate | Weak | Black | Red | Presence | Absence | |
| MRSE | 22 (79%) | 1 (3%) | 5 (18%) | 22 (79%) | 6 (21%) | 22 (79%) | 6 (21%) |
| MRSE + MLSB | 48 (91%) | 2 (4%) | 3 (5%) | 48 (91%) | 5 (9%) | 48 (91%) | 5 (9%) |
| MRSE + MSB | 7 (88%) | 0 | 1 (12%) | 6 (75%) | 2 (25%) | 6 (75%) | 2 (25%) |
| MLSB | 1 (100%) | 0 | 0 | 1 (100%) | 0 | 1 (100%) | 0 |
| MSB | 4 (100%) | 0 | 0 | 4 (100%) | 0 | 4 (100%) | 0 |
| None | 4 (67%) | 0 | 2 (33%) | 4 (67%) | 2 (33%) | 4 (67%) | 2 (33%) |
| Total | 86 (86%) | 3 (3%) | 11 (11%) | 85 (85%) | 15 (15%) | 85 (85%) | 15 (15%) |
| Biofilm Formation | |||||||
|---|---|---|---|---|---|---|---|
| Antibiotics | Christensen Method N (%) | Freeman Method N (%) | icaADB Gene Cluster N (%) | ||||
| Strong | Moderate | Weak | Black | Red | Presence | Lack | |
| Gentamicin | |||||||
| R* | 57 (91%) | 2 (3%) | 4 (6%) | 56 (89%) | 7 (11%) | 56 (89%) | 7 (11%) |
| S** | 29 (78%) | 1 (3%) | 7 (7%) | 29 (78%) | 8 (22%) | 29 (78%) | 8 (22%) |
| Teicoplanin | |||||||
| R | 20 (87%) | 1 (9%) | 2 (4%) | 19 (83%) | 4 (17%) | 19 (83%) | 4 (17%) |
| S | 66 (86%) | 2 (3%) | 9 (12%) | 66 (86%) | 11 (14%) | 66 (86%) | 11 (14%) |
| B | p-Value | Exp(B) | 95% CI | |
|---|---|---|---|---|
| Resistance mechanism 0 | 0.369 | |||
| Resistance mechanism MLSB | −0.50 | 0.573 | 0.61 | 0.11–3.47 |
| Resistance mechanism MSB | 0.43 | 0.665 | 1.53 | 0.22–10.65 |
| Resistance mechanism MRSE | −0.22 | 0.811 | 0.80 | 0.13–4.90 |
| Resistance to gentamycin | −0.68 | 0.305 | 0.51 | 0.14–1.86 |
| Resistance to teicoplanin | 0.49 | 0.486 | 1.63 | 0.41–6.47 |
| Constant | 1.87 | 0.148 | 6.49 |
| B | p-Value | Exp(B) | 95% CI | |
|---|---|---|---|---|
| Resistance mechanism 0 | 0.284 | |||
| Resistance mechanism MLSB | −1.39 | 0.224 | 4.01 | 0.43–37.52 |
| Resistance mechanism MSB | −0.57 | 0.641 | 1.78 | 0.16–19.77 |
| Resistance mechanism MRSE | −0.09 | 0.921 | 1.09 | 0.17–6.97 |
| Resistance to gentamycin | −0.91 | 0.177 | 2.48 | 0.67–9.24 |
| Resistance to teicoplanin | 0.16 | 0.831 | 0.85 | 0.19–3.78 |
| Constant | 3.05 | 0.044 | 0.05 |
| B | p-Value | Exp(B) | 95% CI | |
|---|---|---|---|---|
| Resistance mechanism 0 | 0.369 | |||
| Resistance mechanism MLSB | −0.50 | 0.573 | 0.61 | 0.11–3.47 |
| Resistance mechanism MSB | 0.43 | 0.665 | 1.53 | 0.22–10.65 |
| Resistance mechanism MRSE | −0.22 | 0.811 | 0.80 | 0.13–4.90 |
| Resistance to gentamycin | −0.68 | 0.305 | 0.51 | 0.14–1.86 |
| Resistance to teicoplanin | 0.49 | 0.486 | 1.63 | 0.41–6.47 |
| Constant | 1.87 | 0.148 | 6.49 |
| Average OD Value | Biofilm Production |
|---|---|
| OD ≤ ODc * | Non-biofilm producer |
| ODc < OD ≤ 2×ODc | Weak biofilm producer |
| 2×ODc < OD ≤ 4×ODc | Moderate biofilm producer |
| 4×ODc < OD | Strong biofilm producer |
| Primer | Sequence (5′-3′) | Size of the Product | [20] |
| SA1 | AATCTTTGTCGGTACACGATATTCTTCACG | 108 bp | |
| SA2 | CGTAATGAGATTTCAGTAGATAATACAACA | ||
| SE1 | ATCAAAAAGTTGGCGAACCTTTTCA | 124 bp | |
| SE2 | CAAAAGAGCGTGGAGAAAAGTATCA | ||
| SH1 | GGTCGCTTAGTCGGAACAAT | 271 bp | |
| SH2 | CACGAGCAATCTCATCACCT | ||
| MRS1 | TAGAAATGACTGAACGTCCG | 154 bp | |
| MRS2 | TTGCGATCAATGTTACCGTAG |
| Primer | Sequence (5′-3′) | Size of the Product | [21] |
| ermA1 | TATCTTATCGTTGAGAAGGGATT | 139 bp | |
| ermA2 | CTACACTTGGCTTAGGATGAAA | ||
| ermB1 | CTATCTGATTGTTGAAGAAGGATT | 142 bp | |
| ermB2 | GTTTACTCTTGGTTTAGGATGAAA | ||
| ermC1 | CTTGTTGATCACGATAATTTCC | 190 bp | |
| ermC2 | ATCTTTTAGCAAACCCGTATTC | ||
| msrA1 | TCCAATCATAGCACAAAATC | 163 bp | |
| msrA2 | AATTCCCTCTATTTGGTGGT | ||
| mef1 | AGTATCTTAATCACTAGTGC | 348 bp | |
| mef2 | TTCTTCTGGTACAAAAGTGG |
| Primer | Sequence (5′-3′) | Size of the Product |
|---|---|---|
| icaADB-F | TTATCAATGCCGCAGTTGTC | 546 bp |
| icaADB-R | GTTTAACGCGAGTGCGCTAT |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Skiba-Kurek, I.; Nowak, P.; Empel, J.; Tomczak, M.; Klepacka, J.; Sowa-Sierant, I.; Żak, I.; Pomierny, B.; Karczewska, E. Evaluation of Biofilm Formation and Prevalence of Multidrug-Resistant Strains of Staphylococcus epidermidis Isolated from Neonates with Sepsis in Southern Poland. Pathogens 2021, 10, 877. https://doi.org/10.3390/pathogens10070877
Skiba-Kurek I, Nowak P, Empel J, Tomczak M, Klepacka J, Sowa-Sierant I, Żak I, Pomierny B, Karczewska E. Evaluation of Biofilm Formation and Prevalence of Multidrug-Resistant Strains of Staphylococcus epidermidis Isolated from Neonates with Sepsis in Southern Poland. Pathogens. 2021; 10(7):877. https://doi.org/10.3390/pathogens10070877
Chicago/Turabian StyleSkiba-Kurek, Iwona, Paweł Nowak, Joanna Empel, Magdalena Tomczak, Joanna Klepacka, Iwona Sowa-Sierant, Iwona Żak, Bartosz Pomierny, and Elżbieta Karczewska. 2021. "Evaluation of Biofilm Formation and Prevalence of Multidrug-Resistant Strains of Staphylococcus epidermidis Isolated from Neonates with Sepsis in Southern Poland" Pathogens 10, no. 7: 877. https://doi.org/10.3390/pathogens10070877
APA StyleSkiba-Kurek, I., Nowak, P., Empel, J., Tomczak, M., Klepacka, J., Sowa-Sierant, I., Żak, I., Pomierny, B., & Karczewska, E. (2021). Evaluation of Biofilm Formation and Prevalence of Multidrug-Resistant Strains of Staphylococcus epidermidis Isolated from Neonates with Sepsis in Southern Poland. Pathogens, 10(7), 877. https://doi.org/10.3390/pathogens10070877







