Detection of Prions in Brain Homogenates and CSF Samples Using a Second-Generation RT-QuIC Assay: A Useful Tool for Retrospective Analysis of Archived Samples
Abstract
1. Introduction
2. Results
2.1. Establishment of the Second-Generation RT-QuIC Assay
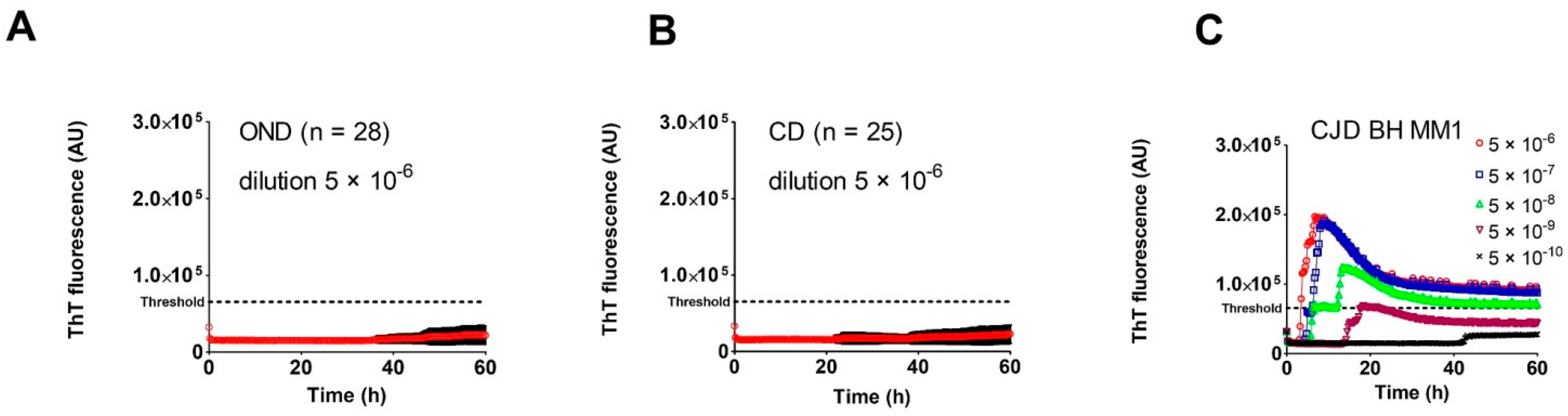
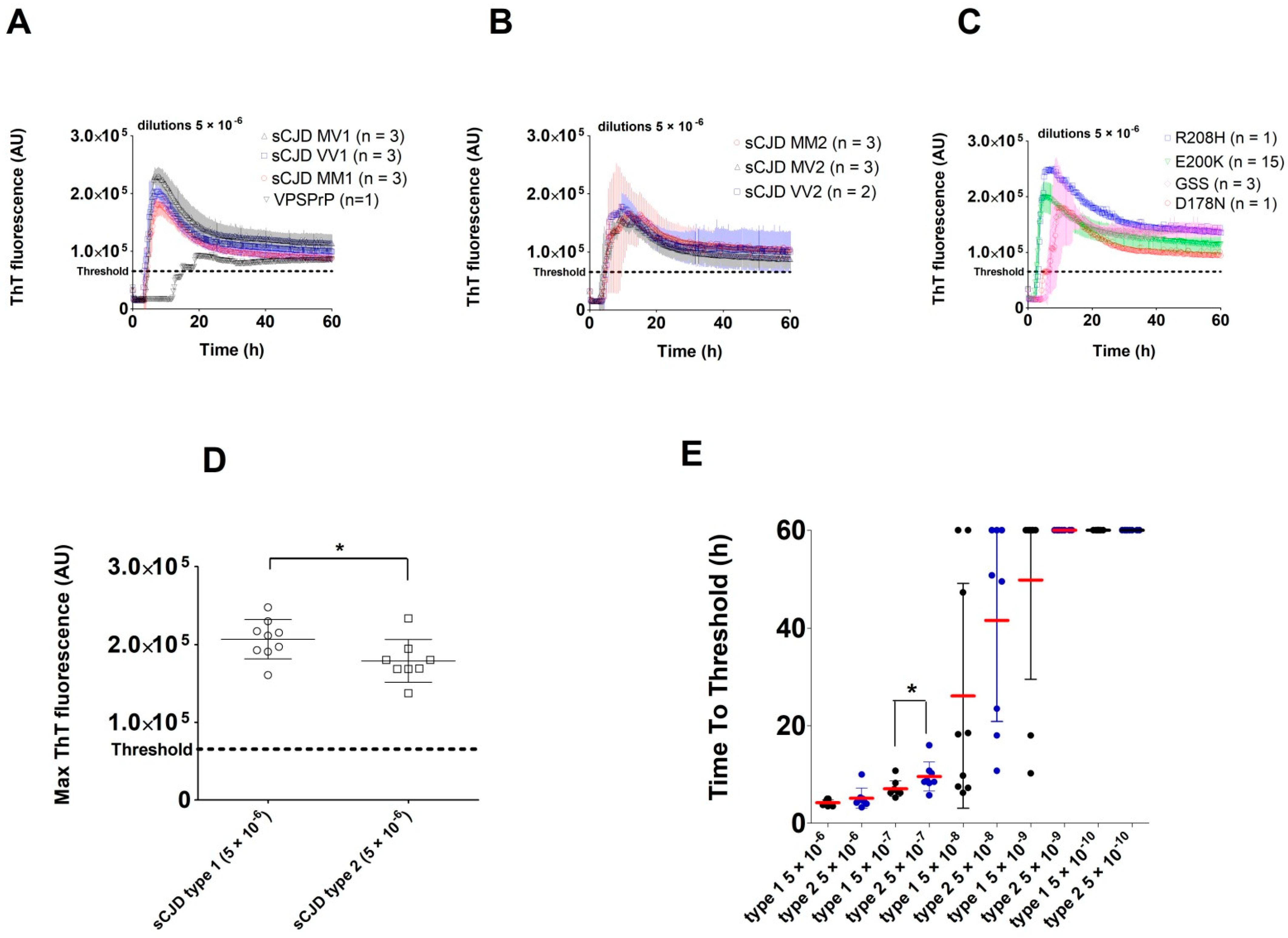
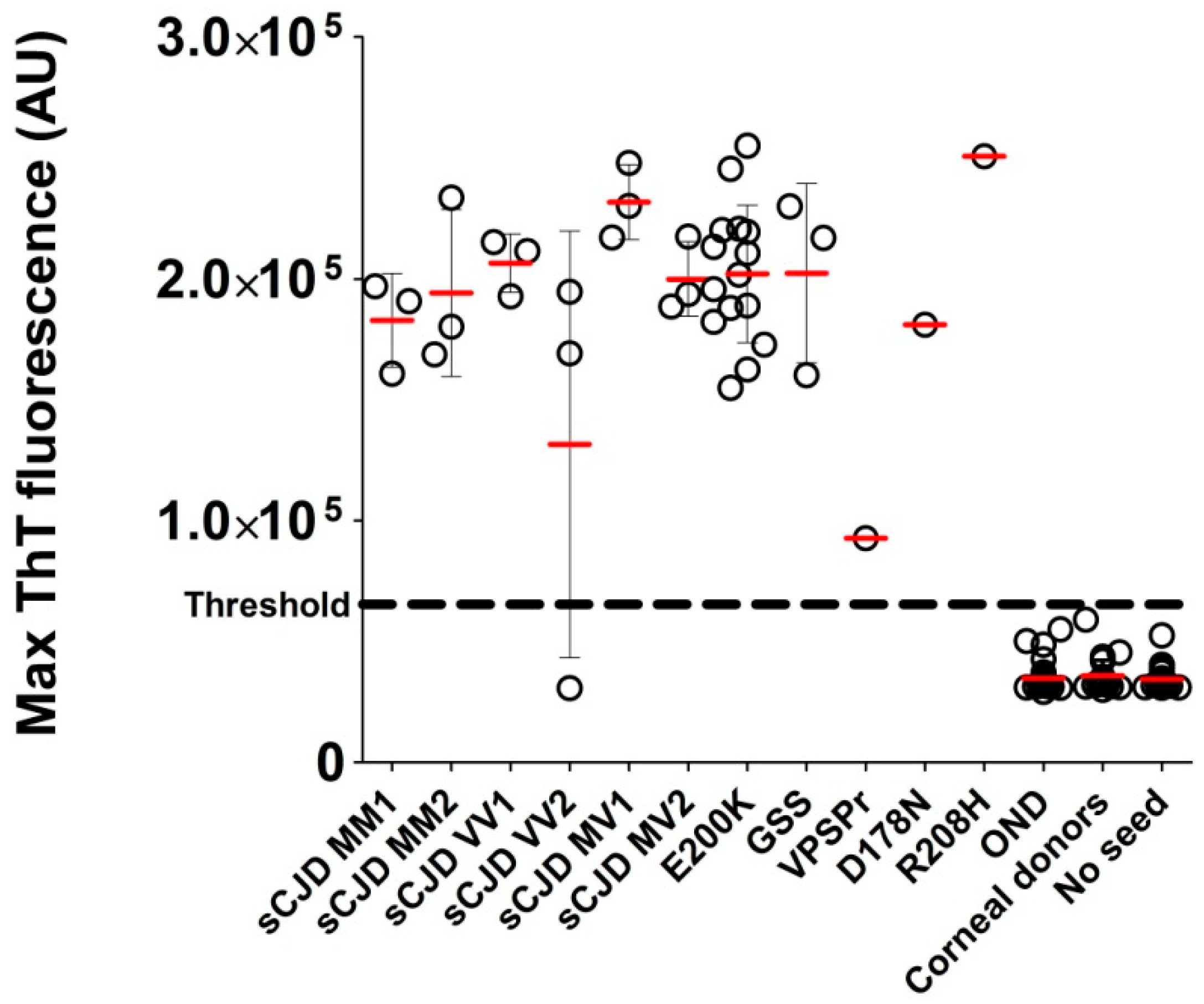
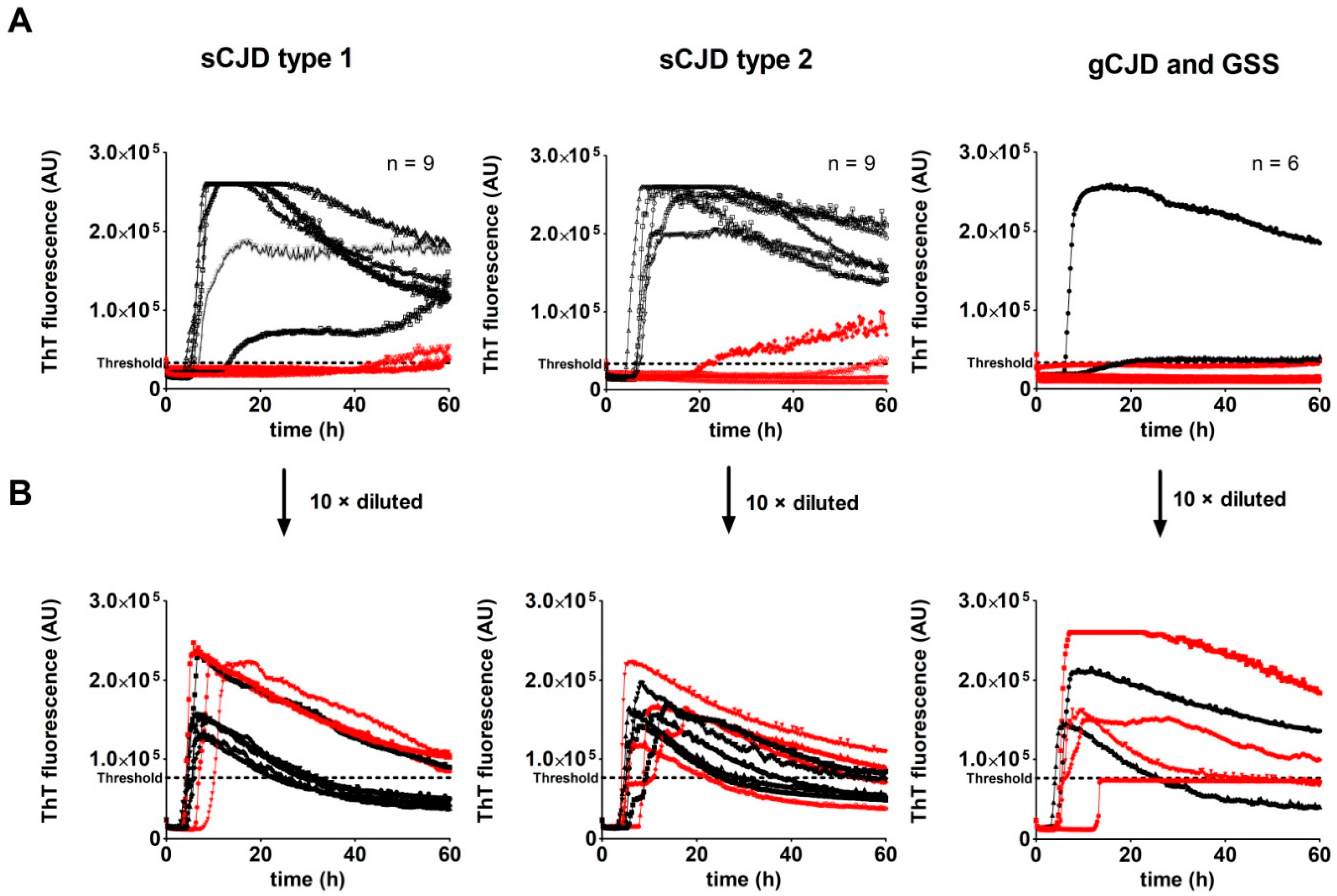
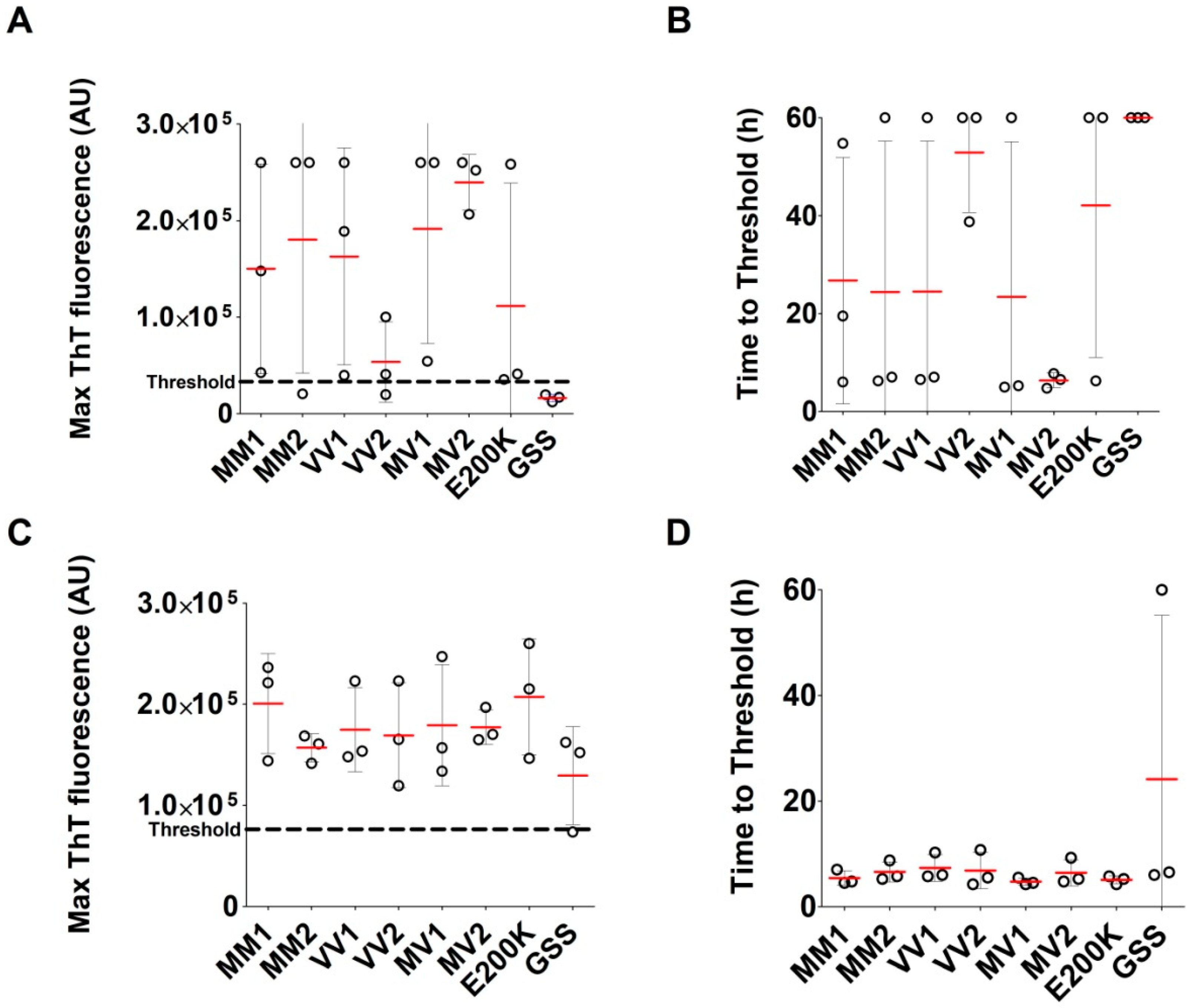
2.2. Analysis of Archived Brain Homogenates Using RT-QuIC
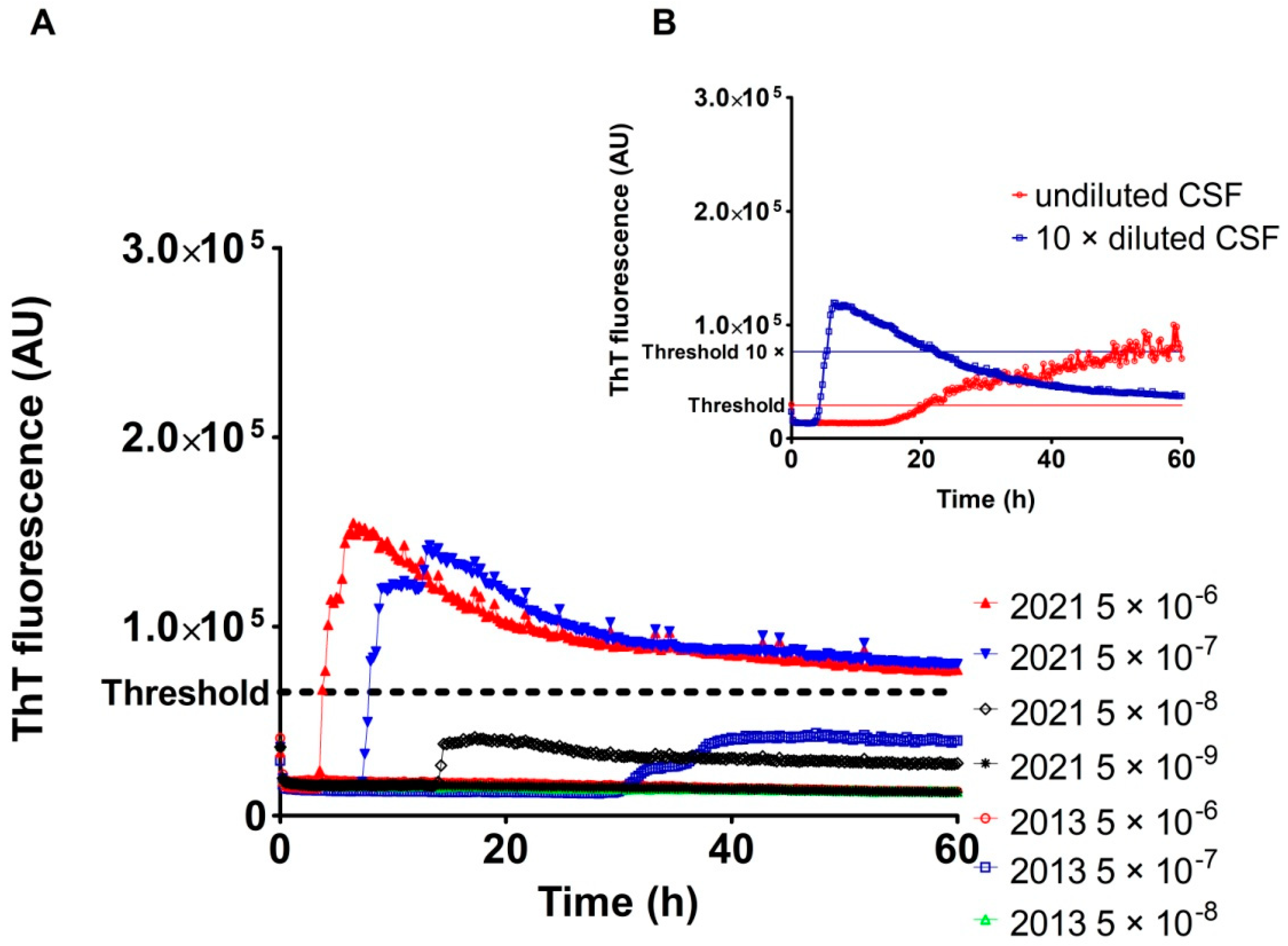
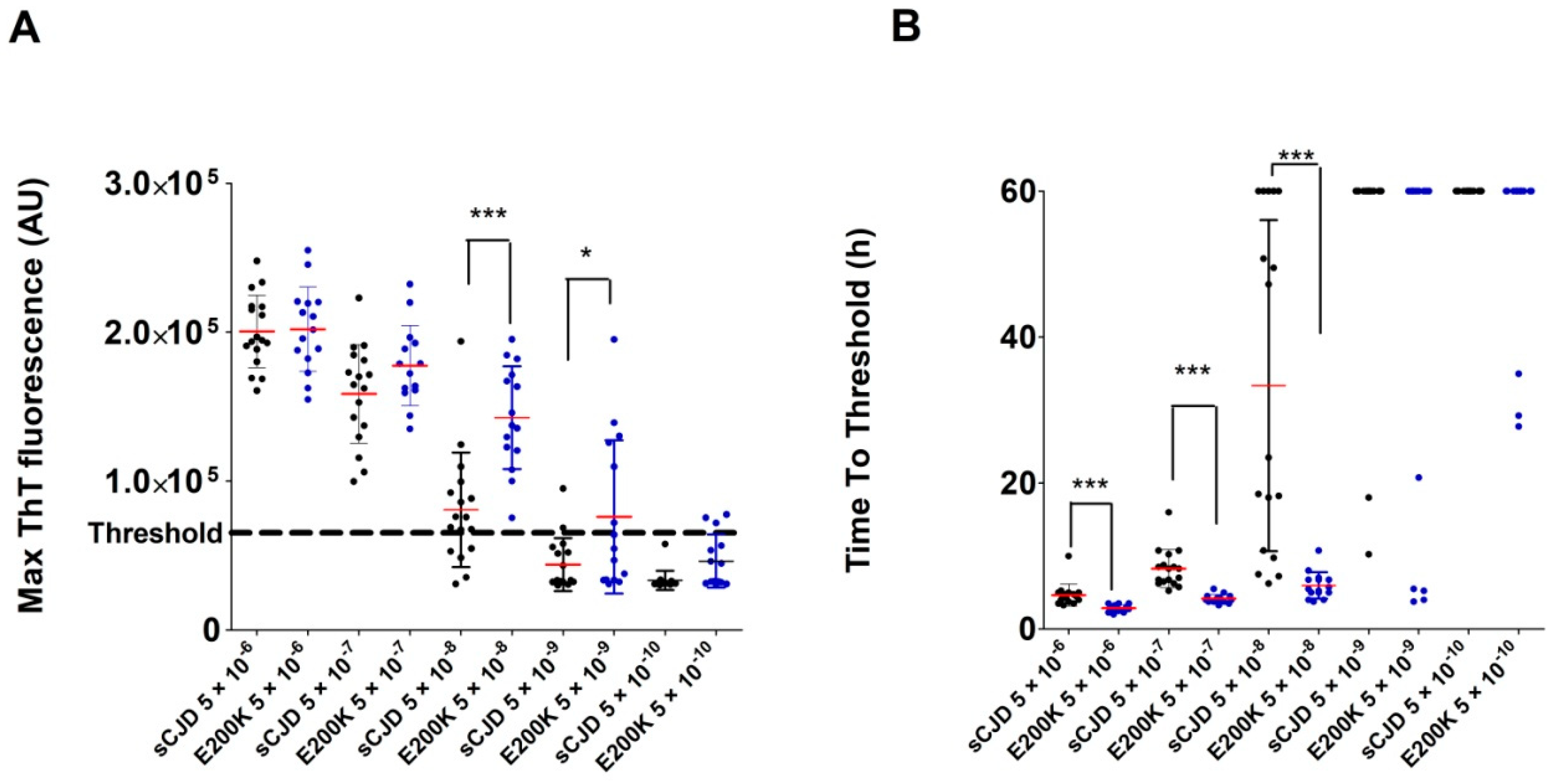
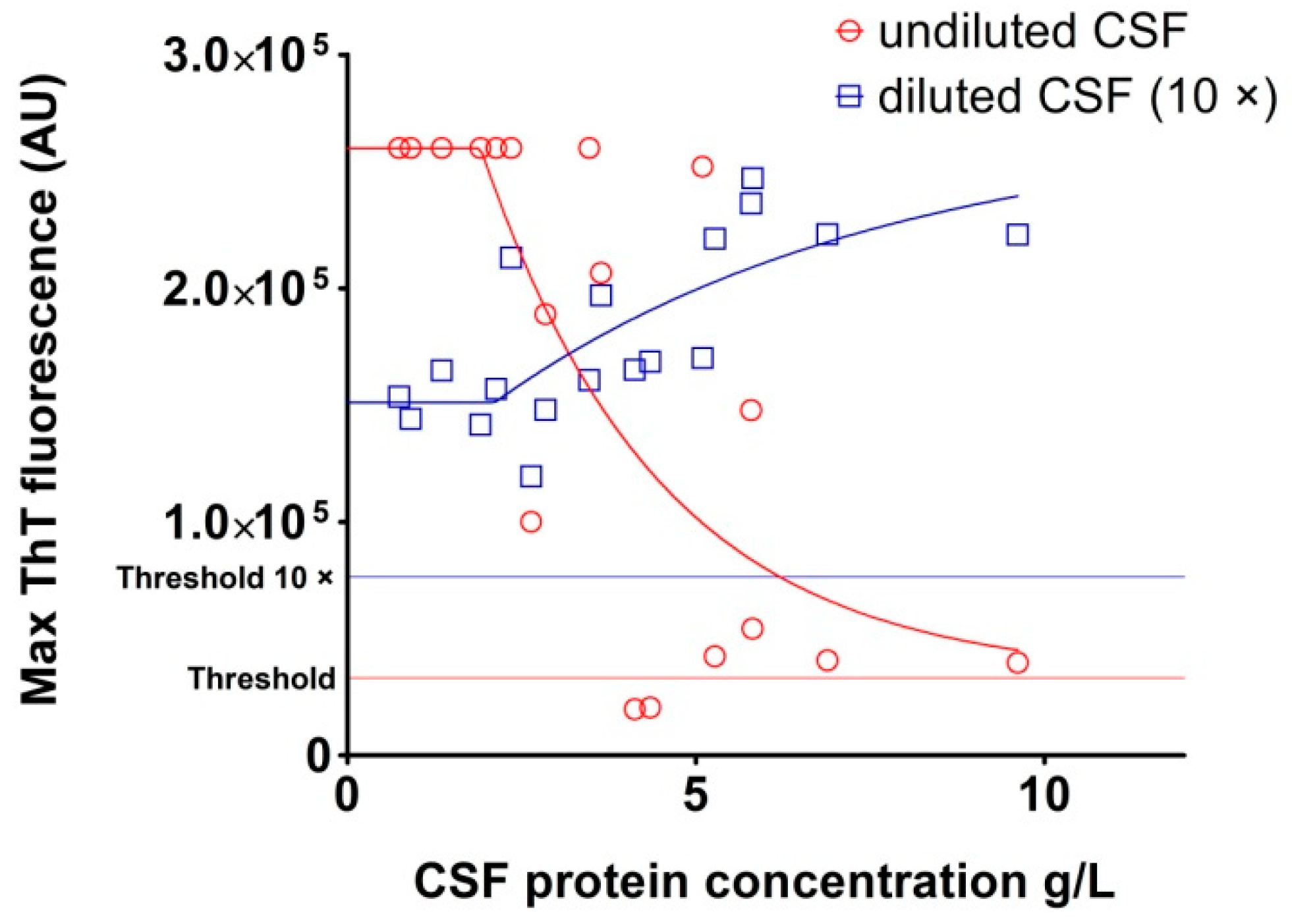
2.3. Analysis of Archived CSF Samples with the RT-QuIC Assay
3. Discussion
4. Materials and Methods
4.1. Ethics Statement
4.2. Prion Disease Samples
4.3. Production of Recombinant Prion Protein for the RT-QuIC Assay
4.4. Second-Generation RT-QuIC
4.5. CSF Protein Concentration Measurement
4.6. Data Analysis
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Goldfarb, L.G.; Brown, P. The transmissible spongiform encephalopathies. Annu. Rev. Med. 1995, 46, 57–65. [Google Scholar] [CrossRef] [PubMed]
- Stahl, N.; Prusiner, S.B. Prions and prion proteins. FASEB J. 1991, 5, 2799–2807. [Google Scholar] [CrossRef]
- Hermann, P.; Appleby, B.; Brandel, J.-P.; Caughey, B.; Collins, S.; Geschwind, M.D.; Green, A.; Haïk, S.; Kovacs, G.G.; Ladogana, A.; et al. Biomarkers and diagnostic guidelines for sporadic Creutzfeldt-Jakob disease. Lancet Neurol. 2021, 20, 235–246. [Google Scholar] [CrossRef]
- Saborio, G.P.; Permanne, B.; Soto, C. Sensitive detection of pathological prion protein by cyclic amplification of protein misfolding. Nature 2001, 411, 810–813. [Google Scholar] [CrossRef] [PubMed]
- Atarashi, R.; Sano, K.; Satoh, K.; Nishida, N. Real-time quaking-induced conversion: A highly sensitive assay for prion detection. Prion 2011, 5, 150–153. [Google Scholar] [CrossRef] [PubMed]
- Atarashi, R.; Satoh, K.; Sano, K.; Fuse, T.; Yamaguchi, N.; Ishibashi, D.; Matsubara, T.; Nakagaki, T.; Yamanaka, H.; Shirabe, S.; et al. Ultrasensitive human prion detection in cerebrospinal fluid by real-time quaking-induced conversion. Nat. Med. 2011, 17, 175–178. [Google Scholar] [CrossRef] [PubMed]
- Wilham, J.M.; Orrú, C.D.; Bessen, R.A.; Atarashi, R.; Sano, K.; Race, B.; Meade-White, K.D.; Taubner, L.M.; Timmes, A.; Caughey, B. Rapid end-point quantitation of prion seeding activity with sensitivity comparable to bioassays. PLoS Pathog. 2010, 6, e1001217. [Google Scholar] [CrossRef] [PubMed]
- Groveman, B.R.; Orrú, C.D.; Hughson, A.G.; Bongianni, M.; Fiorini, M.; Imperiale, D.; Ladogana, A.; Pocchiari, M.; Zanusso, G.; Caughey, B. Extended and direct evaluation of RT-QuIC assays for Creutzfeldt-Jakob disease diagnosis. Ann. Clin. Transl. Neurol. 2017, 4, 139–144. [Google Scholar] [CrossRef]
- Orrú, C.D.; Groveman, B.R.; Hughson, A.G.; Zanusso, G.; Coulthart, M.B.; Caughey, B. Rapid and sensitive RT-QuIC detection of human Creutzfeldt-Jakob disease using cerebrospinal fluid. mBio 2015, 6. [Google Scholar] [CrossRef]
- Foutz, A.; Appleby, B.S.; Hamlin, C.; Liu, X.; Yang, S.; Cohen, Y.; Chen, W.; Blevins, J.; Fausett, C.; Wang, H.; et al. Diagnostic and prognostic value of human prion detection in cerebrospinal fluid. Ann. Neurol. 2017, 81, 79–92. [Google Scholar] [CrossRef]
- Franceschini, A.; Baiardi, S.; Hughson, A.G.; McKenzie, N.; Moda, F.; Rossi, M.; Capellari, S.; Green, A.; Giaccone, G.; Caughey, B.; et al. High diagnostic value of second generation CSF RT-QuIC across the wide spectrum of CJD prions. Sci. Rep. 2017, 7, 10655. [Google Scholar] [CrossRef]
- Dong, T.T.; Satoh, K. The Latest Research on RT-QuIC Assays-A Literature Review. Pathogens 2021, 10, 305. [Google Scholar] [CrossRef] [PubMed]
- Green, A.J.E. RT-QuIC: A new test for sporadic CJD. Pract. Neurol. 2019, 19, 49–55. [Google Scholar] [CrossRef] [PubMed]
- Sano, K.; Satoh, K.; Atarashi, R.; Takashima, H.; Iwasaki, Y.; Yoshida, M.; Sanjo, N.; Murai, H.; Mizusawa, H.; Schmitz, M.; et al. Early detection of abnormal prion protein in genetic human prion diseases now possible using real-time QUIC assay. PLoS ONE 2013, 8, e54915. [Google Scholar] [CrossRef]
- Park, J.H.; Choi, Y.-G.; Lee, Y.-J.; Park, S.-J.; Choi, H.-S.; Choi, K.-C.; Choi, E.-K.; Kim, Y.-S. Real-Time Quaking-Induced Conversion Analysis for the Diagnosis of Sporadic Creutzfeldt-Jakob Disease in Korea. J. Clin. Neurol. 2016, 12, 101–106. [Google Scholar] [CrossRef]
- Bongianni, M.; Orrù, C.; Groveman, B.R.; Sacchetto, L.; Fiorini, M.; Tonoli, G.; Triva, G.; Capaldi, S.; Testi, S.; Ferrari, S.; et al. Diagnosis of Human Prion Disease Using Real-Time Quaking-Induced Conversion Testing of Olfactory Mucosa and Cerebrospinal Fluid Samples. JAMA Neurol. 2017, 74, 155–162. [Google Scholar] [CrossRef]
- Orrú, C.D.; Groveman, B.R.; Foutz, A.; Bongianni, M.; Cardone, F.; McKenzie, N.; Culeux, A.; Poleggi, A.; Grznarova, K.; Perra, D.; et al. Ring trial of 2nd generation RT-QuIC diagnostic tests for sporadic CJD. Ann. Clin. Transl. Neurol. 2020, 7, 2262–2271. [Google Scholar] [CrossRef]
- Rhoads, D.D.; Wrona, A.; Foutz, A.; Blevins, J.; Glisic, K.; Person, M.; Maddox, R.A.; Belay, E.D.; Schonberger, L.B.; Tatsuoka, C.; et al. Diagnosis of prion diseases by RT-QuIC results in improved surveillance. Neurology 2020, 95, e1017–e1026. [Google Scholar] [CrossRef] [PubMed]
- Barbosa, B.; Castrillo, B.B.; Alvim, R.P.; de Brito, M.H.; Gomes, H.R.; Brucki, S.M.; Smid, J.; Nitrini, R.; Landemberger, M.C.; Martins, V.R.; et al. Second-Generation RT-QuIC Assay for the Diagnosis of Creutzfeldt-Jakob Disease Patients in Brazil. Front. Bioeng. Biotechnol. 2020, 8, 929. [Google Scholar] [CrossRef]
- Xiao, K.; Yang, X.H.; Zou, W.Q.; Dong, X.P.; Qi, S.H. Assessment of the Sensitivity and Specificity of the Established Real-time Quaking-induced Conversion (RT-QuIC) Technique in Chinese CJD Surveillance. Biomed. Environ. Sci. 2020, 33, 620–622. [Google Scholar]
- Xiao, K.; Shi, Q.; Zhou, W.; Dong, X.P. Different post-mortem brain regions from three Chinese FFI patients induce different reactive profiles both in the first and second generation RT-QuIC assays. Prion 2020, 14, 163–169. [Google Scholar] [CrossRef] [PubMed]
- Kolářová, K.; Marešová, M.; Manďáková, Z.; Kynčl, J. Prion diseases with a focus on Creutzfeldt-Jakob disease, a summary of the incidence of Creutzfeldt-Jakob disease in the Czech Republic over the last 17 years, 2000–2017. Epidemiol. Mikrobiol. Imunol. 2018, 67, 155–160. [Google Scholar] [PubMed]
- Rohan, Z.; Rusina, R.; Marešová, M.; Matěj, R. Human prion diseases in the Czech Republic. Epidemiol. Mikrobiol. Imunol. 2015, 64, 115–120. [Google Scholar] [PubMed]
- Janoutová, J.; Siváková, L.; Máslová, L.; Hozák, A.; Vařechová, K.; Janout, V. A report of 10 cases of familial Creutzfeld-Jakob disease. Epidemiol. Mikrobiol. Imunol. 2016, 65, 145–148. [Google Scholar]
- Cramm, M.; Schmitz, M.; Karch, A.; Zafar, S.; Varges, D.; Mitrova, E.; Schroeder, B.; Raeber, A.; Kuhn, F.; Zerr, I. Characteristic CSF prion seeding efficiency in humans with prion diseases. Mol. Neurobiol. 2015, 51, 396–405. [Google Scholar] [CrossRef] [PubMed]
- Tesar, A.; Matej, R.; Kukal, J.; Johanidesova, S.; Rektorova, I.; Vyhnalek, M.; Keller, J.; Eliasova, I.; Parobkova, E.; Smetakova, M.; et al. Clinical Variability in P102L Gerstmann-Sträussler-Scheinker Syndrome. Ann. Neurol. 2019, 86, 643–652. [Google Scholar] [CrossRef]
- Green, A.; Sanchez-Juan, P.; Ladogana, A.; Cuadrado-Corrales, N.; Sánchez-Valle, R.; Mitrová, E.; Stoeck, K.; Sklaviadis, T.; Kulczycki, J.; Heinemann, U.; et al. CSF analysis in patients with sporadic CJD and other transmissible spongiform encephalopathies. Eur. J. Neurol. 2007, 14, 121–124. [Google Scholar] [CrossRef]
- Fiorini, M.; Iselle, G.; Perra, D.; Bongianni, M.; Capaldi, S.; Sacchetto, L.; Ferrari, S.; Mombello, A.; Vascellari, S.; Testi, S.; et al. High Diagnostic Accuracy of RT-QuIC Assay in a Prospective Study of Patients with Suspected sCJD. Int. J. Mol. Sci. 2020, 21, 880. [Google Scholar] [CrossRef]
- Mok, T.H.; Nihat, A.; Luk, C.; Sequeira, D.; Batchelor, M.; Mead, S.; Collinge, J.; Jackson, G.S. Bank vole prion protein extends the use of RT-QuIC assays to detect prions in a range of inherited prion diseases. Sci. Rep. 2021, 11, 5231. [Google Scholar] [CrossRef]
- Panigaj, M.; Glier, H.; Wildová, M.; Holada, K. Expression of prion protein in mouse erythroid progenitors and differentiating murine erythroleukemia cells. PLoS ONE 2011, 6, e24599. [Google Scholar] [CrossRef]
| Group | Type | n (BH)/(CSF) | AST (Years ± sd) | Mean Age (Years) | t-tau (Pos./Tested) | 14-3-3 (Pos./Tested) |
|---|---|---|---|---|---|---|
| Sporadic TSE | MM1 | 3/3 | 2 ± 0 | 71 | 3/3 (100%) | 2/3 (66.7%) |
| MM2 | 3/3 | 6.7 ± 0.6 | 65 | 1/1 (100%) | 2/3 (66.7%) | |
| MV1 | 3/3 | 3.3 ± 0.6 | 63 | 2/2 (100%) | 1/2 (50%) | |
| MV2 | 3/3 | 4.3 ± 2.5 | 70 | 3/3 (100%) | 2/3 (66.7%) | |
| VV1 | 3/3 | 2 ± 0.8 | 64 | 3/3 (100%) | 2/3 (66.7%) | |
| VV2 | 3/3 | 6.3 ± 3.6 | 64 | 2/2 (100%) | 2/2 (100%) | |
| VPSPr (VV) | 1/0 | 2 ± 0 | 73 | NT | NT | |
| Genetic TSE | E200K | 15/3 | 9 ± 3.5 | 61 | 7/8 (87.5%) | 6/12 (50%) |
| GSS | 3/3 | 8 ± 3 | 48 | 2/2 (100%) | 0/2 (0%) | |
| D178N (MV) | 1/0 | 13 ± 0 | 59 | NT | 1/1 (100%) | |
| R208H (VV) | 1/0 | 10 ± 0 | 62 | 1/1 (100%) | 1/1 (100%) | |
| OND | FTLD-tau | 5/5 | 3.2 ± 2.2 | 71 | 0/3 (0%) | 0/3 (0%) |
| FTLD-TDP | 5/5 | 3.6 ± 1.7 | 64 | 0/2 (0%) | 0/3 (0%) | |
| AD | 5/5 | 4.6 ± 2.3 | 70 | NT | 1/2 (50%) | |
| AD + others | 4/4 | 2 ± 0 | 79 | 1/1 (100%) | NT | |
| Huntington dis. | 4/5 | 5.8 ± 2.2 | 72 | 0/1 (0%) | NT | |
| PD | 5/5 | 6 ± 2.9 | 70 | 1/1 (100%) | NT | |
| Healthy corneal donors | - | 25/0 | - | - | NT | NT |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Moško, T.; Galušková, S.; Matěj, R.; Brůžová, M.; Holada, K. Detection of Prions in Brain Homogenates and CSF Samples Using a Second-Generation RT-QuIC Assay: A Useful Tool for Retrospective Analysis of Archived Samples. Pathogens 2021, 10, 750. https://doi.org/10.3390/pathogens10060750
Moško T, Galušková S, Matěj R, Brůžová M, Holada K. Detection of Prions in Brain Homogenates and CSF Samples Using a Second-Generation RT-QuIC Assay: A Useful Tool for Retrospective Analysis of Archived Samples. Pathogens. 2021; 10(6):750. https://doi.org/10.3390/pathogens10060750
Chicago/Turabian StyleMoško, Tibor, Soňa Galušková, Radoslav Matěj, Magdalena Brůžová, and Karel Holada. 2021. "Detection of Prions in Brain Homogenates and CSF Samples Using a Second-Generation RT-QuIC Assay: A Useful Tool for Retrospective Analysis of Archived Samples" Pathogens 10, no. 6: 750. https://doi.org/10.3390/pathogens10060750
APA StyleMoško, T., Galušková, S., Matěj, R., Brůžová, M., & Holada, K. (2021). Detection of Prions in Brain Homogenates and CSF Samples Using a Second-Generation RT-QuIC Assay: A Useful Tool for Retrospective Analysis of Archived Samples. Pathogens, 10(6), 750. https://doi.org/10.3390/pathogens10060750






