Genetic and Transcriptomic Variations for Amoxicillin Resistance in Helicobacter pylori under Cryopreservation
Abstract
1. Introduction
2. Materials and Methods
2.1. Bacterial Strains and Culture Conditions
2.2. Selection of Unstable Amoxicillin-Resistant H. pylori Strains In Vitro
2.3. Polymerase Chain Reaction (PCR) and DNA Sequence Analysis
2.4. RNA Extraction and Library Construction
2.5. RNA Sequencing and Data Pretreatment
2.6. Differential Expression Analysis and Functional Analysis
2.7. Quantitative Real-Time PCR
3. Results
3.1. Mutation of PBP1, β-lactamase, Efflux Pump and Membrane Protein in Amoxicillin Resistant H. pylori NX24r
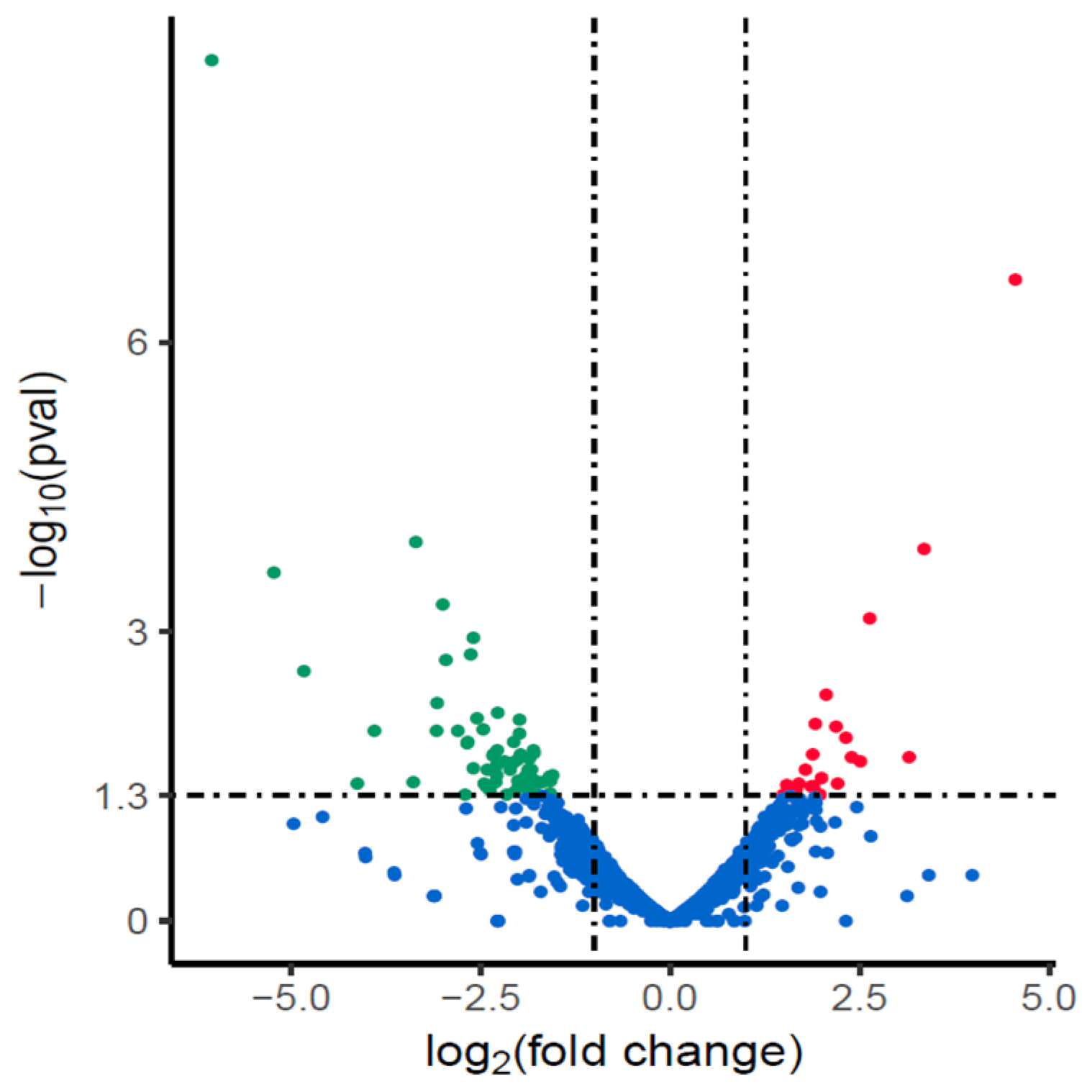
3.2. Changes of Transcription in NX24r after Amoxicillin Screening
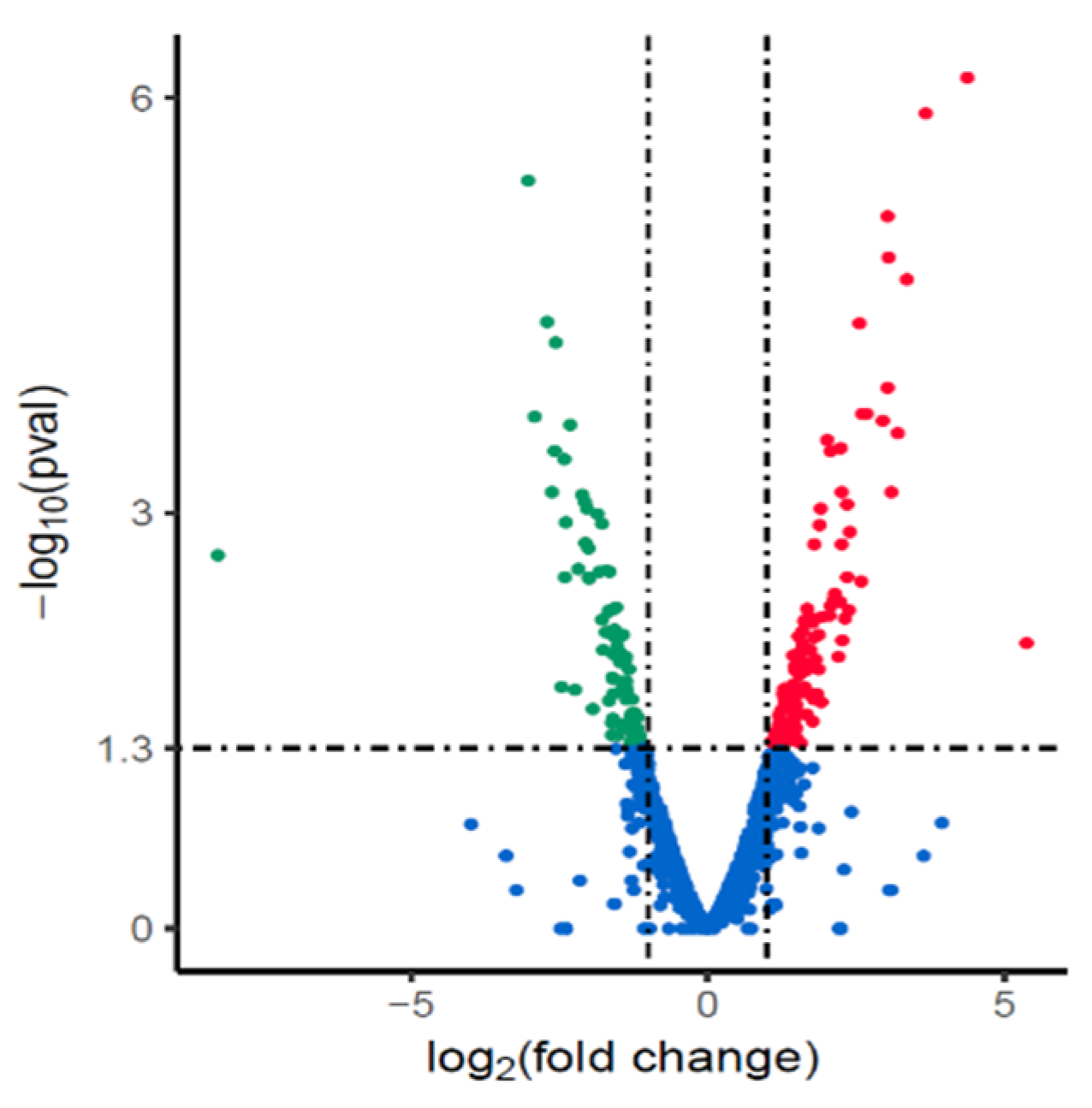
3.3. Changes in Gene Expression of Cryopreserved H. pylori Isolate NX24f Compared with NX24r
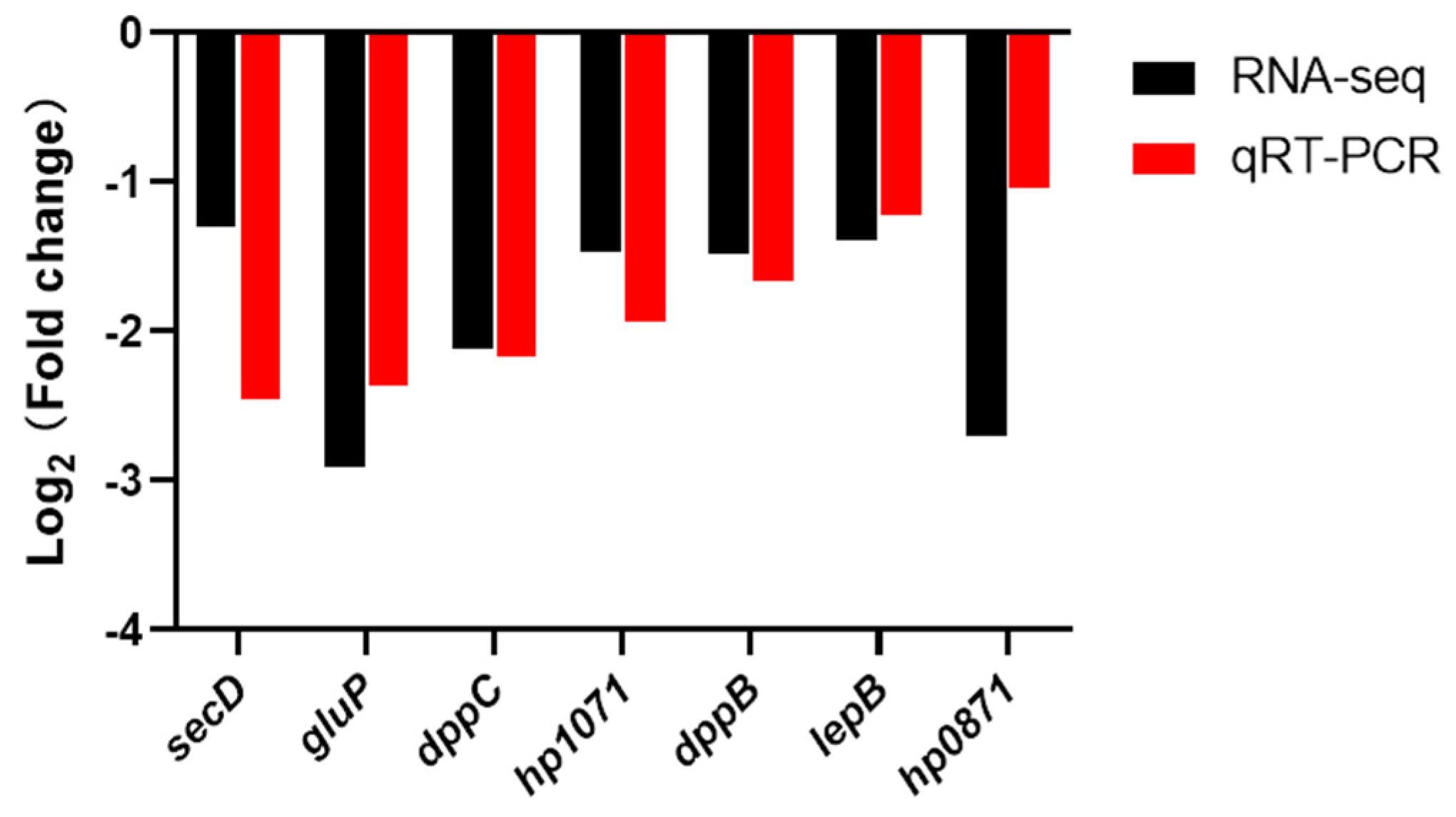
3.4. Verifying the Expression Level of Plasma Membrane Genes by qRT-PCR
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Tai, W.C.; Liang, C.M.; Kuo, C.M.; Huang, P.Y.; Wu, C.K.; Yang, S.C.; Kuo, Y.H.; Lin, M.T.; Lee, C.H.; Hsu, C.N.; et al. A 14 day esomeprazole- and amoxicillin-containing high-dose dual therapy regimen achieves a high eradication rate as first-line anti-Helicobacter pylori treatment in Taiwan: A prospective randomized trial. J. Antimicrob. Chemother. 2019, 74, 1718–1724. [Google Scholar] [CrossRef]
- Chen, M.J.; Wu, M.S.; Chen, C.C.; Chen, C.C.; Fang, Y.J.; Bair, M.J.; Chang, C.Y.; Lee, J.Y.; Hsu, W.F.; Luo, J.C.; et al. Impact of amoxicillin resistance on the efficacy of amoxicillin-containing regimens for Helicobacter pylori eradication: Analysis of five randomized trials. J. Antimicrob. Chemother. 2017, 72, 3481–3489. [Google Scholar] [CrossRef]
- Nishizawa, T.; Suzuki, H.; Tsugawa, H.; Muraoka, H.; Matsuzaki, J.; Hirata, K.; Ikeda, F.; Takahashi, M.; Hibi, T. Enhancement of amoxicillin resistance after unsuccessful Helicobacter pylori eradication. Antimicrob. Agents Chemother. 2011, 55, 3012–3014. [Google Scholar] [CrossRef] [PubMed]
- Shu, X.; Yin, G.; Liu, M.; Peng, K.; Zhao, H.; Jiang, M. Antibiotics resistance of Helicobacter pylori in children with upper gastrointestinal symptoms in Hangzhou, China. Helicobacter 2018, 23, e12481. [Google Scholar] [CrossRef] [PubMed]
- Shinozaki, S.; Nomoto, H.; Kondo, Y.; Sakamoto, H.; Hayashi, Y.; Yamamoto, H.; Lefor, A.K.; Osawa, H. Comparison of vonoprazan and proton pump inhibitors for eradication of Helicobacter pylori. Kaohsiung J. Med. Sci. 2016, 32, 255–260. [Google Scholar] [CrossRef]
- Siddiqui, T.R.; Ahmed, W.; Arif, A.; Bibi, S.; Khan, A. Emerging trends of antimicrobial resistance in Helicobacter pylori isolates obtained from Pakistani patients: The need for consideration of amoxicillin and clarithromycin. J. Pak. Med. Assoc. 2016, 66, 710–716. [Google Scholar]
- Macias-Garcia, F.; Llovo-Taboada, J.; Diaz-Lopez, M.; Baston-Rey, I.; Dominguez-Munoz, J.E. High primary antibiotic resistance of Helicobacter pylori strains isolated from dyspeptic patients: A prevalence cross-sectional study in Spain. Helicobacter 2017, 22, 10. [Google Scholar] [CrossRef]
- Han, S.R.; Bhakdi, S.; Maeurer, M.J.; Schneider, T.; Gehring, S. Stable and unstable amoxicillin resistance in Helicobacter pylori: Should antibiotic resistance testing be performed prior to eradication therapy? J. Clin. Microbiol. 1999, 37, 2740–2741. [Google Scholar] [CrossRef]
- Zhang, Y.Y.; He, L.H.; You, Y.H.; Sun, L.; Han, X.R.; Gong, Y.N.; Zhang, J.Z. The effect of cryopreservation on amoxicillin resistance phenotype of Helicobacter pylori. J. Pathog. Biol. 2019, 14, 497–500. (In Chinese) [Google Scholar]
- Tseng, Y.S.; Wu, D.C.; Chang, C.Y.; Kuo, C.H.; Yang, Y.C.; Jan, C.M.; Su, Y.C.; Kuo, F.C.; Chang, L.L. Amoxicillin resistance with beta-lactamase production in Helicobacter pylori. Eur J. Clin. Invest 2009, 39, 807–812. [Google Scholar] [CrossRef]
- Kwon, Y.H.; Kim, J.Y.; Kim, N.; Park, J.H.; Nam, R.H.; Lee, S.M.; Kim, J.W.; Kim, J.M.; Park, J.Y.; Lee, D.H. Specific mutations of penicillin-binding protein 1A in 77 clinically acquired amoxicillin-resistant Helicobacter pylori strains in comparison with 77 amoxicillin-susceptible strains. Helicobacter 2017, 22. [Google Scholar] [CrossRef]
- Yonezawa, H.; Osaki, T.; Hojo, F.; Kamiya, S. Effect of Helicobacter pylori biofilm formation on susceptibility to amoxicillin, metronidazole and clarithromycin. Microb. Pathog. 2019, 132, 100–108. [Google Scholar] [CrossRef]
- Attaran, B.; Falsafi, T. Ghorbanmehr, N. Effect of biofilm formation by clinical isolates of Helicobacter pylori on the efflux-mediated resistance to commonly used antibiotics. World J. Gastroenterol. 2017, 23, 1163–1170. [Google Scholar] [CrossRef]
- Ge, X.; Cai, Y.; Chen, Z.; Gao, S.; Geng, X.; Li, Y.; Li, Y.; Jia, J.; Sun, Y. Bifunctional Enzyme SpoT Is Involved in Biofilm Formation of Helicobacter pylori with Multidrug Resistance by Upregulating Efflux Pump Hp1174 (gluP). Antimicrob Agents Chemother. 2018, 62, e00957-18. [Google Scholar] [CrossRef]
- Poole, K. Resistance to beta-lactam antibiotics. Cell Mol. Life Sci. 2004, 61, 2200–2223. [Google Scholar]
- DeLoney, C.R. & Schiller, N.L. Characterization of an In vitro-selected amoxicillin-resistant strain of Helicobacter pylori. Antimicrob. Agents Chemother. 2000, 44, 3368–3373. [Google Scholar]
- Dore, M.P.; Graham, D.Y.; Sepulveda, A.R. Different penicillin-binding protein profiles in amoxicillin-resistant Helicobacter pylori. Helicobacter 1999, 4, 154–161. [Google Scholar] [CrossRef]
- Kwon, D.H.; Dore, M.P.; Kim, J.J.; Kato, M.; Lee, M.; Wu, J.Y.; Graham, D.Y. High-level beta-lactam resistance associated with acquired multidrug resistance in Helicobacter pylori. Antimicrob. Agents Chemother. 2003, 47, 2169–2178. [Google Scholar] [CrossRef]
- Kim, B.J.; Kim, J.G. Substitutions in penicillin-binding protein 1 in amoxicillin-resistant Helicobacter pylori strains isolated from Korean patients. Gut Liver 2013, 7, 655–660. [Google Scholar] [CrossRef]
- Gerrits, M.M.; Godoy, A.P.; Kuipers, E.J.; Ribeiro, M.L.; Stoof, J.; Mendonca, S.; van Vliet, A.H.; Pedrazzoli, J., Jr.; Kusters, J.G. Multiple mutations in or adjacent to the conserved penicillin-binding protein motifs of the penicillin-binding protein 1A confer amoxicillin resistance to Helicobacter pylori. Helicobacter 2006, 11, 181–187. [Google Scholar] [CrossRef] [PubMed]
- Bina, J.E.; Alm, R.A.; Uria-Nickelsen, M.; Thomas, S.R.; Trust, T.J.; Hancock, R.E. Helicobacter pylori uptake and efflux: Basis for intrinsic susceptibility to antibiotics in vitro. Antimicrob. Agents Chemother. 2000, 44, 248–254. [Google Scholar] [CrossRef] [PubMed]
- Kutschke, A.; de Jonge, B.L. Compound efflux in Helicobacter pylori. Antimicrob. Agents Chemother. 2005, 49, 3009–3010. [Google Scholar] [CrossRef]
- Co, E.M.; Schiller, N.L. Resistance mechanisms in an in vitro-selected amoxicillin-resistant strain of Helicobacter pylori. Antimicrob. Agents Chemother. 2006, 50, 4174–4176. [Google Scholar] [CrossRef] [PubMed]
- Ohkusa, T.; Miwa, H.; Endo, S.; Okayasu, I.; Sato, N. Helicobacter pylori is a fragile bacteria when stored at low and ultra-low temperatures. J. Gastroenterol. Hepatol. 2004, 19, 200–204. [Google Scholar] [CrossRef]
- Oskouei, D.D.; Bekmen, N.; Ellidokuz, H.; Yilmaz, O. Evaluation of different cryoprotective agents in maintenance of viability of Helicobacter pylori in stock culture media. Braz. J. Microbiol. 2010, 41, 1038–1046. [Google Scholar] [CrossRef]
- Meneghel, J.; Passot, S.; Cenard, S.; Refregiers, M.; Jamme, F.; Fonseca, F. Subcellular membrane fluidity of Lactobacillus delbrueckii subsp. bulgaricus under cold and osmotic stress. Appl. Microbiol. Biotechnol. 2017, 101, 6907–6917. [Google Scholar] [CrossRef]
- Ricke, S.C.; Dawoud, T.M.; Kim, S.A.; Park, S.H.; Kwon, Y.M. Salmonella Cold Stress Response: Mechanisms and Occurrence in Foods. Adv. Appl. Microbiol. 2018, 104, 1–38. [Google Scholar] [PubMed]
- Delcour, A.H. Outer membrane permeability and antibiotic resistance. Biochim. Biophys. Acta. 2009, 1794, 808–816. [Google Scholar] [CrossRef]
- Gautier, J.; Passot, S.; Penicaud, C.; Guillemin, H.; Cenard, S.; Lieben, P.; Fonseca, F. A low membrane lipid phase transition temperature is associated with a high cryotolerance of Lactobacillus delbrueckii subspecies bulgaricus CFL1. J. Dairy Sci. 2013, 96, 5591–5602. [Google Scholar] [CrossRef]
- Huang, Z.; Lv, H.; Ai, Z.; Wang, N.; Xie, X.; Fan, H.; Pan, Z.; Suo, B. Repair mechanism of frozen sublethally damaged Staphylococcus aureus. Acta Microbiologica Sinica. 2015, 55, 1409–1417. (In Chinese) [Google Scholar] [PubMed]
- Geis, G.; Leying, H. Suerbaum, S.; Opferkuch, W. Unusual fatty acid substitution in lipids and lipopolysaccharides of Helicobacter pylori. J. Clin. Microbiol. 1990, 28, 930–932. [Google Scholar] [CrossRef] [PubMed]
- Meneghel, J. Passot, S.; Dupont, S.; Fonseca, F. Biophysical characterization of the Lactobacillus delbrueckii subsp. bulgaricus membrane during cold and osmotic stress and its relevance for cryopreservation. Appl. Microbiol. Biotechnol. 2017, 101, 1427–1441. [Google Scholar] [CrossRef] [PubMed]
- Ghai, I.; Ghai, S. Understanding antibiotic resistance via outer membrane permeability. Infect. Drug Resist. 2018, 11, 523–530. [Google Scholar] [CrossRef]
- Pages, J.M.; James, C.E.; Winterhalter, M. The porin and the permeating antibiotic: A selective diffusion barrier in Gram-negative bacteria. Nat. Rev. Microbiol. 2008, 6, 893–903. [Google Scholar] [CrossRef] [PubMed]
- Eumkeb, G.; Siriwong, S.; Phitaktim, S.; Rojtinnakorn, N.; Sakdarat, S. Synergistic activity and mode of action of flavonoids isolated from smaller galangal and amoxicillin combinations against amoxicillin-resistant Escherichia coli. J. Appl. Microbiol. 2012, 112, 55–64. [Google Scholar] [CrossRef]
- Sawyer, W.S.; Wang, L.; Uehara, T.; Tamrakar, P.; Prathapam, R.; Mostafavi, M.; Metzger, L.E.; Feng, B.; Baxter Rath, C.M. Targeted lipopolysaccharide biosynthetic intermediate analysis with normal-phase liquid chromatography mass spectrometry. PLoS ONE 2019, 14, e0211803. [Google Scholar] [CrossRef]
- Ferrandez, Y.; Condemine, G. Novel mechanism of outer membrane targeting of proteins in Gram-negative bacteria. Mol. Microbiol. 2008, 69, 1349–1357. [Google Scholar] [CrossRef]
- Personne, Y.; Curtis, M.A.; Wareham, D.W.; Waite, R.D. Activity of the type I signal peptidase inhibitor MD3 against multidrug-resistant Gram-negative bacteria alone and in combination with colistin. J. Antimicrob. Chemother. 2014, 69, 3236–3243. [Google Scholar] [CrossRef][Green Version]
- Rapoport, T.A.; Li, L.; Park, E. Structural and Mechanistic Insights into Protein Translocation. Annu. Rev. Cell Dev. Biol. 2017, 33, 369–390. [Google Scholar] [CrossRef] [PubMed]
- Sugai, M.; Wu, H.C. Export of the outer membrane lipoprotein is defective in secD, secE, and secF mutants of Escherichia coli. J. Bacteriol. 1992, 174, 2511–2516. [Google Scholar] [CrossRef]
- Xu, X.; Chen, J.; Huang, X.; Feng, S.; Zhang, X.; She, F.; Wen, Y. The Role of a Dipeptide Transporter in the Virulence of Human Pathogen, Helicobacter pylori. Front. Microbiol. 2021, 12, 633166. [Google Scholar] [CrossRef] [PubMed]
- Rahman, M.M.; Machuca, M.A.; Khan, M.F.; Barlow, C.K.; Schittenhelm, R.B.; Roujeinikova, A. Molecular Basis of Unexpected Specificity of ABC Transporter-Associated Substrate-Binding Protein DppA from Helicobacter pylori. J. Bacteriol. 2019, 201, e00400-19. [Google Scholar] [CrossRef] [PubMed]
- Weinberg, M.V.; Maier, R.J. Peptide transport in Helicobacter pylori: Roles of dpp and opp systems and evidence for additional peptide transporters. J. Bacteriol. 2007, 189, 3392–3402. [Google Scholar] [CrossRef] [PubMed]
- Miftahussurur, M.; Cruz, M.; Subsomwong, P.; Jimenez Abreu, J.A.; Hosking, C.; Nagashima, H.; Akada, J.; Yamaoka, Y. Clarithromycin-Based Triple Therapy is Still Useful as an Initial Treatment for Helicobacter pylori Infection in the Dominican Republic. Am. J. Trop Med. Hyg. 2017, 96, 1050–1059. [Google Scholar] [CrossRef] [PubMed]
- Tomb, J.F.; White, O.; Kerlavage, A.R.; Clayton, R.A.; Sutton, G.G.; Fleischmann, R.D.; Ketchum, K.A.; Klenk, H.P.; Gill, S.; Dougherty, B.A.; et al. The complete genome sequence of the gastric pathogen Helicobacter pylori. Nature 1997, 388, 539–547. [Google Scholar] [CrossRef]
- Limoli, D.H.; Jones, C.J.; Wozniak, D.J. Bacterial Extracellular Polysaccharides in Biofilm Formation and Function. Microbiol. Spectr. 2015, 3. [Google Scholar] [CrossRef]
- Stewart, P.S.; Costerton, J.W. Antibiotic resistance of bacteria in biofilms. Lancet 2001, 358, 135–138. [Google Scholar] [CrossRef]
| Gene | Primer | Sequence (5′-3′) |
|---|---|---|
| pbp1 | pbp1-F | TGCATAAAGGCATTAGACAATCAAG |
| pbp1-R | GCTATCCGCCCTCCTACGGT | |
| TEM | TEM-F | ATAAAATTCTTGAAGACGAAA |
| TEM-R | GACAGTTACCAATGCTTAATCA | |
| hefC | hefC-F | ATGTATAAAACAGCGATTAATCGTCCTATTACGAC |
| hefC-R | TCATTCTAAAGTTTTTTGGTTTTGATAAAACCGCTT | |
| hopC | hopC-F | ATGATAAAGAAAAATAGAACGCTGTTTCTTAGT |
| hopC-R | TTAGAATGAATACCCATAAGACCAATAAACG | |
| hopB | hopB-F | ATGAAACAAAATTTAAAGCCATTCAAAATGAT |
| hopB-R | TTAGAAGGCGTAGCCATAGACC |
| Gene | Primer | Sequence (5′–3′) |
|---|---|---|
| hp1010 | HP1010_F | GCGCGTTAGTCGTTTATGGCGTTT |
| HP1010_R | AGCGCTCAAAGGGTTGTAATTGCC | |
| hp0300 | HP0300_F | CGCTCCTTGGATGCTTGTT |
| HP0300_R | CATGATGCCATCGCCTACC | |
| hp0576 | HP0576_F | GGGAGGGATAAACACCACCA |
| HP0576_R | CGGCATACCCATTCCTAAAA | |
| hp0871 | HP0871_F | GTTTGTACGCATTAGGCACTTCTT |
| HP0871_R | GTTTGAATGGGCATAAATACTTGG | |
| hp1071 | HP1071_F | GTGAGCAATATCCGCTACCCTA |
| HP1071_R | TGCCATAAATCAAATACAACCC | |
| hp1550 | HP1550_F | ATCTGTAAGCATTTCGCCATCT |
| HP1550_R | AAATTAGGCAGCGTGTTGTTGT | |
| hp1174 | HP1174_F | CCGCTGGTAATCCCTTTGTA |
| HP1174_R | CTTGCATTATCGCCCATTTT | |
| hp0299 | HP0299_F | AGGAGATCCGGCGTTAGTGA |
| HP0299_R | GCGTCAATAGCGGCTTGATT |
| Isolates | MIC of Amoxicillin (mg/L) | PBP1 | HefC | HopB | HopC |
|---|---|---|---|---|---|
| NX24 initial | <0.016 | ||||
| NX24r | 256 | I370T, E428K, T556S | M337K, L378F, D976V | / | / |
| NX24f | 5 | I370T, E428K, T556S | M337K, L378F, D976V | / | / |
| Gene ID | log2FC | Gene Name | Description |
|---|---|---|---|
| HP1174 | −2.915 | gluP | glucose/galactose transporter |
| HP0871 | −2.703 | hp0871 | CDP-diacylglycerol pyrophosphatase |
| HP1168 | −2.625 | cstA | carbon starvation protein |
| HP0300 | −2.121 | dppC | dipeptide ABC transporter permease |
| HP0226 | −2.034 | hp0226 | membrane protein |
| HP0942 | −1.823 | hp0942 | D-alanine/glycine permease |
| HP0724 | −1.575 | hp0724 | anaerobic C4-dicarboxylate transporter |
| HP1069 | −1.557 | ftsH | cell division protein |
| HP0770 | −1.506 | flhB | flagellar biosynthesis protein |
| HP0299 | −1.485 | dppB | dipeptide ABC transporter permease |
| HP1071 | −1.461 | hp1071 | CDP-diacylglycerol--serine O-phosphatidyltransferase |
| HP0576 | −1.384 | lepB | signal peptidase I |
| HP1571 | −1.369 | rlpA | rare lipoprotein A |
| HP0943 | −1.326 | dadA | D-amino acid dehydrogenase |
| HP0888 | −1.296 | hp0888 | iron chelatin transport ATP-binding protein |
| HP1270 | −1.295 | hp1270 | NADH-quinone oxidoreductase subunit K |
| HP1550 | −1.295 | secD | preprotein translocase subunit |
| HP1041 | −1.233 | flhA | flagellar biosynthesis protein |
| HP0791 | −1.137 | hp0791 | cadmium, zinc and cobalt-transporting ATPase |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Han, X.; Zhang, Y.; He, L.; Fan, R.; Sun, L.; Fan, D.; Gong, Y.; Chen, X.; You, Y.; Zhao, F.; et al. Genetic and Transcriptomic Variations for Amoxicillin Resistance in Helicobacter pylori under Cryopreservation. Pathogens 2021, 10, 676. https://doi.org/10.3390/pathogens10060676
Han X, Zhang Y, He L, Fan R, Sun L, Fan D, Gong Y, Chen X, You Y, Zhao F, et al. Genetic and Transcriptomic Variations for Amoxicillin Resistance in Helicobacter pylori under Cryopreservation. Pathogens. 2021; 10(6):676. https://doi.org/10.3390/pathogens10060676
Chicago/Turabian StyleHan, Xiurui, Yiyao Zhang, Lihua He, Ruyue Fan, Lu Sun, Dongjie Fan, Yanan Gong, Xiaoli Chen, Yuanhai You, Fei Zhao, and et al. 2021. "Genetic and Transcriptomic Variations for Amoxicillin Resistance in Helicobacter pylori under Cryopreservation" Pathogens 10, no. 6: 676. https://doi.org/10.3390/pathogens10060676
APA StyleHan, X., Zhang, Y., He, L., Fan, R., Sun, L., Fan, D., Gong, Y., Chen, X., You, Y., Zhao, F., Zhang, M., & Zhang, J. (2021). Genetic and Transcriptomic Variations for Amoxicillin Resistance in Helicobacter pylori under Cryopreservation. Pathogens, 10(6), 676. https://doi.org/10.3390/pathogens10060676





