1. Introduction
Tritrichomonas foetus is a flagellate single-cell parasitic protozoan responsible for trichomonosis, which is a venereal disease in the urogenital tract of cattle [
1]. Due to the introduction of artificial insemination, the occurrence of
T. foetus infection disappeared in most European countries, including Poland, which has been
T. foetus-free since 1997. Nevertheless, trichomonosis is a notifiable disease and is one of the diseases listed by the World Organization for Animal Health (OIE). Moreover, there is a duty to examine samples from cattle used for artificial insemination and those exported to other countries [
2,
3]. Due to the unspecified symptoms, it is very difficult to diagnose bovine trichomonosis based on clinical signs only. Therefore, it is necessary to use laboratory techniques. The standard diagnostic methods are microscopic observation (with or without prior cultivation on media) and molecular tests. In the case of microscopy, trichomonads may be viewed on a wet mount or stained slide. In order to increase and support cells growth,
T. foetus can be cultured in vitro in media, including the most commonly used Diamond medium. However, correct
T. foetus microscopic identification based only on the morphology of the parasite and its characteristic “rolling motion” also has its limitations, such as relatively low sensitivity and accuracy. Moreover, a lack of specificity can be caused by the presence of other intestinal or coprophilic trichomonad protozoa that are not
T. foetus, which are present in samples from cattle [
4]; therefore various molecular methods have been developed to accurately identify
T. foetus in samples from cattle.
The major advantages of molecular tests are their high sensitivity and the possibility of DNA amplification, even from dead cells. However, cross-contamination and carry over of amplicons may occur which can lead to the misinterpretation of results. Moreover, PCR can also amplify fragments from other microorganisms, and thus false-positive results have been noted in diagnoses of bovine trichomonosis [
5,
6].
PCR, according to Felleisen et al. [
2], is an important molecular tool for the identification of the DNA of
T. foetus and its used worldwide, especially in governmental and reference laboratories. Thus, the occurrence of false-positive results is related to the loss of Poland’s status as a country free from trichomonosis.
In this context, we present a case of an unspecific reaction with Honigbergiella-like DNA identified during a routine microscopic examination and identified using molecular methods. Therefore, it is important to share the research involved in the diagnosis of trichomonosis, especially in relation to the possible use of non-specific PCR products.
2. Results
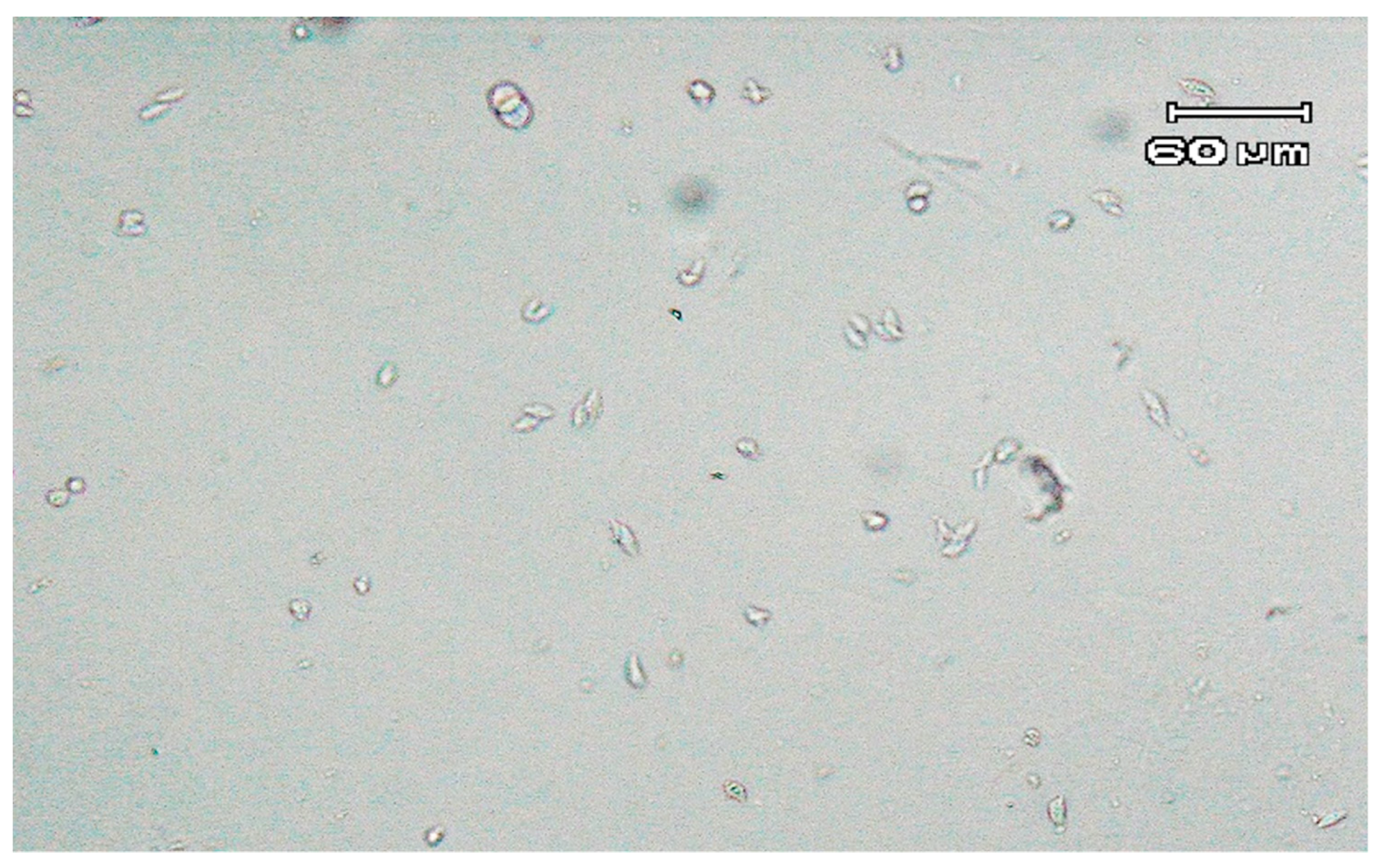
Direct detection of parasites by microscopy revealed the presence of two single trichomonad-shaped microorganisms. Further microscopic observation of the modified Diamond’s medium, conducted between two and seven days, revealed only a few trichomonad-like cells. However, after one week of incubation, examination of the culture showed a higher concentration of forms resembling trichomonads. The majority of the detected parasites had an almond pyriform shape (
Figure 1). Moreover, microorganisms exhibited a jerky, rolling motion.
Conventional PCR according to Felleisen et al. [
2] resulted in the production of a band characteristic of
T. foetus (347 bp) (
Figure 2).
However, based on the results obtained with Sanger sequencing and a subsequent blast analysis, the presence of
T. foetus in a sample from a bull was excluded. Analysis via BLAST sequence similarity searching revealed that the closest homology of the identified microorganisms (named
Honigbergiella-like) was with
Honigbergiella sp. ATCC (American Type Culture Collection) 50321 (GenBank™ accession number AY319274.1), resulting in an identity of 99.62 %. Multiple sequence alignments of
Honigbergiella-like with
T. foetus ATCC 30924 and
Honigbergiella sp. ATCC 50321 sequences taken from GenBank™ were assessed and are shown in
Figure 3. The sequence of the detected parasite was submitted to GenBank under accession number MW233858.
3. Discussion
Trichomonosis is a venereal disease of cattle and can cause seriously economic losses. Therefore, identification of
T. foetus in Polish bovine specimens would increase the risk of reintroducing this dangerous parasite into the cattle population. Based on the results from traditional routine microscopic examination with cell culture and PCR, we assumed that we had identified trichomonads. However, sequencing indicated that there were microorganisms other than
T. foetus. After subsequent blast analysis we found that the closest homology of the identified microorganisms was with
Honigbergiella sp. ATCC 50321 (GenBank™ accession number AY319274.1) with 99.62% identity.
Honigergiella sp. is closely related to
T. foetus because its belongs to the same clad, Parabasalia. However,
Honigbergiella sp., also known as
Pseudotrichomonas keilini, is still poorly understood. According to Hampl et al. [
7], this microorganism switched recently between a free-living habitat in fresh waters and endobiosis. Similarly, closely related to the
Honigbergiella sp. Microorganism,
Honigbergiella ruminantium possibly switched recently from a free-living to a commensal way of life. However, our blast analysis revealed that identity between
Honigbergiella-like DNA and
H. ruminantium was 97%. Thus higher identity was found between
Honigbergiella-like DNA and
Honigbergiella sp. Furthermore, according to Hampl et al. [
7], it is possible that
Honigbergiella sp. may also be present in feces from cattle. Thus, we cannot exclude the fact that
Honigbergiella-like is a non-pathogenic commensal. Therefore, it is important to avoid fecal contamination of samples which may be confused with
T. foetus by removal of material from around the external genitalia of cattle.
The ‘gold standard’ diagnostic method for bovine trichomonosis is a direct microscopic examination and in vitro cultivation in commercial media. However, both methods have limitations, due to the possibility of a false-positive result occurring and the relatively low effectiveness rate of 81–91% for male specimens [
4]. In our study, we identified a few microorganisms with the same morphology and characteristic jerky movements as
T. foetus. Noteworthily, the examination of Diamond’s medium revealed the highest concentration of parasites after seven days of incubation at 37 °C, whereas, according to Lun et al. [
8],
T. foetus should rapidly die after the fifth day in this medium. A probable explanation of this fact is that
Honigbergiella sp. is closely related to that identified in our study by sequencing
Honigbergiella-like DNA in optimal growth conditions at 25 °C. Thus, on the basis of on our findings, we assumed that, in the case of
Honigbergiella-like DNA, the peak concentration is reached later than the maximum growth of
T. foetus.
Currently, molecular tests are used as a good alternative for the diagnosis of trichomonosis. Among several PCRs developed in the last 25 years, conventional PCR, according to Felleisen et al. [
2], has been applied successfully in many laboratories and has high specificity for the detection of trichomonosis. However, to the best of our knowledge, we are the first to identify a cross-reaction in a sample from a bull by conventional PCR [
2]. In our survey, we found that TFR3 and TFR4 primers amplified a fragment of
Honigbergiella-like DNA, although there were several mismatches in the target gene that did not affect the reaction. Similar unspecific results with
T. foetus in PCR were obtained by Frey et al. [
5] where
Simplicimonas-like DNA was identified in vaginal swabs of female cattle. Nevertheless, in this study, real-time PCR based on detection via fluorescence resonance energy transfer (FRET) was used as a molecular tool for
T. foetus identification. In this study, the authors received positive results from reactions in 25/34 samples and the signals occurred mostly between cycles 36 and 40, while a few occurred in cycle 33. It is worth noting that the positive control was regularly detected at around cycle 26. Furthermore, the melting curves of the amplification products of the
Simplicimonas-like DNA were different from the positive control of the reaction. The melting peaks of the reaction with
Simplicimonas-like DNA appeared at about 59 °C, whereas the positive control had its melting peak at about 65 °C. Moreover, the sequencing analysis showed that the product of the PCR reaction (
Simplicimonas-like DNA) had a 91% homology to
Simplicimonas sp. A similar reaction was performed by Schommer et al. [
6], where a minor groove-binder DNA probe (TaqMan
® MGB) was used, and this reaction also amplified
Simplicimonas sp.