The First Report of a Methicillin-Resistant Staphylococcus aureus Isolate Harboring Type IV SCCmec in Thailand
Abstract
1. Introduction
2. Materials and Methods
2.1. Bacterial Isolate
2.2. Phenotypic Study of Antimicrobial Susceptibility Testing
2.3. The SCCmec Pattern
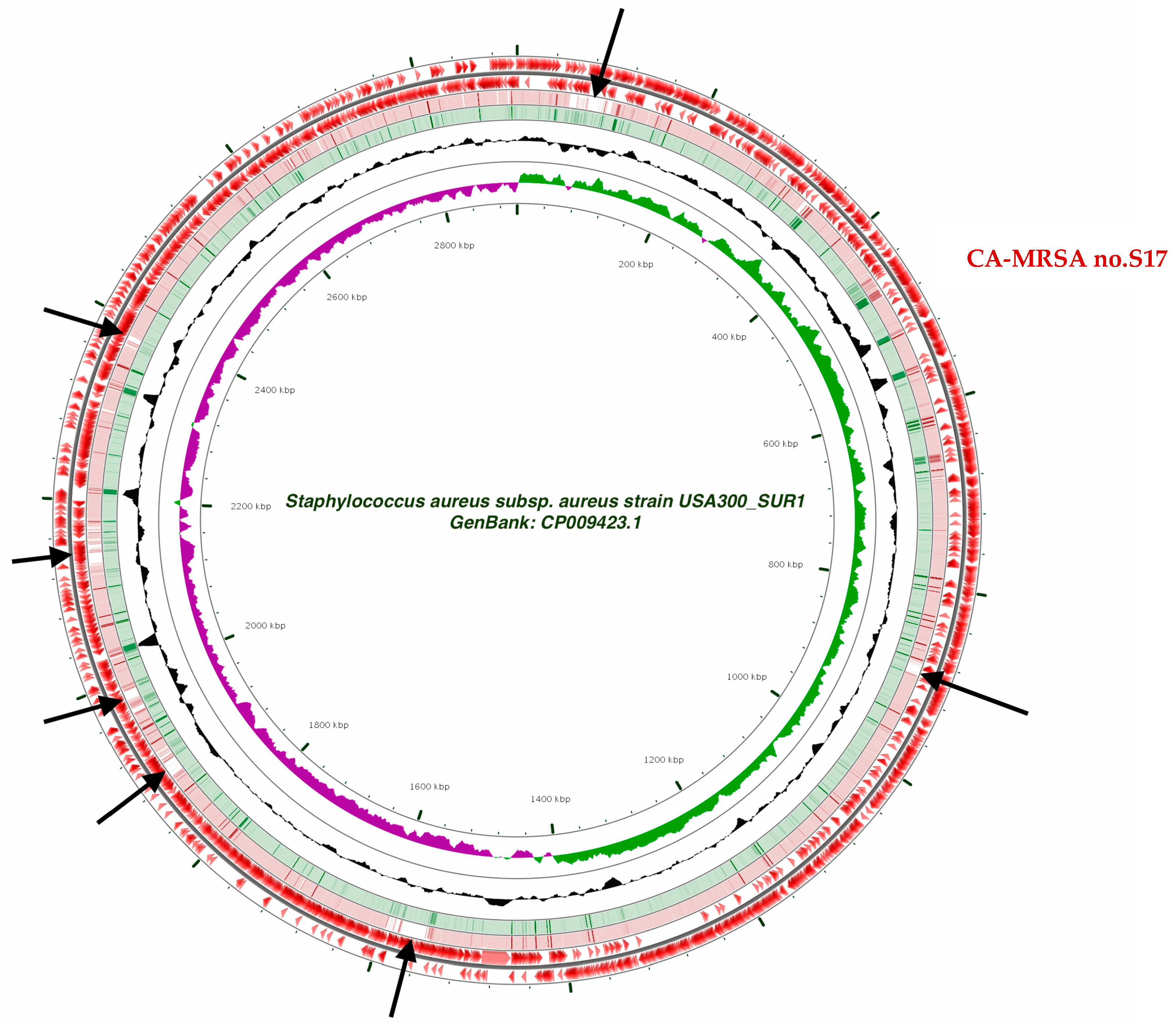
2.4. Whole-Genome Sequencing and Data Analysis
2.5. Genome Sequence Data in Sequence Read Archive (SRA)
3. Results
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Purrello, S.M.; Garau, J.; Giamarellos, E.; Mazzei, T.; Pea, F.; Soriano, A.; Stefani, S. Methicillin-resistant Staphylococcus aureus infections: A review of the currently available treatment options. J. Glob. Antimicrob. Resist. 2016, 7, 178–186. [Google Scholar] [CrossRef] [PubMed]
- Nickerson, E.K.; West, T.E.; Day, N.P.; Peacock, S.J. Staphylococcus aureus disease and drug resistance in resource-limited countries in south and east Asia. Lancet Infect. Dis. 2009, 9, 130–135. [Google Scholar] [CrossRef]
- Tong, S.Y.; Davis, J.S.; Eichenberger, E.; Holland, T.L.; Fowler, V.G., Jr. Staphylococcus aureus infections: Epidemiology, pathophysiology, clinical manifestations, and management. Clin. Microbiol. Rev. 2015, 28, 603–661. [Google Scholar] [CrossRef] [PubMed]
- Marrelli, M.; Tatullo, M.; Dipalma, G.; Inchingolo, F. Oral infection by Staphylococcus aureus in patients affected by White Sponge Nevus: A description of two cases occurred in the same family. Int. J. Med. Sci. 2012, 9, 47–50. [Google Scholar] [CrossRef]
- Chaiwarith, R.; Pacharasupal, P.; Sirisanthana, T. Epidemiology, clinical characteristics and treatment outcomes of healthcare- associated methicillin-resistant Staphylococcus aureus BLOODSTREAM infections at Chiang Mai University Hospital: A retrospective study. Southeast Asian J. Trop. Med. Public Health 2014, 45, 897–905. [Google Scholar]
- De Kraker, M.E.; Davey, P.G.; Grundmann, H.; Burden Study Group. Mortality and hospital stay associated with resistant Staphylococcus aureus and Escherichia coli bacteremia: Estimating the burden of antibiotic resistance in Europe. PLoS Med. 2011, 8, e1001104. [Google Scholar] [CrossRef]
- Lakhundi, S.; Zhang, K. Methicillin-Resistant Staphylococcus aureus: Molecular Characterization, Evolution, and Epidemiology. Clin. Microbiol. Rev. 2018, 31, e00020-18. [Google Scholar] [CrossRef] [PubMed]
- Huh, K.; Chung, D.R. Changing epidemiology of community-associated methicillin-resistant Staphylococcus aureus in the Asia-Pacific region. Expert Rev. Anti Infect. Ther. 2016, 14, 1007–1022. [Google Scholar] [CrossRef]
- Mekviwattanawong, S.; Srifuengfung, S.; Chokepaibulkit, K.; Lohsiriwat, D.; Thamlikitkul, V. Epidemiology of Staphylococcus aureus infections and the prevalence of infection caused by community-acquired methicillin-resistant Staphylococcus aureus in hospitalized patients at Siriraj Hospital. J. Med. Assoc. Thai. 2006, 89 (Suppl. S5), S106–S117. [Google Scholar]
- Santimaleeworagun, W.; Jitwasinkul, T.; Preechachuawong, P.; Samret, W. Mono sulfamethoxazole/trimethoprim and vancomycin combination antimicrobial activity against methicillin-resistant Staphylococcus aureus. Southeast Asian J. Trop. Med. Public Health 2020, 51, 115–123. [Google Scholar]
- Clinical and Laboratory Standards Institute. Performance Standards for Antimicrobial Susceptibility Testing: Twenty-Fifth Informational Supplement; Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2015; p. 177. [Google Scholar]
- Zhang, K.; McClure, J.A.; Elsayed, S.; Louie, T.; Conly, J.M. Novel multiplex PCR assay for characterization and concomitant subtyping of staphylococcal cassette chromosome mec types I to V in methicillin-resistant Staphylococcus aureus. J. Clin. Microbiol. 2005, 43, 5026–5033. [Google Scholar] [CrossRef] [PubMed]
- Larsen, M.V.; Cosentino, S.; Rasmussen, S.; Friis, C.; Hasman, H.; Marvig, R.L.; Jelsbak, L.; Sicheritz-Ponten, T.; Ussery, D.W.; Aarestrup, F.M.; et al. Multilocus sequence typing of total-genome-sequenced bacteria. J. Clin. Microbiol. 2012, 50, 1355–1361. [Google Scholar] [CrossRef] [PubMed]
- Center for Genomic Epidemiology. Prediction of Bacterial Species Using a Fast K-mer Algorithm. Available online: https://cge.cbs.dtu.dk/services/KmerFinder/ (accessed on 14 November 2020).
- Bartels, M.D.; Petersen, A.; Worning, P.; Nielsen, J.B.; Larner-Svensson, H.; Johansen, H.K.; Andersen, L.P.; Jarlov, J.O.; Boye, K.; Larsen, A.R.; et al. Comparing whole-genome sequencing with Sanger sequencing for spa typing of methicillin-resistant Staphylococcus aureus. J. Clin. Microbiol. 2014, 52, 4305–4308. [Google Scholar] [CrossRef] [PubMed]
- Zankari, E.; Hasman, H.; Cosentino, S.; Vestergaard, M.; Rasmussen, S.; Lund, O.; Aarestrup, F.M.; Larsen, M.V. Identification of acquired antimicrobial resistance genes. J. Antimicrob. Chemother. 2012, 67, 2640–2644. [Google Scholar] [CrossRef]
- Grant, J.R.; Stothard, P. The CGView Server: A comparative genomics tool for circular genomes. Nucleic Acids Res. 2008, 36, W181–W184. [Google Scholar] [CrossRef]
- Khan, A.; Wilson, B.; Gould, I.M. Current and future treatment options for community-associated MRSA infection. Expert Opin. Pharm. 2018, 19, 457–470. [Google Scholar] [CrossRef]
- Elston, J.W.; Barlow, G.D. Community-associated MRSA in the United Kingdom. J. Infect. 2009, 59, 149–155. [Google Scholar] [CrossRef]
- Park, S.H.; Park, C.; Yoo, J.H.; Choi, S.M.; Choi, J.H.; Shin, H.H.; Lee, D.G.; Lee, S.; Kim, J.; Choi, S.E.; et al. Emergence of community-associated methicillin-resistant Staphylococcus aureus strains as a cause of healthcare-associated bloodstream infections in Korea. Infect. Control Hosp. Epidemiol. 2009, 30, 146–155. [Google Scholar] [CrossRef]
- Saeed, K.; Gould, I.; Esposito, S.; Ahmad-Saeed, N.; Ahmed, S.S.; Alp, E.; Bal, A.M.; Bassetti, M.; Bonnet, E.; Chan, M.; et al. Corrigendum to ‘Panton-Valentine Leucocidin (PVL) Staphylococcus aureus a position statement from the International Society of Chemotherapy’ [International Journal of Antimicrobial Agents 51/1 (2018) 16-25]. Int. J. Antimicrob. Agents 2018, 52, 125. [Google Scholar] [CrossRef]
- Kreienbuehl, L.; Charbonney, E.; Eggimann, P. Community-acquired necrotizing pneumonia due to methicillin-sensitive Staphylococcus aureus secreting Panton-Valentine leukocidin: A review of case reports. Ann. Intensive Care 2011, 1, 52. [Google Scholar] [CrossRef]
- Yamasaki, O.; Kaneko, J.; Morizane, S.; Akiyama, H.; Arata, J.; Narita, S.; Chiba, J.; Kamio, Y.; Iwatsuki, K. The association between Staphylococcus aureus strains carrying panton-valentine leukocidin genes and the development of deep-seated follicular infection. Clin. Infect. Dis. 2005, 40, 381–385. [Google Scholar] [CrossRef]
- Lina, G.; Piemont, Y.; Godail-Gamot, F.; Bes, M.; Peter, M.O.; Gauduchon, V.; Vandenesch, F.; Etienne, J. Involvement of Panton-Valentine leukocidin-producing Staphylococcus aureus in primary skin infections and pneumonia. Clin. Infect. Dis. 1999, 29, 1128–1132. [Google Scholar] [CrossRef]
- Moran, G.J.; Krishnadasan, A.; Gorwitz, R.J.; Fosheim, G.E.; McDougal, L.K.; Carey, R.B.; Talan, D.A.; EMERGEncy ID Net Study Group. Methicillin-resistant S. aureus infections among patients in the emergency department. N. Engl. J. Med. 2006, 355, 666–674. [Google Scholar] [CrossRef] [PubMed]
- Nakaminami, H.; Ito, A.; Sakanashi, D.; Suematsu, H.; Yamagishi, Y.; Mikamo, H.; Noguchi, N. Genetic diversity of pvl-positive community-onset methicillin-resistant Staphylococcus aureus isolated at a university hospital in Japan. J. Infect. Chemother. 2017, 23, 856–858. [Google Scholar] [CrossRef]
- Asiimwe, B.B.; Baldan, R.; Trovato, A.; Cirillo, D.M. Molecular epidemiology of Panton-Valentine Leukocidin-positive community-acquired methicillin resistant Staphylococcus aureus isolates in pastoral communities of rural south western Uganda. BMC Infect. Dis. 2017, 17, 24. [Google Scholar] [CrossRef] [PubMed]
- Hewagama, S.; Spelman, T.; Woolley, M.; McLeod, J.; Gordon, D.; Einsiedel, L. The Epidemiology of Staphylococcus aureus and Panton-Valentine Leucocidin (pvl) in Central Australia, 2006–2010. BMC Infect. Dis. 2016, 16, 1–6. [Google Scholar] [CrossRef]
- Loewen, K.; Schreiber, Y.; Kirlew, M.; Bocking, N.; Kelly, L. Community-associated methicillin-resistant Staphylococcus aureus infection: Literature review and clinical update. Can. Fam. Physician 2017, 63, 512–520. [Google Scholar] [PubMed]
- Chongtrakool, P.; Ito, T.; Ma, X.X.; Kondo, Y.; Trakulsomboon, S.; Tiensasitorn, C.; Jamklang, M.; Chavalit, T.; Song, J.H.; Hiramatsu, K. Staphylococcal cassette chromosome mec (SCCmec) typing of methicillin-resistant Staphylococcus aureus strains isolated in 11 Asian countries: A proposal for a new nomenclature for SCCmec elements. Antimicrob. Agents Chemother. 2006, 50, 1001–1012. [Google Scholar] [CrossRef]
- Lulitanond, A.; Chanawong, A.; Sribenjalux, P.; Wilailuckana, C.; Kaewkes, W.; Vorachit, M.; Ito, T.; Hiramatsu, K. Preliminary report of SCCmec-types and antimicrobial susceptibilities of methicillin-resistant Staphylococcus aureus isolates from a university hospital in Thailand. Southeast. Asian J. Trop Med. Public Health 2010, 41, 920–927. [Google Scholar]
- Sinlapasorn, S.; Lulitanond, A.; Angkititrakul, S.; Chanawong, A.; Wilailuckana, C.; Tavichakorntrakool, R.; Chindawong, K.; Seelaget, C.; Krasaesom, M.; Chartchai, S.; et al. SCCmec IX in meticillin-resistant Staphylococcus aureus and meticillin-resistant coagulase-negative staphylococci from pigs and workers at pig farms in Khon Kaen, Thailand. J. Med. Microbiol. 2015, 64, 1087–1093. [Google Scholar] [CrossRef]
- Larsen, J.; Imanishi, M.; Hinjoy, S.; Tharavichitkul, P.; Duangsong, K.; Davis, M.F.; Nelson, K.E.; Larsen, A.R.; Skov, R.L. Methicillin-resistant Staphylococcus aureus ST9 in pigs in Thailand. PLoS ONE 2012, 7, e31245. [Google Scholar] [CrossRef] [PubMed]
- Witte, W. Community-acquired methicillin-resistant Staphylococcus aureus: What do we need to know? Clin. Microbiol. Infect. 2009, 15 (Suppl. S7), 17–25. [Google Scholar] [CrossRef] [PubMed]
- Yeap, A.D.; Woods, K.; Dance, D.A.B.; Pichon, B.; Rattanavong, S.; Davong, V.; Phetsouvanh, R.; Newton, P.N.; Shetty, N.; Kearns, A.M. Molecular Epidemiology of Staphylococcus aureus Skin and Soft Tissue Infections in the Lao People’s Democratic Republic. Am. J. Trop. Med. Hyg. 2017, 97, 423–428. [Google Scholar] [CrossRef] [PubMed]
| Resistance Gene | Identity | Template Length | Contig | Position in Contig | Predicted Phenotype | Accession Number |
|---|---|---|---|---|---|---|
| β-lactam | ||||||
| blaZ | 100 | 849/849 | NODE_15_length_14257_cov_356.776825 | 7028–7876 | β-lactam resistance | JBTH01000015 |
| mecA | 100 | 2007/2007 | NODE_1_length_202378_cov_183.284088 | 83,894–85,900 | β-lactam resistance | BX571856 |
| Macrolide | ||||||
| Inu(A) | 99.79 | 486/486 | NODE_34_length_2491_cov_187.230835 | 2007–2492 | Lincosamide resistance | M14039 |
| Tetracycline | ||||||
| tet(K) | 99.86 | 1380/1380 | NODE_6_length_4546_cov_373.607574 | 2335–3714 | Tetracycline resistance | U38656 |
| Trimethoprim | ||||||
| dfrG | 100 | 498/498 | NODE_1_length_202378_cov_183.284088 | 90,645–91,142 | Trimethoprim resistance | AB205645 |
| Virulence Factor | Identity | Template Length | Contig | Position in Contig | Protein Function | Accession Number |
|---|---|---|---|---|---|---|
| eta | 100 | 843/843 | NODE_140_length_521104_cov_145.043243 | 517966..518808 | Exfoliative toxin A | AP008953.1 |
| hlgA | 99.89 | 930/930 | NODE_83_length_193242_cov_168.671219 | 110976..111905 | Gamma-hemolysin chain II precursor | AP014942.1 |
| hlgB | 100 | 977/977 | NODE_83_length_193242_cov_168.671219 | 113422..114398 | Gamma-hemolysin component B precursor | AP014942.1 |
| hlgC | 99.79 | 948/948 | NODE_83_length_193242_cov_168.671219 | 112473..113420 | Gamma-hemolysin component C | AP014653.1 |
| lukD | 99.8 | 984/984 | NODE_118_length_478021_cov_147.302170 | 469250..470233 | Leukocidin D component | AP014653.1 |
| lukE | 99.89 | 936/936 | NODE_118_length_478021_cov_147.302170 | 470235..471170 | Leukocidin E component | BA000018.3 |
| lukE | 99.89 | 936/936 | NODE_118_length_478021_cov_147.302170 | 470235..471170 | Leukocidin E component | CP001781.1 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Santimaleeworagun, W.; Preechachuawong, P.; Samret, W.; Jitwasinkul, T. The First Report of a Methicillin-Resistant Staphylococcus aureus Isolate Harboring Type IV SCCmec in Thailand. Pathogens 2021, 10, 430. https://doi.org/10.3390/pathogens10040430
Santimaleeworagun W, Preechachuawong P, Samret W, Jitwasinkul T. The First Report of a Methicillin-Resistant Staphylococcus aureus Isolate Harboring Type IV SCCmec in Thailand. Pathogens. 2021; 10(4):430. https://doi.org/10.3390/pathogens10040430
Chicago/Turabian StyleSantimaleeworagun, Wichai, Praewdow Preechachuawong, Wandee Samret, and Tossawan Jitwasinkul. 2021. "The First Report of a Methicillin-Resistant Staphylococcus aureus Isolate Harboring Type IV SCCmec in Thailand" Pathogens 10, no. 4: 430. https://doi.org/10.3390/pathogens10040430
APA StyleSantimaleeworagun, W., Preechachuawong, P., Samret, W., & Jitwasinkul, T. (2021). The First Report of a Methicillin-Resistant Staphylococcus aureus Isolate Harboring Type IV SCCmec in Thailand. Pathogens, 10(4), 430. https://doi.org/10.3390/pathogens10040430






