Indwelling Device-Associated Biofilms in Critically Ill Cancer Patients—Study Protocol
Abstract
1. Introduction
2. Expected Results
3. Discussion
4. Materials and Methods
4.1. Objectives
- Identification of pathogens involved in ID biofilm formation (ET, CVC, AC, UC) in critically ill cancer patients;
- Comparison of four IDs derived from biofilm-associated pathogens collected from the same patient;
- Comparison of the biofilm-associated pathogens with those identified in currently used biological samples (tracheal aspirate/bronchoalveolar lavage, blood culture, urinary culture and surgical wound swab) collected from the same patient;
- Examination of correlations between different parameters: biofilm-associated pathogens and patient clinical and biological data, such as nasal, pharyngeal, rectal and skin pathogen screening data, will be assessed, as well as risk factors, such as neutropenia, chemo/radiotherapy, corticosteroid treatment and previous anti-infective therapy, ID exposure time and biological markers of inflammation. The diagnosed infection will also be used as an index and includes respiratory tract infection, urinary tract infection, bloodstream infection, surgical site infection and sepsis of unknown origin. Finally, the severity scores will also be assessed as follows: Sequential [Sepsis-Related] Organ Failure Assessment (SOFA) score and Acute Physiology and Chronic Health Evaluation II (APACHE II) score. The ICU and hospital LOS and the patient outcome, which are categorized as survival/death will be also included in the analysis.
4.2. Study Design
4.3. Settings
4.4. Eligibility Criteria
4.4.1. Inclusion Criteria
- Signed informed consent;
- Age ≥18 years;
- Suspected/proven sepsis/septic shock;
- APACHE II score ≥10;
- Predictable invasive ventilatory support ≥48 h;
- Patient estimated survival ≥4 days.
4.4.2. Exclusion Criteria
- Patient/legal representative refusal;
- Age <18 years;
- Chronic psychiatric/neurological disease with impaired decision-making capacity;
- Pregnancy;
- Invasive ventilatory support <2 days;
- Death in less than 4 days following ICU admission.
4.5. Ethical Aspects
4.6. Description of the Used Methods
4.6.1. Standard of Care in the ICU
4.6.2. Swab Sampling
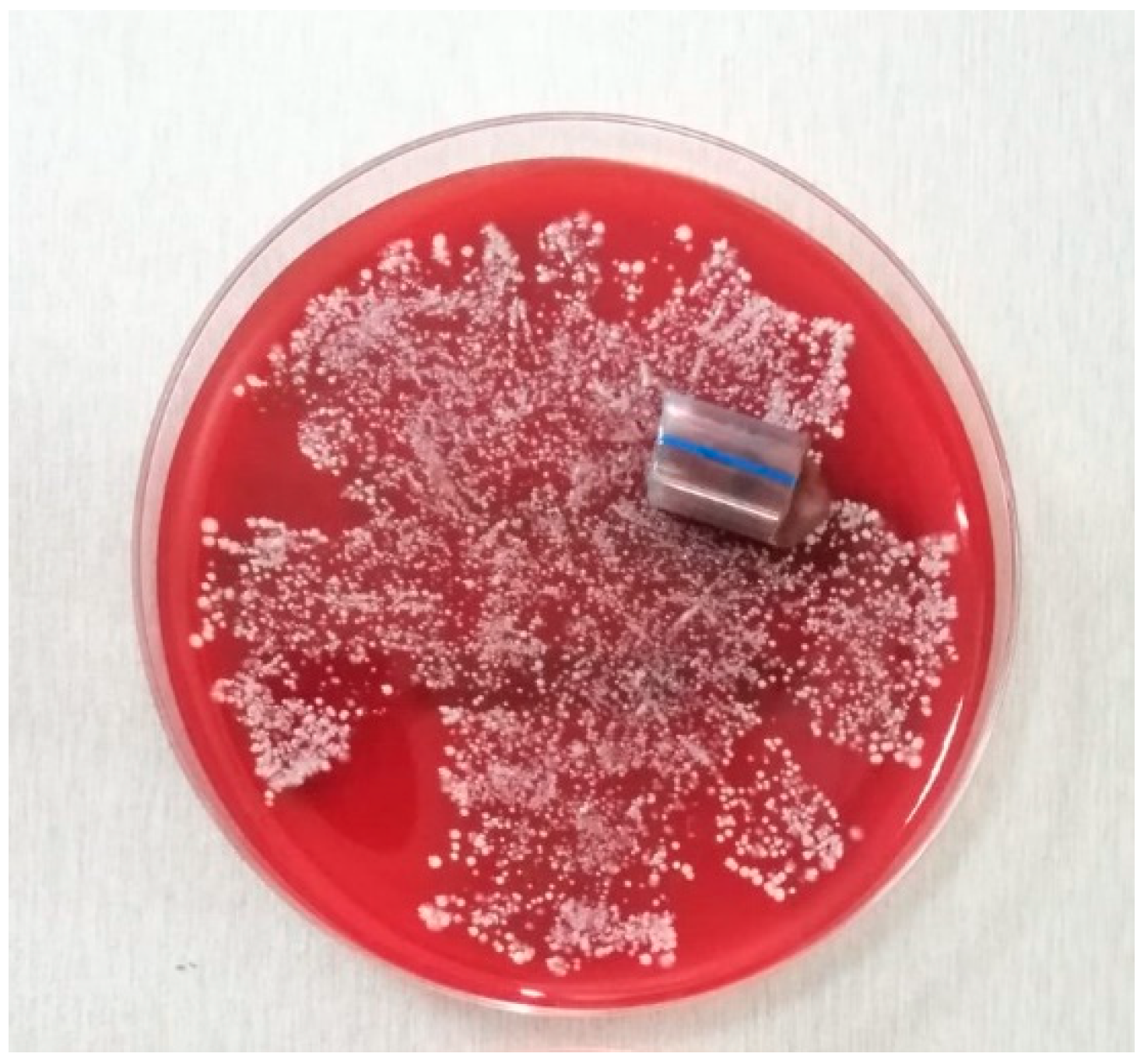
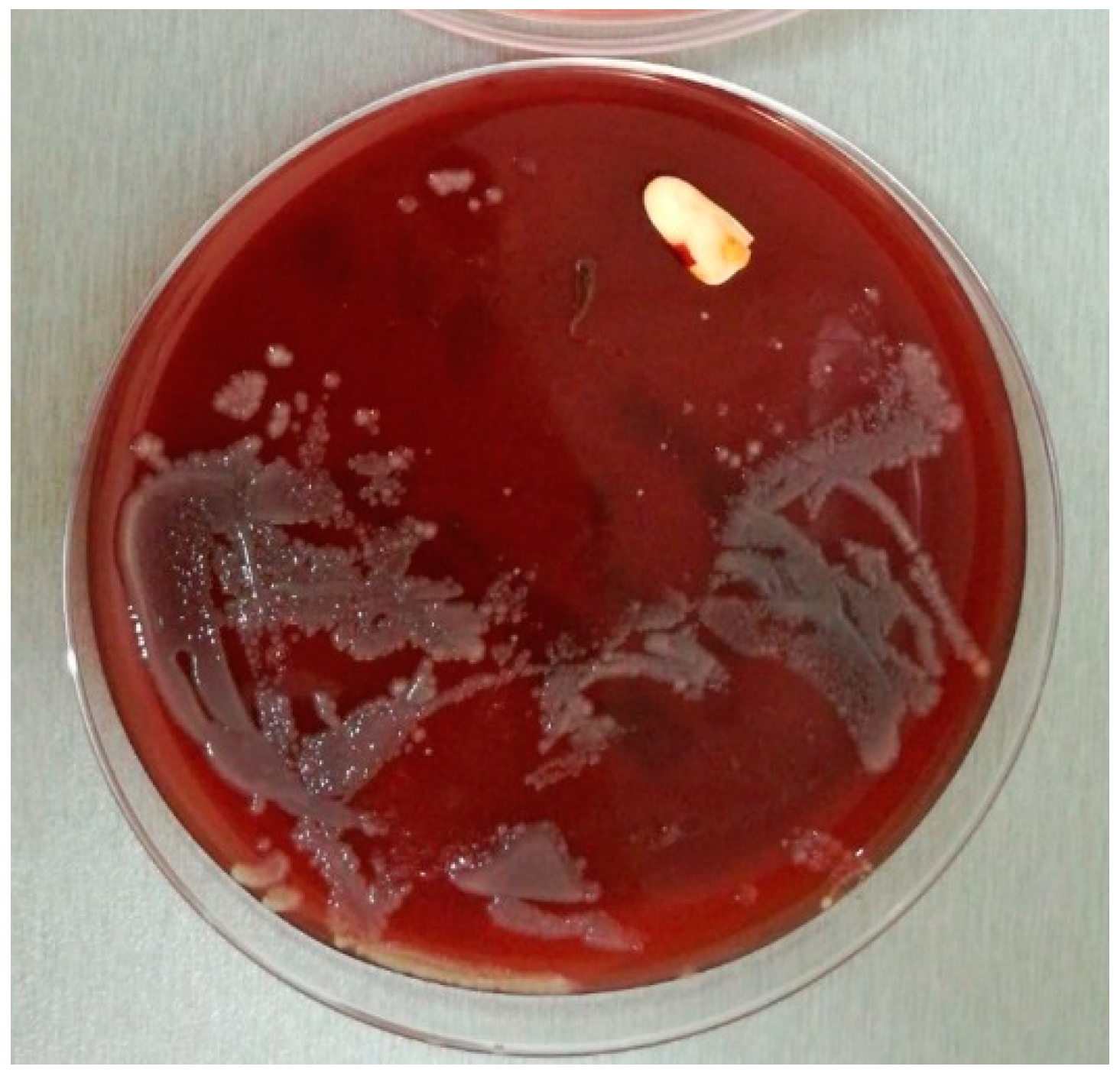
4.6.3. Biofilm Sampling and Transport
4.6.4. Standard of Care in the Microbiology Laboratory
4.6.5. Biofilm Microbiological Processing and Analysis
4.6.6. Biofilm NGS Processing and Analysis
4.7. Data Collection and Follow-Up
- Patient data, which involves inclusion and exclusion criteria, demographic data (age, gender, weight, height), hospital admission (day, reason), ICU admission (day, type, admission source, primary admission diagnosis), patient chronic illnesses and co-morbid conditions and ICU admission scores (APACHE II, quick SOFA and SOFA), immunosuppression risk factors, ICU/hospital LOS, 28-Day status;
- Sepsis monitoring, which includes suspected/proven infection criteria, scoring (daily SOFA score), colonization status, microbiological monitoring, anti-infective treatment and source control and 1st, 3rd and 7th ICU day evaluation of sepsis evolution (shock criteria, inflammatory markers, multiple organ dysfunctions, multi-organ support therapy);
- Monitoring of each ID, which includes insertion date, ID manipulation (endotracheal aspiration, bronchoalveolar lavage, measurement of intra-abdominal pressure, arterial blood/central venous blood/urine sampling), date of extraction, results of biofilm pathogen identification by culture growth and NGS and antimicrobial resistance profile.
4.8. Strategies to Ensure Adequate Enrolment and Protocol Compliance
4.9. Statistical Analysis
4.10. Sample Size Power Calculation
4.11. Strengths and Limitations of the Study Protocol
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Trial Status
References
- Hajj, J.; Blaine, N.; Salavaci, J.; Jacoby, D. The “Centrality of Sepsis”: A Review on Incidence, Mortality, and Cost of Care. Healthcare 2018, 6, 90. [Google Scholar] [CrossRef] [PubMed]
- Abou Dagher, G.; El Khuri, C.; Chehadeh, A.A.H.; Chami, A.; Bachir, R.; Zebian, D.; Bou Chebl, R. Are patients with cancer with sepsis and bacteraemia at a higher risk of mortality? A retrospective chart review of patients presenting to a tertiary care centre in Lebanon. BMJ Open 2017, 7, e013502. [Google Scholar] [CrossRef]
- Kochanek, M.; Schalk, E.; von Bergwelt-Baildon, M.; Beutel, G.; Buchheidt, D.; Hentrich, M.; Henze, L.; Kiehl, M.; Liebregts, T.; von Lilienfeld-Toal, M.; et al. Management of sepsis in neutropenic cancer patients: 2018 guidelines from the Infectious Diseases Working Party (AGIHO) and Intensive Care Working Party (iCHOP) of the German Society of Hematology and Medical Oncology (DGHO). Ann. Hematol. 2019, 98, 1051–1069. [Google Scholar] [CrossRef] [PubMed]
- Haque, M.; Sartelli, M.; McKimm, J.; Abu Bakar, M. Health care-associated infections—An overview. Infect. Drug Resist. 2018, 11, 2321–2333. [Google Scholar] [CrossRef] [PubMed]
- Cabarkapa, I.; Levic, J.; Djuragic, O. Biofilm. In Microbial Pathogens and Strategies for Combating Them: Science, Technology and Education; Méndez-Vilas, A., Ed.; Formatex Research Center: Badajoz, Spain, 2013; Volume 4, pp. 42–52. [Google Scholar]
- Høiby, N.; Bjarnsholt, T.; Moser, C.; Bassi, G.L.; Coenye, T.; Donelli, G.; Hall-Stoodley, L.; Holá, V.; Imbert, C.; Kirketerp-Møller, K.; et al. ESCMID Study Group for Biofilms and Consulting External Expert Werner Zimmerli. ESCMID guideline for the diagnosis and treatment of biofilm infections 2014. Clin. Microbiol. Infect. 2015, 21 (Suppl. 1), S1–S25. [Google Scholar] [CrossRef]
- Bauer, T.T.; Torres, A.; Ferrer, R.; Heyer, C.M.; Schultze-Werninghaus, G.; Rasche, K. Biofilm formation in endotracheal tubes. Association between pneumonia and the persistence of pathogens. Monaldi Arch. Chest Dis. 2002, 57, 84–87. [Google Scholar] [PubMed]
- Hall-Stoodley, L.; Stoodley, P.; Kathju, S.; Høiby, N.; Moser, C.; Costerton, J.W.; Moter, A.; Bjarnsholt, T. Towards diagnostic guidelines for biofilm-associated infections. FEMS Immunol. Med. Microbiol. 2012, 65, 127–145. [Google Scholar] [CrossRef]
- Treter, J.; Macedo, A.J. Catheters: A suitable surface for biofilm formation. In Science against Microbial Pathogens: Communicating Current Research and Technological Advances; Méndez-Vilas, A., Ed.; Formatex Research Center: Badajoz, Spain, 2011; Volume 3, pp. 835–842. [Google Scholar]
- Mukherjee, S.; Bassler, B.L. Bacterial quorum sensing in complex and dynamically changing environments. Nat. Rev. Microbiol. 2019, 17, 371–382. [Google Scholar] [CrossRef] [PubMed]
- Pérez-Velázquez, J.; Gölgeli, M.; García-Contreras, R. Mathematical Modelling of Bacterial Quorum Sensing: A Review. Bull. Math Biol. 2016, 78, 1585–1639. [Google Scholar] [CrossRef]
- Khatoon, Z.; McTiernan, C.D.; Suuronen, E.J.; Mah, T.F.; Alarcon, E.I. Bacterial biofilm formation on implantable devices and approaches to its treatment and prevention. Heliyon 2018, 4, e01067. [Google Scholar] [CrossRef] [PubMed]
- Licker, M.; Moldovan, R.; Hogea, E.; Muntean, D.; Horhat, F.; Baditoiu, L.; Rogobete, A.; Tirziu, E.; Zambori, C. Microbial biofilm in human health—An updated theoretical and practical insight. Rev. Romana Med. Lab. 2017, 25, 9–26. [Google Scholar] [CrossRef][Green Version]
- Ehrlich, G.D.; DeMeo, P.; Palmer, M.; Sauber, T.J.; Altman, D.; Altman, G.; Sotereanos, N.; Conti, S.; Baratz, M.; Maale, G.; et al. Culture-Negative Infections in Orthopedic Surgery. In Culture Negative Orthopedic Biofilm Infections; Ehrlich, G.D., DeMeo, P.J., Costerton, J.W., Winkler, H., Eds.; Springer Series on Biofilms 7: Berlin, Germany, 2012; Volume 7, pp. 17–27. [Google Scholar] [CrossRef]
- Sakr, Y.; Jaschinski, U.; Wittebole, X.; Szakmany, T.; Lipman, J.; Namendys-Silva, S.A.; Martin-Loeches, I.; Leone, M.; Lupu, M.N.; Vincent, J.L.; et al. Sepsis in intensive care unit patients: Worldwide data from the intensive care over nations audit. Open Forum Infect. Dis. 2018, 5, 1–9. [Google Scholar] [CrossRef]
- Voidazan, S.; Albu, S.; Toth, R.; Grigorescu, B.; Rachita, A.; Moldovan, I. Healthcare associated infections—A new pathology in medical practice? Int. J. Environ. Res. Public Health 2020, 17, 760. [Google Scholar] [CrossRef] [PubMed]
- Khan, I.D.; Basu, A.; Kiran, S.; Trivedi, S.; Pandit, P.; Chattoraj, A. Device-Associated Healthcare-Associated Infections (DA-HAI) and the caveat of multiresistance in a multidisciplinary intensive care unit. Med. J. Armed Forces India 2017, 73, 222–231. [Google Scholar] [CrossRef] [PubMed]
- Singer, M.; Deutschman, C.S.; Seymour, C.W.; Shankar-Hari, M.; Annane, D.; Bauer, M.; Bellomo, R.; Bernard, G.R.; Chiche, J.D.; Coopersmith, C.M.; et al. The Third International Consensus Definitions for Sepsis and Septic Shock (Sepsis-3). JAMA 2016, 315, 801–810. [Google Scholar] [CrossRef]
- Levy, M.M.; Evans, L.E.; Rhodes, A. The Surviving Sepsis Campaign Bundle: 2018 Update. Crit. Care Med. 2018, 46, 997–1000. [Google Scholar] [CrossRef]
- Vincent, J.L.; Brealey, D.; Libert, N.; Abidi, N.E.; O’Dwyer, M.; Zacharowski, K.; Mikaszewska-Sokolewicz, M.; Schrenzel, J.; Simon, F.; Wilks, M.; et al. Rapid Diagnosis of Infections in the Critically Ill Team. Rapid Diagnosis of Infection in the Critically Ill, a Multicenter Study of Molecular Detection in Bloodstream Infections, Pneumonia, and Sterile Site Infections. Crit. Care Med. 2015, 43, 2283–2291. [Google Scholar] [CrossRef] [PubMed]
- Sinha, M.; Jupe, J.; Mack, H.; Coleman, T.P.; Lawrence, S.M.; Fraley, S.I. Emerging Technologies for Molecular Diagnosis of Sepsis. Clin. Microbiol. Rev. 2018, 31, 1–26. [Google Scholar] [CrossRef] [PubMed]
- Jamal, M.; Ahmad, W.; Andleeb, S.; Jalil, F.; Imran, M.; Nawaz, M.A.; Hussain, T.; Ali, M.; Rafiq, M.; Kamil, M.A. Bacterial biofilm and associated infections. J. Chin. Med. Assoc. 2018, 81, 7–11. [Google Scholar] [CrossRef] [PubMed]
- Opota, O.; Croxatto, A.; Prod’hom, G.; Greub, G. Blood culture-based diagnosis of bacteraemia: State of the art. Clin. Microbiol. Infect. 2015, 21, 313–322. [Google Scholar] [CrossRef] [PubMed]
- Grumaz, S.; Stevens, P.; Grumaz, C.; Decker, S.O.; Weigand, M.A.; Hofer, S.; Brenner, T.; von Haeseler, A.; Sohn, K. Next-generation sequencing diagnostics of bacteremia in septic patients. Genome Med. 2016, 8, 73. [Google Scholar] [CrossRef]
- Opota, O.; Jaton, K.; Greub, G. Microbial diagnosis of bloodstream infection: Towards molecular diagnosis directly from blood. Clin. Microbiol. Infect. 2015, 21, 323–331. [Google Scholar] [CrossRef] [PubMed]
- Souvenir, D.; Anderson, D.E., Jr.; Palpant, S.; Mroch, H.; Askin, S.; Anderson, J.; Claridge, J.; Eiland, J.; Malone, C.; Garrison, M.W.; et al. Blood cultures positive for coagulase-negative staphylococci: Antisepsis, pseudobacteremia, and therapy of patients. J. Clin. Microbiol. 1998, 36, 1923–1926. [Google Scholar] [CrossRef] [PubMed]
- Pien, B.C.; Sundaram, P.; Raoof, N.; Costa, S.F.; Mirrett, S.; Woods, C.W.; Reller, L.B.; Weinstein, M.P. The clinical and prognostic importance of positive blood cultures in adults. Am. J. Med. 2010, 123, 819–828. [Google Scholar] [CrossRef] [PubMed]
- Garcia, R.A.; Spitzer, E.D.; Beaudry, J.; Beck, C.; Diblasi, R.; Gilleeny-Blabac, M.; Haugaard, C.; Heuschneider, S.; Kranz, B.P.; McLean, K.; et al. Multidisciplinary team review of best practices for collection and handling of blood cultures to determine effective interventions for increasing the yield of true-positive bacteremias, reducing contamination, and eliminating false-positive central line-associated bloodstream infections. Am. J. Infect. Control 2015, 43, 1222–1237. [Google Scholar] [CrossRef] [PubMed]
- Gilligan, P.H. Blood culture contamination: A clinical and financial burden. Infect. Control Hosp. Epidemiol. 2013, 34, 22–23. [Google Scholar] [CrossRef] [PubMed]
- Brenner, T.; Decker, S.O.; Grumaz, S.; Stevens, P.; Bruckner, T.; Schmoch, T.; Pletz, M.W.; Bracht, H.; Hofer, S.; Marx, G.; et al. TIFOnet Critical Care Trials Group. Next-generation sequencing diagnostics of bacteremia in sepsis (Next GeneSiS-Trial): Study protocol of a prospective, observational, noninterventional, multicenter, clinical trial. Medicine (Baltimore) 2018, 97, e9868. [Google Scholar] [CrossRef] [PubMed]
- Lee, T.; Pang, S.; Stegger, M.; Sahibzada, S.; Abraham, S.; Daley, D.; Coombs, G. Australian Group on Antimicrobial Resistance. A three-year whole genome sequencing perspective of Enterococcus faecium sepsis in Australia. PLoS ONE 2020, 15, e0228781. [Google Scholar] [CrossRef]
- Decker, S.O.; Sigl, A.; Grumaz, C.; Stevens, P.; Vainshtein, Y.; Zimmermann, S.; Weigand, M.A.; Hofer, S.; Sohn, K.; Brenner, T. Immune-response patterns and next generation sequencing diagnostics for the detection of mycoses in patients with septic shock-Results of a combined clinical and experimental investigation. Int. J. Mol. Sci. 2017, 18, 1796. [Google Scholar] [CrossRef] [PubMed]
- Xiao, W.; Han, P.; Xu, Z.; Huang, M. Pulmonary scedosporiosis in a patient with acute hematopoietic failure: Diagnosis aided by next-generation sequencing. Int. J. Infect. Dis. 2019, 85, 114–116. [Google Scholar] [CrossRef]
- Boers, S.A.; Jansen, R.; Hays, J.P. Understanding and overcoming the pitfalls and biases of next-generation sequencing (NGS) methods for use in the routine clinical microbiological diagnostic laboratory. Eur. J. Clin. Microbiol. Infect. Dis. 2019, 38, 1059–1070. [Google Scholar] [CrossRef]
- Grumaz, C.; Hoffmann, A.; Vainshtein, Y.; Kopp, M.; Grumaz, S.; Stevens, P.; Decker, S.O.; Weigand, M.A.; Hofer, S.; Brenner, T.; et al. Rapid Next-Generation Sequencing-Based Diagnostics of Bacteremia in Septic Patients. J. Mol. Diagn. 2020, 22, 405–418. [Google Scholar] [CrossRef] [PubMed]
- Dai, Y.; Chen, L.; Chang, W.; Lu, H.; Cui, P.; Ma, X. Culture-Negative Streptococcus suis Infection Diagnosed by Metagenomic Next-Generation Sequencing. Front. Public Health 2019, 7, 379. [Google Scholar] [CrossRef] [PubMed]
- Xu, A.; Zhu, H.; Gao, B.; Weng, H.; Ding, Z.; Li, M.; Weng, X.; He, G. Diagnosis of severe community-acquired pneumonia caused by Acinetobacter baumannii through next-generation sequencing: A case report. BMC Infect. Dis. 2020, 20, 45. [Google Scholar] [CrossRef] [PubMed]
- Ishihara, T.; Watanabe, N.; Inoue, S.; Aoki, H.; Tsuji, T.; Yamamoto, B.; Yanagi, H.; Oki, M.; Kryukov, K.; Nakagawa, S.; et al. Usefulness of next-generation DNA sequencing for the diagnosis of urinary tract infection. Drug Discov. Ther. 2020, 14, 42–49. [Google Scholar] [CrossRef] [PubMed]
- Wilson, M.R.; Naccache, S.N.; Samayoa, E.; Biagtan, M.; Bashir, H.; Yu, G.; Salamat, S.M.; Somasekar, S.; Federman, S.; Miller, S.; et al. Actionable diagnosis of neuroleptospirosis by next-generation sequencing. N. Engl. J. Med. 2014, 370, 2408–2417. [Google Scholar] [CrossRef]
- Loss, S.H.; Nunes, D.; Franzosi, O.S.; Salazar, G.S.; Teixeira, C.; Vieira, S. Chronic critical illness: Are we saving patients or creating victims? Rev. Bras. Ter. Intensiva 2017, 29, 87–95. [Google Scholar] [CrossRef] [PubMed]
- Gominet, M.; Compain, F.; Beloin, C.; Lebeaux, D. Central venous catheters and biofilms: Where do we stand in 2017? APMIS 2017, 125, 365–375. [Google Scholar] [CrossRef] [PubMed]
- Camargo, J.F.; Ahmed, A.A.; Lindner, M.S.; Morris, M.I.; Anjan, S.; Anderson, A.D.; Prado, C.E.; Dalai, S.C.; Martinez, O.V.; Komanduri, K.V. Next-generation sequencing of microbial cell-free DNA for rapid noninvasive diagnosis of infectious diseases in immunocompromised hosts. F1000 Res. 2019, 8, 1194. [Google Scholar] [CrossRef]
- Nates, J.L.; Nunnally, M.; Kleinpell, R.; Blosser, S.; Goldner, J.; Birriel, B.; Fowler, C.S.; Byrum, D.; Miles, W.S.; Bailey, H.; et al. ICU admission, discharge, and triage guidelines: A framework to enhance clinical operations, development of institutional policies, and further research. Crit. Care Med. 2016, 44, 1553–1602. [Google Scholar] [CrossRef] [PubMed]
- Lee, C.S.; Montalmont, B.; O’Hara, J.A.; Syed, A.; Chaussard, C.; McGaha, T.L.; Pakstis, D.L.; Lee, J.H.; Shutt, K.A.; Doi, Y. Screening for methicillin-resistant Staphylococcus aureus colonization using sponges. Infect. Control Hosp. Epidemiol. 2015, 36, 28–33. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Fernández-Barat, L.; Ferrer, M.; Sierra, J.M.; Soy, D.; Guerrero, L.; Vila, J.; Li Bassi, G.; Cortadellas, N.; Martínez-Olondris, P.; Rigol, M.; et al. Linezolid limits burden of methicillin-resistant Staphylococcus aureus in biofilm of tracheal tubes. Crit. Care Med. 2012, 40, 2385–2389. [Google Scholar] [CrossRef] [PubMed]
- O’Flaherty, N.; Crowley, B. How to use central venous catheter tip cultures. Arch. Dis. Child. Educ. Pract. Ed. 2015, 100, 69–74. [Google Scholar] [CrossRef] [PubMed]
- Natham, H.; Kondagadapu, S.; Kadiyala, V.; Mohan, A.; Chaudhury, A.; Samantaray, A. Bacterial Colonisation and Antibiotic Sensitivity Profile of Endotracheal Tubes in Mechanically Ventilated Patients. J. Clin. Diagn. Res. 2019, 13, UC01–UC06. [Google Scholar] [CrossRef]
- Djeribi, R.; Bouchloukh, W.; Jouenne, T.; Menaa, B. Characterization of bacterial biofilms formed on urinary catheters. Am. J. Infect. Control 2012, 40, 854–859. [Google Scholar] [CrossRef] [PubMed]
- Clinical and Laboratory Standards Institute (CLSI). Performance Standards for Antimicrobial Susceptibility Testing, 29th ed.; CLSI Supplement M100 2019; Wayne, P.A., Ed.; Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2019; Available online: https://clsi.org/media/2663/m100ed29_sample.pdf (accessed on 6 May 2020).
- Maki, D.G.; Weise, C.E.; Sarafin, H.W. A semiquantitative culture method for identifying intravenous-catheter-related infection. N. Engl. J. Med. 1977, 296, 1305–1309. [Google Scholar] [CrossRef]
- Xu, Y.; Moser, C.; Al-Soud, W.A.; Sørensen, S.; Høiby, N.; Nielsen, P.H.; Thomsen, T.R. Culture-dependent and -independent investigations of microbial diversity on urinary catheters. J. Clin. Microbiol. 2012, 50, 3901–3908. [Google Scholar] [CrossRef] [PubMed]
- Chen, P.; Sun, W.; He, Y. Comparison of the next-generation sequencing (NGS) technology with culture methods in the diagnosis of bacterial and fungal infections. J. Thorac. Dis. 2020, 12, 4924–4929. [Google Scholar] [CrossRef]
- Gupta, S.; Mortensen, M.S.; Schjørring, S.; Trivedi, U.; Vestergaard, G.; Stokholm, J.; Bisgaard, H.; Krogfelt, K.A.; Sørensen, S.J. Amplicon sequencing provides more accurate microbiome information in healthy children compared to culturing. Commun. Biol. 2019, 2, 291. [Google Scholar] [CrossRef]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lungu, O.; Grigoras, I.; Dorneanu, O.S.; Lunca, C.; Vremera, T.; Copacianu, S.B.; Ivanov, I.; Iancu, L.S. Indwelling Device-Associated Biofilms in Critically Ill Cancer Patients—Study Protocol. Pathogens 2021, 10, 306. https://doi.org/10.3390/pathogens10030306
Lungu O, Grigoras I, Dorneanu OS, Lunca C, Vremera T, Copacianu SB, Ivanov I, Iancu LS. Indwelling Device-Associated Biofilms in Critically Ill Cancer Patients—Study Protocol. Pathogens. 2021; 10(3):306. https://doi.org/10.3390/pathogens10030306
Chicago/Turabian StyleLungu, Olguta, Ioana Grigoras, Olivia Simona Dorneanu, Catalina Lunca, Teodora Vremera, Stefania Brandusa Copacianu, Iuliu Ivanov, and Luminita Smaranda Iancu. 2021. "Indwelling Device-Associated Biofilms in Critically Ill Cancer Patients—Study Protocol" Pathogens 10, no. 3: 306. https://doi.org/10.3390/pathogens10030306
APA StyleLungu, O., Grigoras, I., Dorneanu, O. S., Lunca, C., Vremera, T., Copacianu, S. B., Ivanov, I., & Iancu, L. S. (2021). Indwelling Device-Associated Biofilms in Critically Ill Cancer Patients—Study Protocol. Pathogens, 10(3), 306. https://doi.org/10.3390/pathogens10030306






