Epitope Binning of Novel Monoclonal Anti F1 and Anti LcrV Antibodies and Their Application in a Simple, Short, HTRF Test for Clinical Plague Detection
Abstract
1. Introduction
2. Results
2.1. Antibody Generation and Screening
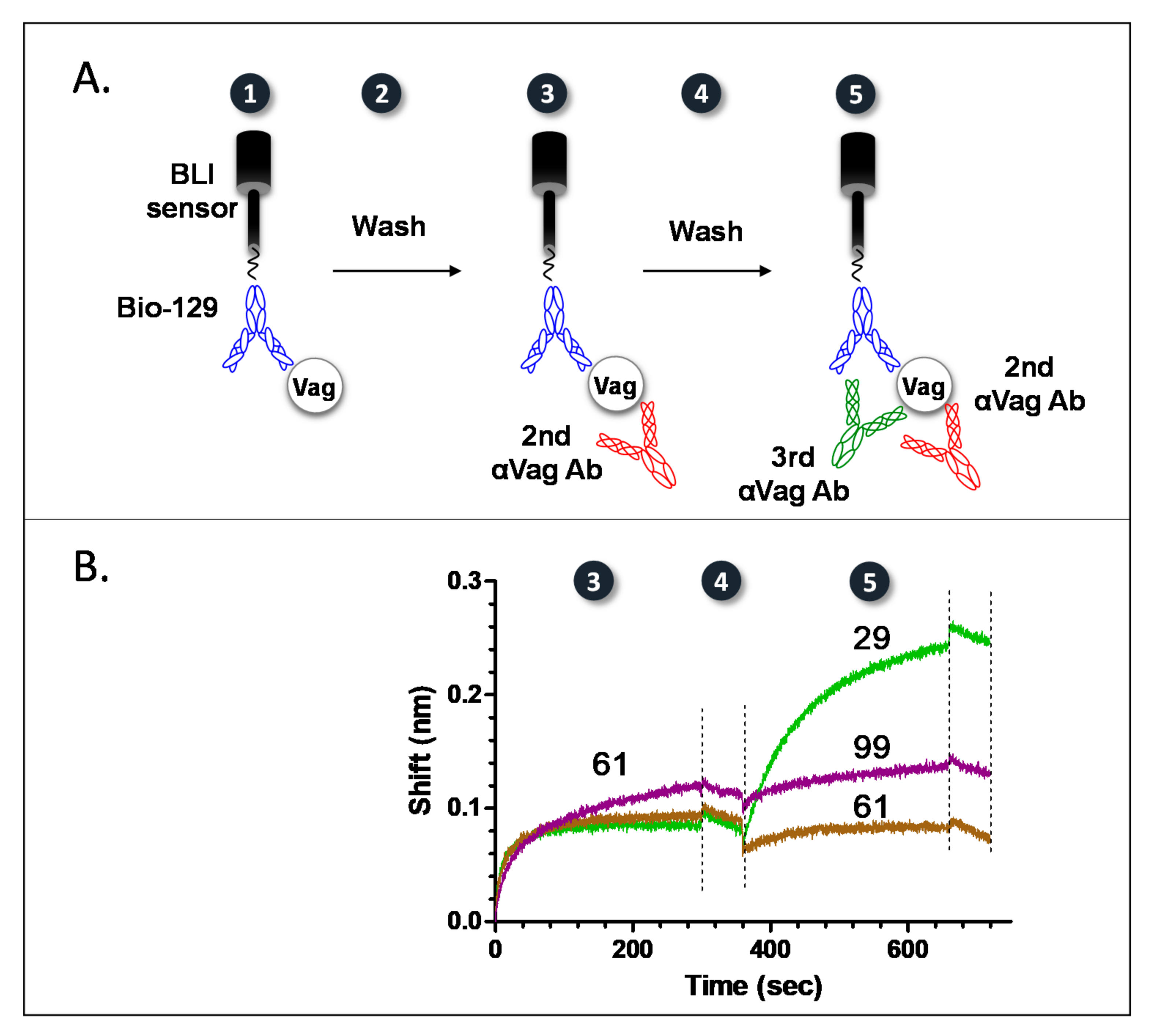
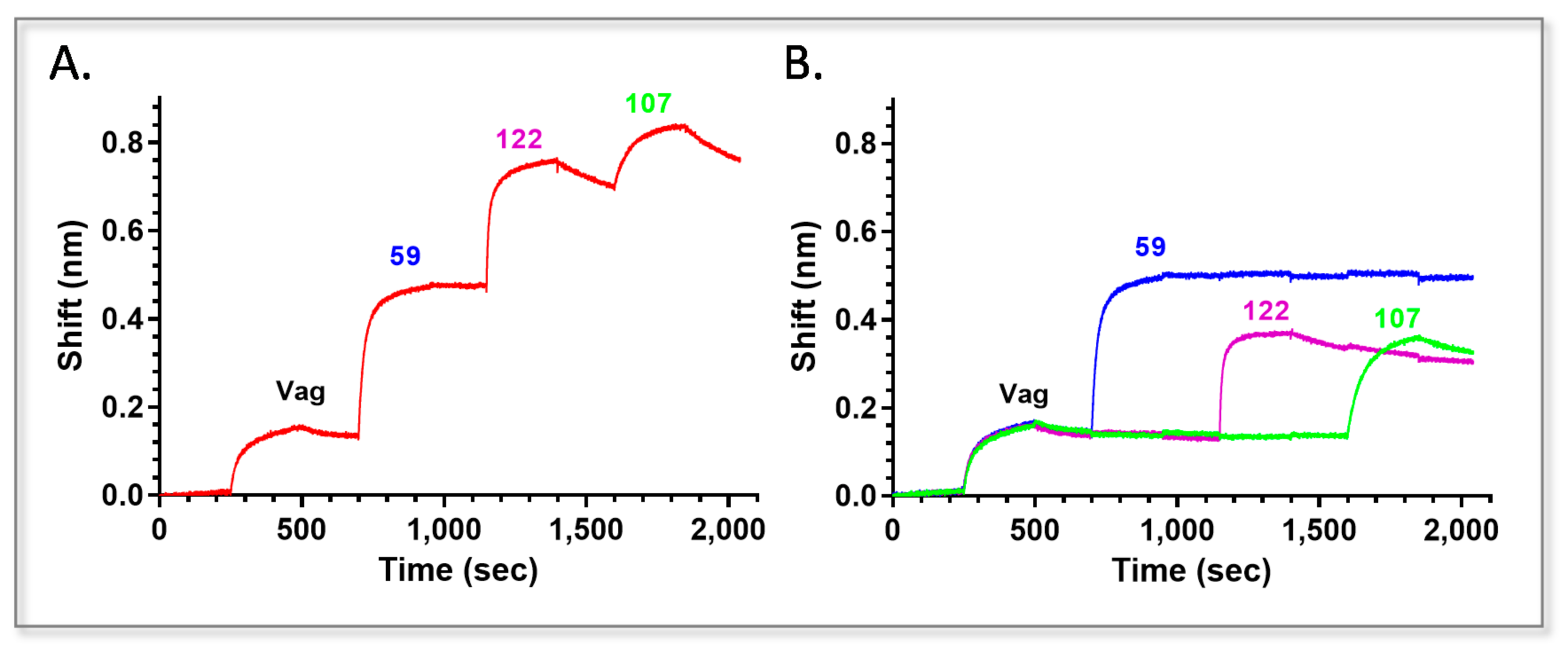
2.2. Epitope Binning of Anti-Vag Antibodies
2.3. Picking the Best Anti Vag Antibody from Each Bin
2.4. Characterization of Anti Vag Antibodies
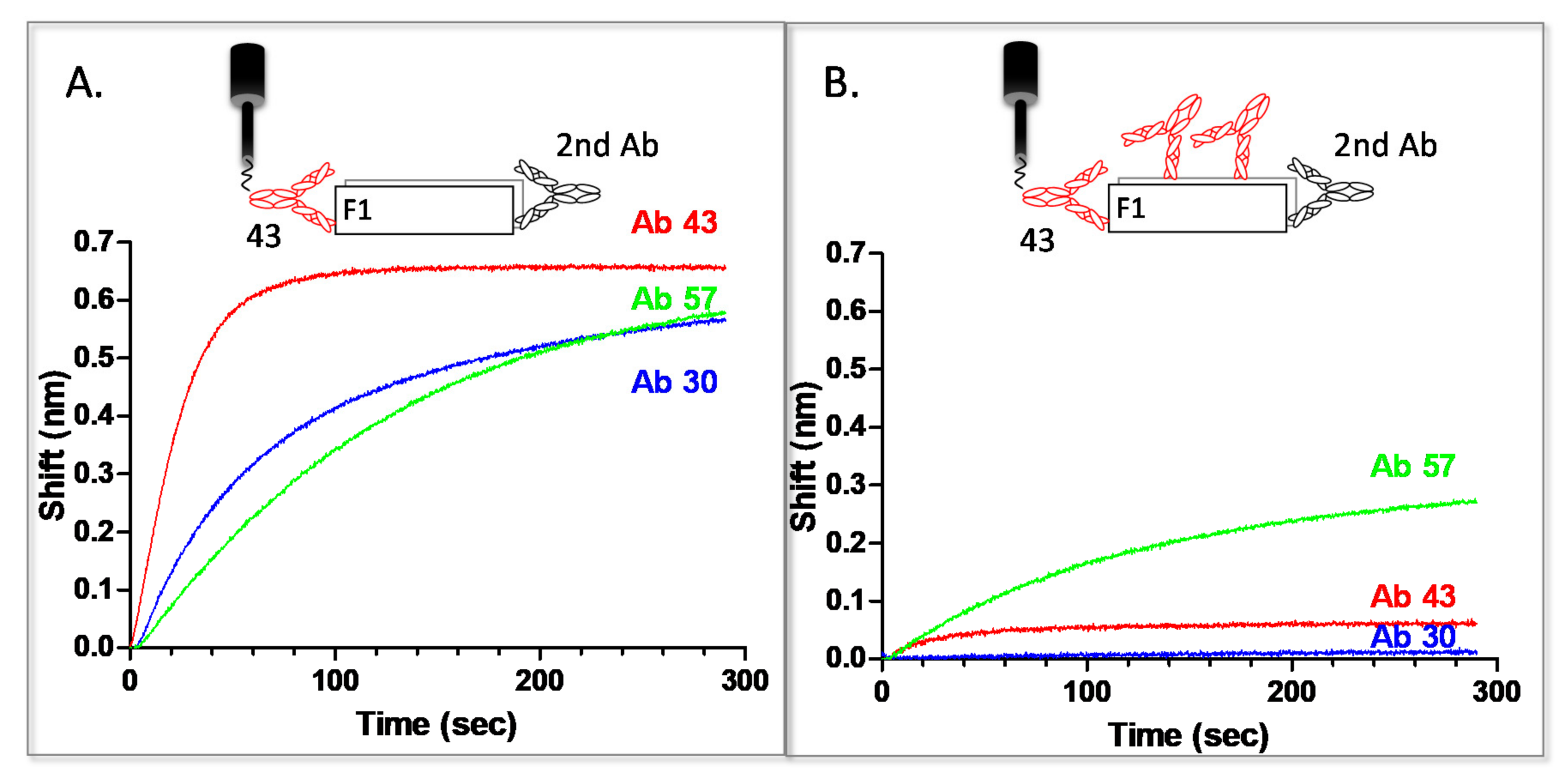
2.5. Epitope Binning of Anti-F1 Antibodies
2.6. HTRF Test for Antigen Detection
2.7. Detection of Disease Biomarkers from Inoculated Blood Cultures
2.8. Universality of Antigen Detection
2.9. Specificity of Antigen Detection
3. Discussion
4. Materials and Methods
4.1. Ethical Statement
4.2. Antigens
4.3. Strains
| Strains | Relevant Characteristics | Reference |
|---|---|---|
| Yersinia strains | ||
| Yersinia pestis | ||
| Kimberley53 (Kim53) | Virulent strain. Biovar Orientalis | [51] |
| Kimberley53 ∆pCD1∆pPCP1 | Spontaneously pPCP1 and pCD1-cured Kimberley53 | [52] |
| Bombay∆pCD1 | Biovar Orientalis | [53] |
| Alexander | Virulent strain. Biovar Orientalis | [54] |
| EV76 | pgm− (Girard’s strain). Biovar Orientalis | [55] |
| IV 75 195 | Virulent strain. | This study |
| KIMD27Δpgm | Biovar Mediaevelis | [56] |
| Yersinia pseudotuberculosis | 484337 | [57] |
| Yersinia enterocolitica | ||
| IP134 | O:3 | [58] |
| ATCC27729 | O:8 | [59] |
| IP383 | O:9 | |
| Other strains | ||
| E. coli | ATCC25922 | |
| P. aeruginosa | ATCC27853 | |
| S. typhimurium | ||
| B. anthracis | Vollum ATCC 14578 (Tox+ Cap+) | Bacillus Genetic Stock center |
| F. tularensis holarctica | ATCC29684 |
4.4. Immunization
4.5. Hybridoma Generation
4.6. Polyclonal Antibodies Generation
4.7. Antibody Labeling
4.8. Biolayer Interferometry
4.9. Preparation of Blood Cultures
4.10. Blood Culture Processing
4.11. HTRF Assays
4.12. Calculation of HTRF Signals
4.13. Safety Considerations
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Cohen, N.; Mechaly, A.; Zahavy, E. Energy transfer efficacy as a function of donor-acceptor distance in a homogenous time resolved fluorescence energy transfer (htrf) immunoassay for protective antigen detection. Luminescence 2012, 27, 543–545. [Google Scholar]
- Cohen, N.; Zahavy, E.; Zichel, R.; Fisher, M. An internal standard approach for homogeneous TR-FRET immunoassays facilitates the detection of bacteria, biomarkers, and toxins in complex matrices. Anal. Bioanal. Chem. 2016, 408, 5179–5188. [Google Scholar] [CrossRef] [PubMed]
- Förster, T. Zwischenmolekulare Energiewanderung und Fluoreszenz. Annalen der Physik 1948, 437, 55–75. [Google Scholar] [CrossRef]
- Zahavy, E.; Fisher, M.; Bromberg, A.; Olshevsky, U. Detection of frequency resonance energy transfer pair on double-labeled microsphere and Bacillus anthracis spores by flow cytometry. Appl. Environ. Microbiol. 2003, 69, 2330–2339. [Google Scholar] [CrossRef]
- Abdiche, Y.N.; Lindquist, K.C.; Stone, D.M.; Rajpal, A.; Pons, J. Label-free epitope binning assays of monoclonal antibodies enable the identification of antigen heterogeneity. J. Immunol. Methods 2012, 382, 101–116. [Google Scholar] [CrossRef]
- Abdiche, Y.N.; Miles, A.; Eckman, J.; Foletti, D.; Van Blarcom, T.J.; Yeung, Y.A.; Pons, J.; Rajpal, A. High-throughput epitope binning assays on label-free array-based biosensors can yield exquisite epitope discrimination that facilitates the selection of monoclonal antibodies with functional activity. PLoS ONE 2014, 9, e92451. [Google Scholar] [CrossRef]
- Estep, P.; Reid, F.; Nauman, C.; Liu, Y.; Sun, T.; Sun, J.; Xu, Y. High throughput solution-based measurement of antibody-antigen affinity and epitope binning. MAbs 2013, 5, 270–278. [Google Scholar] [CrossRef]
- Jia, X.C.; Raya, R.; Zhang, L.; Foord, O.; Walker, W.L.; Gallo, M.L.; Haak-Frendscho, M.; Green, L.L.; Davis, C.G. A novel method of Multiplexed Competitive Antibody Binning for the characterization of monoclonal antibodies. J. Immunol. Methods 2004, 288, 91–98. [Google Scholar] [CrossRef]
- Liu, S.; Zhang, H.; Dai, J.; Hu, S.; Pino, I.; Eichinger, D.J.; Lyu, H.; Zhu, H. Characterization of monoclonal antibody’s binding kinetics using oblique-incidence reflectivity difference approach. MAbs 2015, 7, 110–119. [Google Scholar] [CrossRef]
- Miller, P.L.; Wolfert, R.L.; Diedrich, G. Epitope binning of murine monoclonal antibodies by a multiplexed pairing assay. J. Immunol. Methods 2011, 365, 118–125. [Google Scholar] [CrossRef]
- Scott, N.; Qazi, O.; Wright, M.J.; Fairweather, N.F.; Deonarain, M.P. Characterization of a panel of anti-tetanus toxin single-chain Fvs reveals cooperative binding. Mol. Immunol. 2010, 47, 1931–1941. [Google Scholar] [CrossRef]
- Perry, R.D.; Fetherston, J.D. Yersinia pestis—Etiological agent of plague. Clin. Microbiol. Rev. 1997, 10, 35–66. [Google Scholar] [CrossRef] [PubMed]
- Inglesby, T.V.; Dennis, D.T.; Henderson, D.A.; Bartlett, J.G.; Ascher, M.S.; Eitzen, E.; Fine, A.D.; Friedlander, A.M.; Hauer, J.; Koerner, J.F.; et al. Plague as a biological weapon: Medical and public health management. Working Group on Civilian Biodefense. JAMA 2000, 283, 2281–2290. [Google Scholar] [CrossRef] [PubMed]
- Chanteau, S.; Rahalison, L.; Ratsitorahina, M.; Mahafaly; Rasolomaharo, M.; Boisier, P.; O’Brien, T.; Aldrich, J.; Keleher, A.; Morgan, C.; et al. Early diagnosis of bubonic plague using F1 antigen capture ELISA assay and rapid immunogold dipstick. Int. J. Med. Microbiol. 2000, 290, 279–283. [Google Scholar] [CrossRef]
- Splettstoesser, W.D.; Rahalison, L.; Grunow, R.; Neubauer, H.; Chanteau, S. Evaluation of a standardized F1 capsular antigen capture ELISA test kit for the rapid diagnosis of plague. FEMS Immunol. Med. Microbiol. 2004, 41, 149–155. [Google Scholar] [CrossRef][Green Version]
- Williams, J.E.; Arntzen, L.; Tyndal, G.L.; Isaacson, M. Application of enzyme immunoassays for the confirmation of clinically suspect plague in Namibia. Bull. WHO 1986, 64, 745–752. [Google Scholar] [PubMed]
- Rajerison, M.; Melocco, M.; Andrianaivoarimanana, V.; Rahajandraibe, S.; Rakotoarimanana, F.; Spiegel, A.; Ratsitorahina, M.; Baril, L. Performance of plague rapid diagnostic test compared to bacteriology: A retrospective analysis of the data collected in Madagascar. BMC Infect. Dis. 2020, 20, 90. [Google Scholar] [CrossRef]
- Flashner, Y.; Fisher, M.; Tidhar, A.; Mechaly, A.; Gur, D.; Gideon, H.; Zahavy, E.; Mamroud, E.; Cohen, S. The search for early markers of plague: Evidence for accumulation of soluble Yersinia pestis LcrV in bubonic and pneumonic mouse models of disease. FEMS Immunol. Med. Microbiol. 2010, 59, 197–206. [Google Scholar] [CrossRef]
- Anisimov, A.P.; Lindler, L.E.; Pier, G.B. Intraspecific diversity of Yersinia pestis. Clin. Microbiol. Rev. 2004, 17, 434–464. [Google Scholar] [CrossRef]
- Davies, K.J.; Fritz, D.L.; Pitt, M.L.M.; Welkos, S.L.; Worsham, P.L.; Friedlander, A.M. Pathology of experimental pneumonic plague produced by F1-positive and F1-negative Yersinia pestis in african green monkeys. Arch. Pathol. 1996, 120, 156–163. [Google Scholar]
- Williams, J.E.; Harrison, D.N.; Quan, T.J.; Mullins, J.L.; Barnes, A.M.; Cavanaugh, D.C. Atypical plague bacilli isolated from rodents, fleas, and man. Am. J. Public Health 1978, 68, 262–264. [Google Scholar] [CrossRef]
- Winter, C.C.; Cherry, W.B.; Moody, M.D. An unusual strain of Pasteurella pestis isolated from a fatal human case of plague. B World Health Organ. 1960, 23, 408–409. [Google Scholar]
- Sittner, A.; Mechaly, A.; Vitner, E.; Aftalion, M.; Levy, Y.; Levy, H.; Mamroud, E.; Fisher, M. Improved production of monoclonal antibodies against the LcrV antigen of Yersinia pestis using FACS-aided hybridoma selection. J. Biol. Meth. 2018, 5, e100. [Google Scholar] [CrossRef] [PubMed]
- Lad, L.; Clancy, S.; Kovalenko, M.; Liu, C.; Hui, T.; Smith, V.; Pagratis, N. High-throughput kinetic screening of hybridomas to identify high-affinity antibodies using bio-layer interferometry. J. Biomol. Screen 2015, 20, 498–507. [Google Scholar] [CrossRef] [PubMed]
- Ylera, F.; Harth, S.; Waldherr, D.; Frisch, C.; Knappik, A. Off-rate screening for selection of high-affinity anti-drug antibodies. Anal. Biochem. 2013, 441, 208–213. [Google Scholar] [CrossRef] [PubMed]
- Andrews, G.P.; Heath, D.G.; Anderson, G.W., Jr.; Welkos, S.L.; Friedlander, A.M. Fraction 1 capsular antigen (F1) purification from Yersinia pestis CO92 and from an Escherichia coli recombinant strain and efficacy against lethal plague challenge. Infect. Immun. 1996, 64, 2180–2187. [Google Scholar] [CrossRef]
- Zavialov, A.V.; Berglund, J.; Pudney, A.F.; Fooks, L.J.; Ibrahim, T.M.; MacIntyre, S.; Knight, S.D. Structure and biogenesis of the capsular F1 antigen from Yersinia pestis: Preserved folding energy drives fiber formation. Cell 2003, 113, 587–596. [Google Scholar] [CrossRef]
- Cohen, N.; Mechaly, A.; Mazor, O.; Fisher, M.; Zahavy, E. Rapid homogenous time-resolved fluorescence (HTRF) immunoassay for anthrax detection. J. Fluoresc. 2014, 24, 795–801. [Google Scholar] [CrossRef]
- Kopylov, P.K.; Svetoch, T.E.; Ivanov, S.A.; Kombarova, T.I.; Perovskaya, O.N.; Titareva, G.M.; Anisimov, A.P. Characteristics of the Chromatographic Cleaning and Protectiveness of the LcrV Isoform of Yersinia pestis. Appl. Biochem. Microbiol. 2019, 55, 524–533. [Google Scholar] [CrossRef]
- Cao, L.K.; Anderson, G.P.; Ligler, F.S.; Ezzell, J. Detection of Yersinia pestis fraction 1 antigen with a fiber optic biosensor. J. Clin. Microbiol. 1995, 33, 336–341. [Google Scholar] [CrossRef] [PubMed]
- Chanteau, S.; Rabarijaona, L.; Hager, J.; Boisier, P.; Burans, J.; Rasolomaharo, M. F1 antigenemia in bubonic plague patients, a marker of gravity and efficacy of treatment. Trans. R Soc. Trop. Med. Hyg. 1998, 92, 572–573. [Google Scholar] [CrossRef]
- Gomes-Solecki, M.J.; Savitt, A.G.; Rowehl, R.; Glass, J.D.; Bliska, J.B.; Dattwyler, R.J. LcrV capture enzyme-linked immunosorbent assay for detection of Yersinia pestis from human samples. Clin. Diagn. Lab. Immunol. 2005, 12, 339–346. [Google Scholar] [CrossRef]
- Hill, J.; Copse, C.; Leary, S.; Stagg, A.J.; Williamson, E.D.; Titball, R.W. Synergistic protection of mice against plague with monoclonal antibodies specific for the F1 and V antigens of Yersinia pestis. Infect. Immun. 2003, 71, 2234–2238. [Google Scholar] [CrossRef]
- Ivashchenko, T.A.; Belova, E.V.; Dentovskaia, S.V.; Belkova, S.A.; Balakhonov, S.V.; Ignatov, S.G.; Shemiakin, I.G. Development and testing of an enzyme immunoassay-based monoclonal test system for the detection of the Yersinia pestis V antigen. Prikl. Biokhimiia Mikrobiol. 2014, 50, 211–218. [Google Scholar]
- Lillo, A.M.; Ayriss, J.E.; Shou, Y.; Graves, S.W.; Bradbury, A.R.; Pavlik, P. Development of phage-based single chain Fv antibody reagents for detection of Yersinia pestis. PLoS ONE 2011, 6, e27756. [Google Scholar] [CrossRef] [PubMed]
- Quenee, L.E.; Berube, B.J.; Segal, J.; Elli, D.; Ciletti, N.A.; Anderson, D.; Schneewind, O. Amino acid residues 196-225 of LcrV represent a plague protective epitope. Vaccine 2010, 28, 1870–1876. [Google Scholar] [CrossRef][Green Version]
- Read, T.; Olkhov, R.V.; Williamson, E.D.; Shaw, A.M. Kinetic epitope mapping of monoclonal antibodies raised against the Yersinia pestis virulence factor LcrV. Biosens. Bioelectron. 2015, 65, 47–53. [Google Scholar] [CrossRef]
- Tomaso, H.; Thullier, P.; Seibold, E.; Guglielmo, V.; Buckendahl, A.; Rahalison, L.; Neubauer, H.; Scholz, H.C.; Splettstoesser, W.D. Comparison of hand-held test kits, immunofluorescence microscopy, enzyme-linked immunosorbent assay, and flow cytometric analysis for rapid presumptive identification of Yersinia pestis. J. Clin. Microbiol. 2007, 45, 3404–3407. [Google Scholar] [CrossRef]
- Xiao, X.; Zhu, Z.; Dankmeyer, J.L.; Wormald, M.M.; Fast, R.L.; Worsham, P.L.; Cote, C.K.; Amemiya, K.; Dimitrov, D.S. Human anti-plague monoclonal antibodies protect mice from Yersinia pestis in a bubonic plague model. PLoS ONE 2010, 5, e13047. [Google Scholar] [CrossRef]
- Chanteau, S.; Rahalison, L.; Ralafiarisoa, L.; Foulon, J.; Ratsitorahina, M.; Ratsifasoamanana, L.; Carniel, E.; Nato, F. Development and testing of a rapid diagnostic test for bubonic and pneumonic plague. Lancet 2003, 361, 211–216. [Google Scholar] [CrossRef]
- Cohen, O.; Mechaly, A.; Sabo, T.; Alcalay, R.; Aloni-Grinstein, R.; Seliger, N.; Kronman, C.; Mazor, O. Characterization and epitope mapping of the polyclonal antibody repertoire elicited by ricin holotoxin-based vaccination. Clin. Vaccine Immunol. 2014, 21, 1534–1540. [Google Scholar] [CrossRef] [PubMed]
- Noy-Porat, T.; Rosenfeld, R.; Ariel, N.; Epstein, E.; Alcalay, R.; Zvi, A.; Kronman, C.; Ordentlich, A.; Mazor, O. Isolation of Anti-Ricin protective antibodies exibiting high affinity from immunized non-human primates. Toxins 2016, 8, 64. [Google Scholar] [CrossRef] [PubMed]
- Mechaly, A.; Alcalay, R.; Noy-Porat, T.; Epstein, E.; Gal, Y.; Mazor, O. Novel Phage Display-Derived Anti-Abrin Antibodies Confer Post-Exposure Protection against Abrin Intoxication. Toxins 2018, 10, 80. [Google Scholar] [CrossRef]
- Meyer, M.H.; Stehr, M.; Bhuju, S.; Krause, H.J.; Hartmann, M.; Miethe, P.; Singh, M.; Keusgen, M. Magnetic biosensor for the detection of Yersinia pestis. J. Microbiol. Methods 2007, 68, 218–224. [Google Scholar] [CrossRef]
- Nikitin, P.I.; Vetoshko, P.M.; Ksenevich, T.I. New type of biosensor based on magnetic nanoparticle detection. J. Magn. Magn. Mater. 2007, 311, 445–449. [Google Scholar] [CrossRef]
- Mechaly, A.; Marx, S.; Levy, O.; Yitzhaki, S.; Fisher, M. Highly Stable Lyophilized Homogeneous Bead-Based Immunoassays for On-Site Detection of Bio Warfare Agents from Complex Matrices. Anal. Chem. 2016, 88, 6283–6291. [Google Scholar] [CrossRef]
- Levy, Y.; Flashner, Y.; Tidhar, A.; Zauberman, A.; Aftalion, M.; Lazar, S.; Gur, D.; Shafferman, A.; Mamroud, E. T cells play an essential role in anti-F1 mediated rapid protection against bubonic plague. Vaccine 2011, 29, 6866–6873. [Google Scholar] [CrossRef]
- Holtzman, T.; Levy, Y.; Marcus, D.; Flashner, Y.; Mamroud, E.; Cohen, S.; Fass, R. Production and purification of high molecular weight oligomers of Yersinia pestis F1 capsular antigen released by high cell density culture of recombinant Escherichia coli cells carrying the caf1 operon. Microb. Cell Factories 2006, 5, P98. [Google Scholar] [CrossRef]
- Leary, S.E.; Williamson, E.D.; Griffin, K.F.; Russell, P.; Eley, S.M.; Titball, R.W. Active immunization with recombinant V antigen from Yersinia pestis protects mice against plague. Infect. Immun. 1995, 63, 2854–2858. [Google Scholar] [CrossRef]
- Petsch, D.; Anspach, F.B. Endotoxin removal from protein solutions. J. Biotechnol. 2000, 76, 97–119. [Google Scholar] [CrossRef]
- Hertman, I.; Ben-Gurion, R. A study on pesticin biosynthesis. J. Gen. Microbiol. 1959, 21, 135–143. [Google Scholar] [CrossRef]
- Vagima, Y.; Zauberman, A.; Levy, Y.; Gur, D.; Tidhar, A.; Aftalion, M.; Shafferman, A.; Mamroud, E. Circumventing Y. pestis Virulence by Early Recruitment of Neutrophils to the Lungs during Pneumonic Plague. PLoS Pathog. 2015, 11, e1004893. [Google Scholar] [CrossRef]
- Ben-Gurion, R.; Hertman, I. Bacteriocin-like material produced by Pasteurella pestis. J. Gen. Microbiol. 1958, 19, 289–297. [Google Scholar] [CrossRef] [PubMed]
- Markenson, J.; Ben-Efraim, S. Behavior in vivo of strains of Pasteurella pestis in relation to their efficacy as live vaccines. Annales de L’institut Pasteur 1969, 117, 196–212. [Google Scholar] [PubMed]
- Zauberman, A.; Gur, D.; Levy, Y.; Aftalion, M.; Vagima, Y.; Tidhar, A.; Chitlaru, T.; Mamroud, E. Postexposure Administration of a Yersinia pestis Live Vaccine for Potentiation of Second-Line Antibiotic Treatment Against Pneumonic Plague. J. Infect. Dis. 2019, 220, 1147–1151. [Google Scholar] [CrossRef] [PubMed]
- Garcia, E.; Nedialkov, Y.A.; Elliott, J.; Motin, V.L.; Brubaker, R.R. Molecular characterization of KatY (antigen 5), a thermoregulated chromosomally encoded catalase-peroxidase of Yersinia pestis. J. Bacteriol. 1999, 181, 3114–3122. [Google Scholar] [CrossRef] [PubMed]
- Shwimmer, A.; Freed, M.; Blum, S.; Khatib, N.; Weissblit, L.; Friedman, S.; Elad, D. Mastitis caused by Yersinia pseudotuberculosis in Israeli dairy cattle and public health implications. Zoonoses Public Health 2007, 54, 353–357. [Google Scholar] [CrossRef]
- Saraka, D.; Savin, C.; Kouassi, S.; Cissé, B.; Koffi, E.; Cabanel, N.; Brémont, S.; Faye-Kette, H.; Dosso, M.; Carniel, E. Yersinia enterocolitica, a Neglected Cause of Human Enteric Infections in Côte d’Ivoire. PLoS Negl. Trop. Dis. 2017, 11, e0005216. [Google Scholar] [CrossRef] [PubMed]
- Zauberman, A.; Velan, B.; Mamroud, E.; Flashner, Y.; Shafferman, A.; Cohen, S. Disparity between Yersinia pestis and Yersinia enterocolitica O:8 in YopJ/YopP-dependent functions. Adv. Exp. Med. Biol. 2007, 603, 312–320. [Google Scholar] [CrossRef]
- Kohler, G.; Milstein, C. Continuous cultures of fused cells secreting antibody of predefined specificity. Nature 1975, 256, 495–497. [Google Scholar] [CrossRef] [PubMed]
- Levy, Y.; Flashner, Y.; Zauberman, A.; Tidhar, A.; Aftalion, M.; Lazar, S.; Gur, D.; Cohen, S.; Shafferman, A.; Mamroud, E. Protection Against Plague Afforded by Treatment with Polyclonal αLcrV and αF1 Antibodies. In The Challenge of Highly Pathogenic Microorganism; Shafferman, A., Ordentlich, A., Velan, B., Eds.; Springer: Dordrecht, The Netherlands, 2010; pp. 269–274. [Google Scholar]
- Mechaly, A.; Vitner, E.; Levy, H.; Weiss, S.; Bar-David, E.; Gur, D.; Koren, M.; Cohen, H.; Cohen, O.; Mamroud, E.; et al. Simultaneous immunodetection of anthrax, plague and tularemia from blood cultures utilizing multiplexed suspension arrays. J. Clin. Microbiol. 2018. [Google Scholar] [CrossRef] [PubMed]
- Knepp, A.M.; Grunbeck, A.; Banerjee, S.; Sakmar, T.P.; Huber, T. Direct measurement of thermal stability of expressed CCR5 and stabilization by small molecule ligands. Biochemistry 2011, 50, 502–511. [Google Scholar] [CrossRef] [PubMed]
- ICH Guideline ICHHT Validation of Analytical Procedures: Methodology; EMEA: Amsterdam, The Netherlands, 1996.


| Ab | Kon (1/M·s) | Koff (1/s) | KD (nM) |
|---|---|---|---|
| 59 | 1.4 ± 0.3 × 106 | 3.6 ± 0.4 × 10−4 | 0.3 ± 0.2 |
| 122 | 1.2 ± 0.03 × 106 | 1.1 ± 0.4 × 10−3 | 0.9 ± 0.2 |
| 38 | 1.6 ± 0.09 × 106 | 4.4 ± 0.8 × 10−3 | 2.8 ± 0.5 |
| 107 | 1.9 ± 0.5 × 105 | 6.0 ± 0.3 × 10−4 | 3.1 ± 1.0 |
| Y. pestis Strains | Biovar | Bacterial Concentration (cfu/mL) | F1 | Vag | Estimated Antigen Concentration | |
|---|---|---|---|---|---|---|
| ΔF F1 | ΔF Vag | F1 (ng/mL) | Vag (ng/mL) | |||
| EV 76 | Orientalis | 2.5 × 107 | 28.1 | 19.2 | 876 | 485 |
| Kimberly53 * | 4.7 × 106 | 90 | 10 | 1000 | 248 | |
| 2.0 × 107 | 274.4 | 9.8 | 1504 | 242 | ||
| Kim53ΔpCD1ΔpPCP1** | 2.0 × 107 | 2262 | - | 15,637 | - | |
| BombayΔpCD1 ** | 1.3 × 108 | 233.9 | - | 1216 | - | |
| Alexander * | 2.3 × 106 | 14.4 | 4.9 | 388 | 115 | |
| IV 75 195 * | 1.2 × 106 | 2.7 | 1.1 | 73 | 15 | |
| KIMD27Δpgm | Mediaevalis | 1.4 × 108 | 4.1 | 0.6 | 21 | 2 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mechaly, A.; Vitner, E.B.; Levy, Y.; Gur, D.; Barlev-Gross, M.; Sittner, A.; Koren, M.; Levy, H.; Mamroud, E.; Fisher, M. Epitope Binning of Novel Monoclonal Anti F1 and Anti LcrV Antibodies and Their Application in a Simple, Short, HTRF Test for Clinical Plague Detection. Pathogens 2021, 10, 285. https://doi.org/10.3390/pathogens10030285
Mechaly A, Vitner EB, Levy Y, Gur D, Barlev-Gross M, Sittner A, Koren M, Levy H, Mamroud E, Fisher M. Epitope Binning of Novel Monoclonal Anti F1 and Anti LcrV Antibodies and Their Application in a Simple, Short, HTRF Test for Clinical Plague Detection. Pathogens. 2021; 10(3):285. https://doi.org/10.3390/pathogens10030285
Chicago/Turabian StyleMechaly, Adva, Einat B. Vitner, Yinon Levy, David Gur, Moria Barlev-Gross, Assa Sittner, Michal Koren, Haim Levy, Emanuelle Mamroud, and Morly Fisher. 2021. "Epitope Binning of Novel Monoclonal Anti F1 and Anti LcrV Antibodies and Their Application in a Simple, Short, HTRF Test for Clinical Plague Detection" Pathogens 10, no. 3: 285. https://doi.org/10.3390/pathogens10030285
APA StyleMechaly, A., Vitner, E. B., Levy, Y., Gur, D., Barlev-Gross, M., Sittner, A., Koren, M., Levy, H., Mamroud, E., & Fisher, M. (2021). Epitope Binning of Novel Monoclonal Anti F1 and Anti LcrV Antibodies and Their Application in a Simple, Short, HTRF Test for Clinical Plague Detection. Pathogens, 10(3), 285. https://doi.org/10.3390/pathogens10030285






