First Detection of the West Nile Virus Koutango Lineage in Sandflies in Niger
Abstract
1. Introduction
2. Results
2.1. Arthropod Species
2.2. Virus Detection
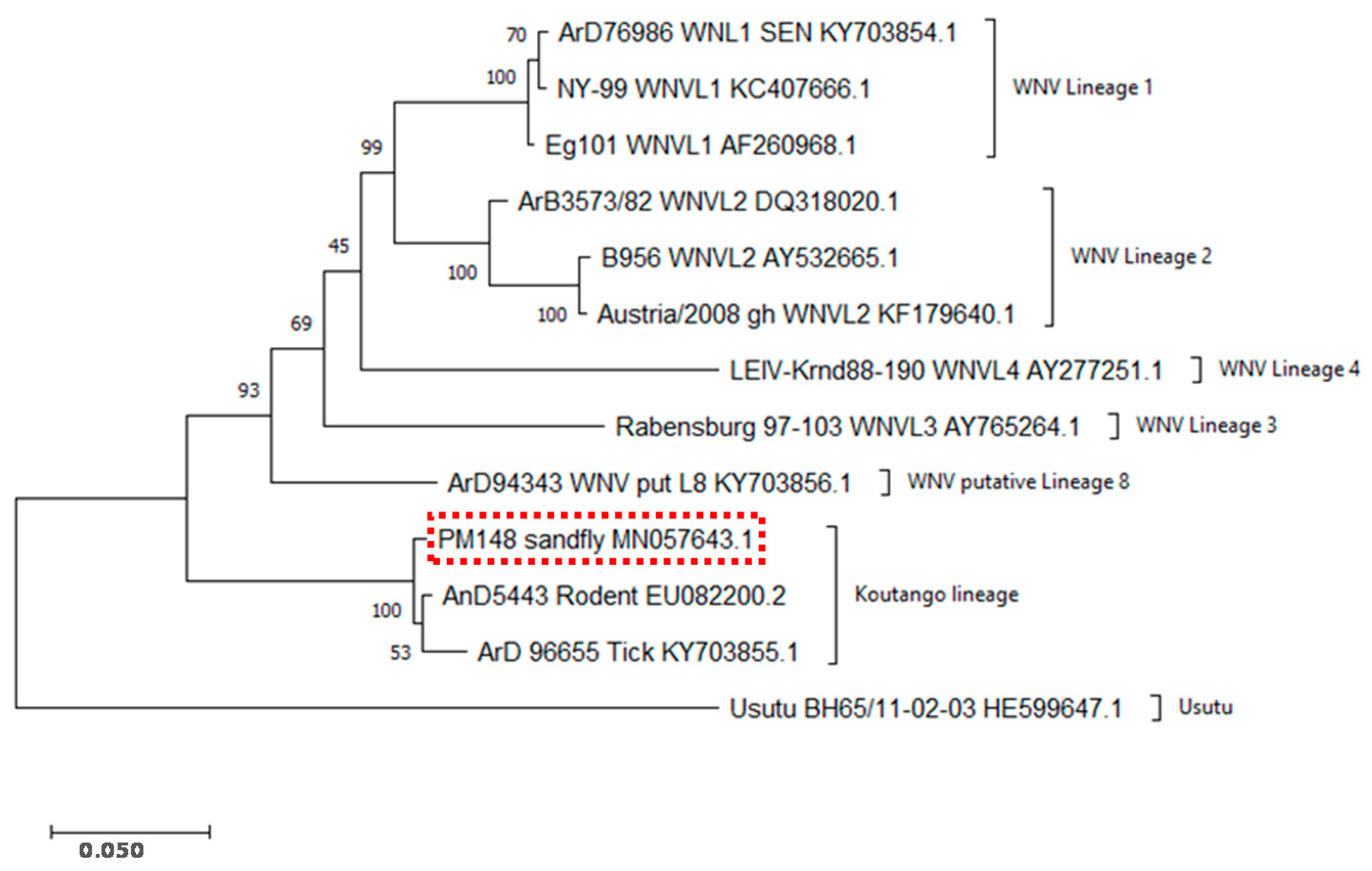
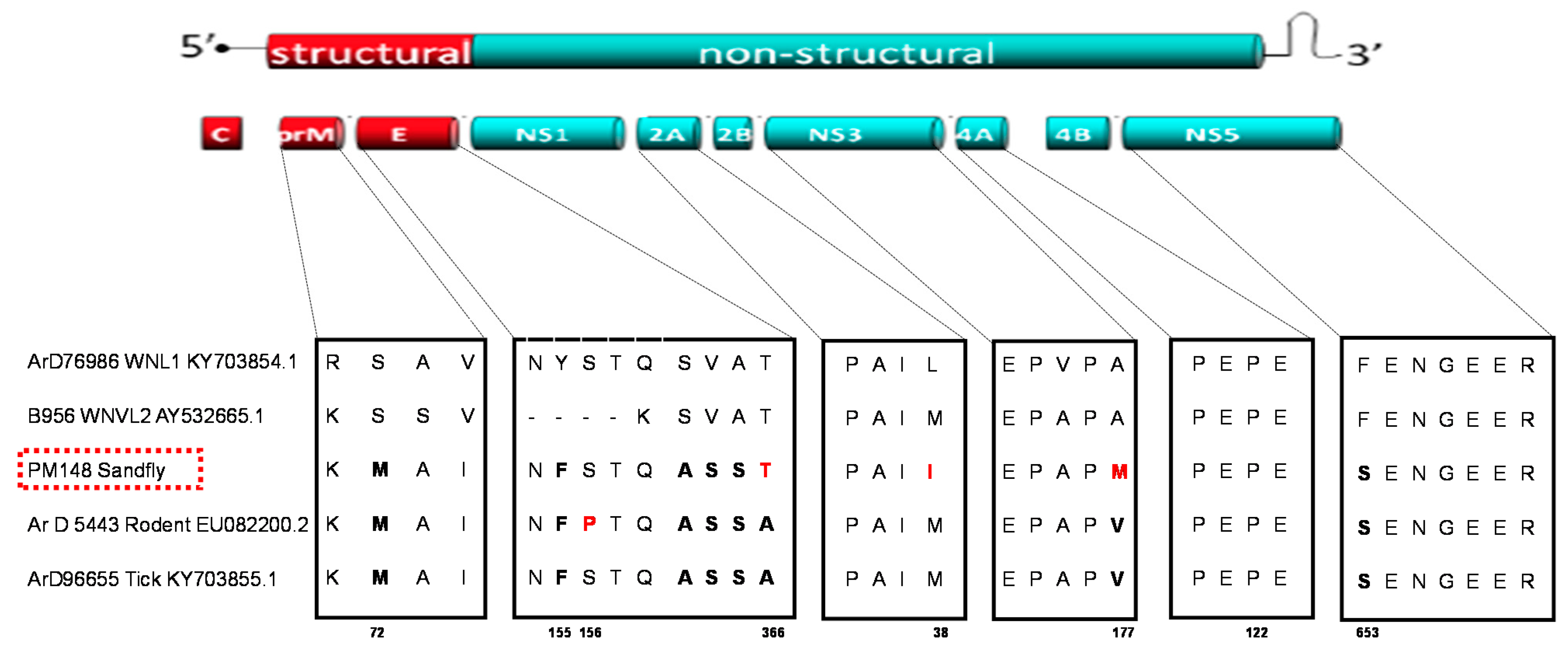
2.3. Sequencing and Evolutionary Analyses
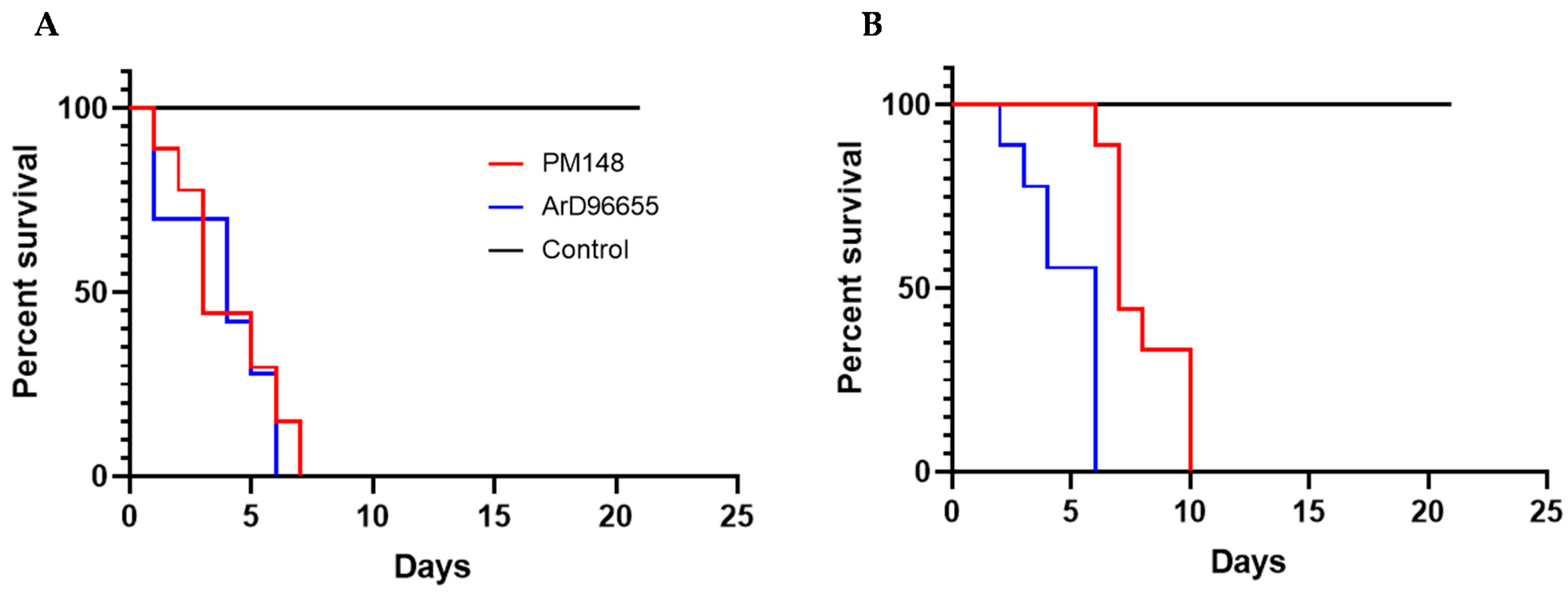
2.4. In Vivo Characterization
3. Discussion
4. Materials and Methods
4.1. Collection and Processing of Arthropods
4.2. Virus Isolation
4.3. RT-PCR and Titration
4.4. Viral Sequencing and Phylogenetic Analyses
4.5. In Vivo Characterization in Mice
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Hubálek, Z.; Halouzka, J. West Nile Fever—A Reemerging Mosquito-Borne Viral Disease in Europe. Emerg. Infect. Dis. 1999, 5, 643–650. [Google Scholar] [CrossRef]
- Rappole, J.H.; Hubálek, Z. Migratory Birds and West Nile Virus. J. Appl. Microbiol. 2003, 94, 47S–58S. [Google Scholar] [CrossRef]
- Hayes, E.B.; Komar, N.; Nasci, R.S.; Montgomery, S.P.; O’Leary, D.R.; Campbell, G.L. Epidemiology and Transmission Dynamics of West Nile Virus Disease. Emerg. Infect. Dis. 2005, 11, 1167–1173. [Google Scholar] [CrossRef]
- Kulasekera, V.L.; Kramer, L.; Nasci, R.S.; Mostashari, F.; Cherry, B.; Trock, S.C.; Glaser, C.; Miller, J.R. West Nile Virus Infection in Mosquitoes, Birds, Horses, and Humans, Staten Island, New York, 2000. Emerg. Infect. Dis. 2001, 7, 722–725. [Google Scholar] [CrossRef]
- DeGroote, J.P.; Sugumaran, R.; Ecker, M. Landscape, Demographic and Climatic Associations with Human West Nile Virus Occurrence Regionally in 2012 in the United States of America. Geospat. Health 2014, 9, 153–168. [Google Scholar] [CrossRef]
- Hernández-Triana, L.M.; Jeffries, C.L.; Mansfield, K.L.; Carnell, G.; Fooks, A.R.; Johnson, N. Emergence of West Nile Virus Lineage 2 in Europe: A Review on the Introduction and Spread of a Mosquito-Borne Disease. Front. Public Health 2014, 2, 271. [Google Scholar] [CrossRef] [PubMed]
- Vilibic-Cavlek, T.; Kaic, B.; Barbic, L.; Pem-Novosel, I.; Slavic-Vrzic, V.; Lesnikar, V.; Kurecic-Filipovic, S.; Babic-Erceg, A.; Listes, E.; Stevanovic, V.; et al. First Evidence of Simultaneous Occurrence of West Nile Virus and Usutu Virus Neuroinvasive Disease in Humans in Croatia during the 2013 Outbreak. Infection 2014, 42, 689–695. [Google Scholar] [CrossRef] [PubMed]
- Cox, S.L.; Campbell, G.D.; Nemeth, N.M. Outbreaks of West Nile Virus in Captive Waterfowl in Ontario, Canada. Avian Pathol. J. WVPA 2015, 44, 135–141. [Google Scholar] [CrossRef] [PubMed]
- Fall, G.; Diallo, M.; Loucoubar, C.; Faye, O.; Sall, A.A. Vector Competence of Culex Neavei and Culex Quinquefasciatus (Diptera: Culicidae) from Senegal for Lineages 1, 2, Koutango and a Putative New Lineage of West Nile Virus. Am. J. Trop. Med. Hyg. 2014, 90, 747–754. [Google Scholar] [CrossRef]
- Murray, K.O.; Mertens, E.; Despres, P. West Nile Virus and Its Emergence in the United States of America. Vet. Res. 2010, 41, 67. [Google Scholar] [CrossRef]
- Anukumar, B.; Sapkal, G.N.; Tandale, B.V.; Balasubramanian, R.; Gangale, D. West Nile Encephalitis Outbreak in Kerala, India, 2011. J. Clin. Virol. 2014, 61, 152–155. [Google Scholar] [CrossRef] [PubMed]
- Bakonyi, T.; Ivanics, E.; Erdélyi, K.; Ursu, K.; Ferenczi, E.; Weissenböck, H.; Nowotny, N. Lineage 1 and 2 Strains of Encephalitic West Nile Virus, Central Europe. Emerg. Infect. Dis. 2006, 12, 618–623. [Google Scholar] [CrossRef] [PubMed]
- Coz, J.; Le Gonidec, G.; Cornet, M.; Valade, M.; Lemoine, M.; Gueye, A. Transmission Experimentale d’un Arbovirus Du Groupe B, Le Virus Koutango Par Aedes aegypti L. Cah. ORSTOM Ser. Ent. Med. Parasitol. 1975, 13, 57–62. [Google Scholar]
- Butenko, A.M.; Semashko, I.V.; Skvortsova, T.M.; Gromashevskiĭ, V.L.; Kondrashina, N.G. Detection of the Koutango virus (Flavivirus, Togaviridae) in Somalia. Med. Parazitol. (Mosk.) 1986, 65–68. Available online: https://pubmed.ncbi.nlm.nih.gov/3018465/ (accessed on 22 February 2021).
- Charrel, R.N.; Brault, A.C.; Gallian, P.; Lemasson, J.-J.; Murgue, B.; Murri, S.; Pastorino, B.; Zeller, H.; de Chesse, R.; de Micco, P.; et al. Evolutionary Relationship between Old World West Nile Virus Strains. Evidence for Viral Gene Flow between Africa, the Middle East, and Europe. Virology 2003, 315, 381–388. [Google Scholar] [CrossRef]
- Hall, R.A.; Scherret, J.H.; Mackenzie, J.S. Kunjin Virus: An Australian Variant of West Nile? Ann. N. Y. Acad. Sci. 2001, 951, 153–160. [Google Scholar] [CrossRef] [PubMed]
- Fall, G.; Di Paola, N.; Faye, M.; Dia, M.; de Melo Freire, C.C.; Loucoubar, C.; de Andrade Zanotto, P.M.; Faye, O. Biological and Phylogenetic Characteristics of West African Lineages of West Nile Virus. PLoS Negl. Trop. Dis. 2017, 11, e0006078. [Google Scholar] [CrossRef]
- Jan, C.; Languillat, G.; Renaudet, J.; Robin, Y. A serological survey of arboviruses in Gabon. Bull. Soc. Pathol. Exot. Fil. 1978, 71, 140–146. [Google Scholar]
- Shope, R.E. Epidemiology of Other Arthropod-Borne Flaviviruses Infecting Humans. Adv. Virus Res. 2003, 61, 373–391. [Google Scholar] [CrossRef]
- de Araujo Lobo, J. Koutango: Under Reported Arboviral Disease in West Africa. Ph.D. Thesis, Louisiana State University, Baton Rouge, LA, USA, 2012. [Google Scholar]
- Pérez-Ramírez, E.; Llorente, F.; Del Amo, J.; Fall, G.; Sall, A.A.; Lubisi, A.; Lecollinet, S.; Vázquez, A.; Jiménez-Clavero, M.Á. Pathogenicity Evaluation of Twelve West Nile Virus Strains Belonging to Four Lineages from Five Continents in a Mouse Model: Discrimination between Three Pathogenicity Categories. J. Gen. Virol. 2017, 98, 662–670. [Google Scholar] [CrossRef]
- Prow, N.A.; Setoh, Y.X.; Biron, R.M.; Sester, D.P.; Kim, K.S.; Hobson-Peters, J.; Hall, R.A.; Bielefeldt-Ohmann, H. The West Nile Virus-like Flavivirus Koutango Is Highly Virulent in Mice Due to Delayed Viral Clearance and the Induction of a Poor Neutralizing Antibody Response. J. Virol. 2014, 88, 9947–9962. [Google Scholar] [CrossRef]
- de Araújo Lobo, J.M.; Christofferson, R.C.; Mores, C.N. Investigations of Koutango Virus Infectivity and Dissemination Dynamics in Aedes Aegypti Mosquitoes. Environ. Health Insights 2014, 8, 9–13. [Google Scholar] [CrossRef]
- Coz, J.; Valade, M.; Cornet, M.; Robin, Y. Transovarian transmission of a Flavivirus, the Koutango virus, in Aedes aegypti L. C. R. Hebd. Seances Acad. Sci. Ser. Sci. Nat. 1976, 283, 109–110. [Google Scholar]
- Lagare, A.; Fall, G.; Ibrahim, A.; Ousmane, S.; Sadio, B.; Abdoulaye, M.; Alhassane, A.; Mahaman, A.E.; Issaka, B.; Sidikou, F.; et al. First Occurrence of Rift Valley Fever Outbreak in Niger, 2016. Vet. Med. Sci. 2019, 5, 70–78. [Google Scholar] [CrossRef]
- Beasley, D.W.C.; Whiteman, M.C.; Zhang, S.; Huang, C.Y.-H.; Schneider, B.S.; Smith, D.R.; Gromowski, G.D.; Higgs, S.; Kinney, R.M.; Barrett, A.D.T. Envelope Protein Glycosylation Status Influences Mouse Neuroinvasion Phenotype of Genetic Lineage 1 West Nile Virus Strains. J. Virol. 2005, 79, 8339–8347. [Google Scholar] [CrossRef] [PubMed]
- Setoh, Y.X.; Prow, N.A.; Hobson-Peters, J.; Lobigs, M.; Young, P.R.; Khromykh, A.A.; Hall, R.A. Identification of Residues in West Nile Virus Pre-Membrane Protein That Influence Viral Particle Secretion and Virulence. J. Gen. Virol. 2012, 93, 1965–1975. [Google Scholar] [CrossRef] [PubMed]
- Van Slyke, G.A.; Ciota, A.T.; Willsey, G.G.; Jaeger, J.; Shi, P.-Y.; Kramer, L.D. Point Mutations in the West Nile Virus (Flaviviridae; Flavivirus) RNA-Dependent RNA Polymerase Alter Viral Fitness in a Host-Dependent Manner in Vitro and in Vivo. Virology 2012, 427, 18–24. [Google Scholar] [CrossRef]
- Parrot, L.; Hornet, J.; Cadenat, J. Note on the Phlebotomines. XLVIII. Phlebotomines of French West Africa. 1. Senegal, Soudan, Niger. Arch. Inst. Pasteur D’Algerie 1945, 23, 232–244. [Google Scholar]
- Le Pont, F.; Robert, V.; Vattier-Bernard, G.; Rispail, P.; Jarry, D. Notes on the phlebotomus of Aïr (Niger). Bull. Soc. Pathol. Exot. 1990 1993, 86, 286–289. [Google Scholar]
- Abonnenc, E.; Dyemkouma, A.; Hamon, J. On the presence of phlebotomus (phlebotomus) orientalis parrot, 1936, in the republic of niger. Bull. Soc. Pathol. Exot. Fil. 1964, 57, 158–164. [Google Scholar]
- Fontenille, D.; Traore-Lamizana, M.; Trouillet, J.; Leclerc, A.; Mondo, M.; Ba, Y.; Digoutte, J.P.; Zeller, H.G. First Isolations of Arboviruses from Phlebotomine Sand Flies in West Africa. Am. J. Trop. Med. Hyg. 1994, 50, 570–574. [Google Scholar] [CrossRef] [PubMed]
- Nabeth, P.; Kane, Y.; Abdalahi, M.O.; Diallo, M.; Ndiaye, K.; Ba, K.; Schneegans, F.; Sall, A.A.; Mathiot, C. Rift Valley Fever Outbreak, Mauritania, 1998: Seroepidemiologic, Virologic, Entomologic, and Zoologic Investigations. Emerg. Infect. Dis. 2001, 7, 1052–1054. [Google Scholar] [CrossRef] [PubMed]
- Ba, Y.; Trouillet, J.; Thonnon, J.; Fontenille, D. Phlebotomine Sandflies Fauna in the Kedougou Area of Senegal, Importance in Arbovirus Transmission. Bull. Soc. Pathol. Exot. 1999, 92, 131–135. [Google Scholar] [PubMed]
- Smithburn, K.C.; Haddow, A.J.; Lumsden, W.H.R. An Outbreak of Sylvan Yellow Fever in Uganda with Aëdes (Stegomyia) Africanus Theobald as Principal Vector and Insect Host of the Virus. Ann. Trop. Med. Parasitol. 1949, 43, 74–89. [Google Scholar] [CrossRef] [PubMed]
- Clerc, Y.; Rodhain, F.; Digoutte, J.P.; Tesh, R.; Heme, G.; Coulanges, P. The Perinet virus, rhabdoviridae, of the vesiculovirus type isolated in Madagascar from Culicidae. Arch. Inst. Pasteur Madag. 1982, 49, 119–129. [Google Scholar]
- Schmidt, J.R.; Schmidt, M.L.; Said, M.I. Phlebotomus Fever in Egypt. Isolation of Phlebotomus Fever Viruses from Phlebotomus Papatasi. Am. J. Trop. Med. Hyg. 1971, 20, 483–490. [Google Scholar] [CrossRef] [PubMed]
- Zhioua, E.; Moureau, G.; Chelbi, I.; Ninove, L.; Bichaud, L.; Derbali, M.; Champs, M.; Cherni, S.; Salez, N.; Cook, S.; et al. Punique Virus, a Novel Phlebovirus, Related to Sandfly Fever Naples Virus, Isolated from Sandflies Collected in Tunisia. J. Gen. Virol. 2010, 91, 1275–1283. [Google Scholar] [CrossRef]
- Saluzzo, J.F.; Adam, F.; Heme, G.; Digoutte, J.P. Isolation of viruses from rodents in Senegal (1983–1985). Description of a new poxvirus. Bull. Soc. Pathol. Exot. Fil. 1986, 79, 323–333. [Google Scholar]
- Clark, G.G.; Seda, H.; Gubler, D.J. Use of the “CDC Backpack Aspirator” for Surveillance of Aedes Aegypti in San Juan, Puerto Rico. J. Am. Mosq. Control. Assoc. 1994, 10, 119–124. [Google Scholar] [PubMed]
- Sudia, W.D.; Chamberlain, R.W. Battery-Operated Light Trap, an Improved Model. J. Am. Mosq. Control Assoc. 1988, 4, 536–538. [Google Scholar]
- Service, M. Mosquito Ecology: Field Sampling Methods, Chapman Hall ed.; Springer: London, UK, 1993. [Google Scholar]
- Edwards, E. Mosquitoes of the Ethiopian Region: III Culicine Adults and Pupae; British Museum Natural History: London, UK, 1941. [Google Scholar]
- Diagne, N.; Fontenille, D.; Konate, L.; Faye, O.; Lamizana, M.T.; Legros, F.; Molez, J.F.; Trape, J.F. Anopheles of Senegal. An annotated and illustrated list. Bull. Soc. Pathol. Exot. 1994, 87, 267–277. [Google Scholar]
- Digoutte, J.P.; Calvo-Wilson, M.A.; Mondo, M.; Traore-Lamizana, M.; Adam, F. Continuous Cell Lines and Immune Ascitic Fluid Pools in Arbovirus Detection. Res. Virol. 1992, 143, 417–422. [Google Scholar] [CrossRef]
- Wagner, D.; de With, K.; Huzly, D.; Hufert, F.; Weidmann, M.; Breisinger, S.; Eppinger, S.; Kern, W.V.; Bauer, T.M. Nosocomial Acquisition of Dengue. Emerg. Infect. Dis. 2004, 10, 1872–1873. [Google Scholar] [CrossRef] [PubMed]
- Weidmann, M.; Faye, O.; Faye, O.; Kranaster, R.; Marx, A.; Nunes, M.R.T.; Vasconcelos, P.F.C.; Hufert, F.T.; Sall, A.A. Improved LNA Probe-Based Assay for the Detection of African and South American Yellow Fever Virus Strains. J. Clin. Virol. 2010, 48, 187–192. [Google Scholar] [CrossRef] [PubMed]
- Faye, O.; Faye, O.; Diallo, D.; Diallo, M.; Weidmann, M.; Sall, A.A. Quantitative Real-Time PCR Detection of Zika Virus and Evaluation with Field-Caught Mosquitoes. Virol. J. 2013, 10, 311. [Google Scholar] [CrossRef] [PubMed]
- Fall, G.; Faye, M.; Weidmann, M.; Kaiser, M.; Dupressoir, A.; Ndiaye, E.H.; Ba, Y.; Diallo, M.; Faye, O.; Sall, A.A. Real-Time RT-PCR Assays for Detection and Genotyping of West Nile Virus Lineages Circulating in Africa. Vector Borne Zoonotic Dis. 2016, 16, 781–789. [Google Scholar] [CrossRef] [PubMed]
- De Madrid, A.T.; Porterfield, J.S. A Simple Micro-Culture Method for the Study of Group B Arboviruses. Bull. World Health Organ. 1969, 40, 113–121. [Google Scholar]
- Jones, D.T.; Taylor, W.R.; Thornton, J.M. The Rapid Generation of Mutation Data Matrices from Protein Sequences. Comput. Appl. Biosci. CABIOS 1992, 8, 275–282. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
| Species | Tchintabaraden | Intoussane | Tasnala | Total |
|---|---|---|---|---|
| Anopheles gambiae | 40 | 52 | 92 | |
| Anopheles rufipes | 1 | 3 | 4 | |
| Cx. antennatus | 1 | 1 | ||
| Cx. ethiopicus | 1 | 3 | 4 | |
| Cx. quinquefasciatus | 14 | 4 | 18 | |
| Cx. perexiguus | 14 | 25 | 39 | |
| Total mosquitoes | 71 | 87 | 158 | |
| Biting midges | 1 | 2 | 3 | |
| Sandflies | 359 | 29 | 10,428 | 10,816 |
| Total arthropods | 431 | 29 | 10,517 | 10,977 |
| Strains | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1.PM148_sandfly_MN057643.1_Koutango_lineage | ||||||||||||
| 2.AnD5443_Rodent_EU082200.2_Koutango_lineage | 0.00584 | |||||||||||
| 3.ArD_96655_Tick_KY703855.1_Koutango_lineage | 0.02121 | 0.00848 | ||||||||||
| 4.ArD76986_SEN_KY703854.1_WNV_Lineage_1 | 0.11371 | 0.11469 | 0.11764 | |||||||||
| 5.Eg101_AF260968.1_WNV_Lineage_1 | 0.13990 | 0.11208 | 0.14274 | 0.00584 | ||||||||
| 6.NY-99_KC407666.1_WNV_Lineage_1 | 0.14174 | 0.11404 | 0.14398 | 0.00467 | 0.00769 | |||||||
| 7.ArB3573/82_DQ318020.1_WNV_Lineage_2 | 0.13645 | 0.11371 | 0.14039 | 0.06095 | 0.07205 | 0.07557 | ||||||
| 8.Austria/2008_gh_KF179640.1_WNV_Lineage_2 | 0.13084 | 0.11310 | 0.12682 | 0.06004 | 0.08429 | 0.08696 | 0.03494 | |||||
| 9.B956_AY532665.1_WNV_Lineage_2 | 0.13294 | 0.11353 | 0.12892 | 0.06196 | 0.08603 | 0.08930 | 0.03663 | 0.00634 | ||||
| 10.ArD94343_KY703856.1_WNV_putative_Lineage_8 | 0.12553 | 0.12752 | 0.12917 | 0.09528 | 0.09432 | 0.09368 | 0.09176 | 0.09147 | 0.09156 | |||
| 11.LEIV-Krnd88-190_AY277251.1_WNV_Lineage_4 | 0.18058 | 0.15722 | 0.18384 | 0.12070 | 0.13969 | 0.14094 | 0.13333 | 0.13487 | 0.13640 | 0.14568 | ||
| 12.Rabensburg_97-103_AY765264.1_WNV_Lineage_3 | 0.16703 | 0.14386 | 0.17689 | 0.10107 | 0.12331 | 0.12359 | 0.11968 | 0.12823 | 0.13222 | 0.12224 | 0.16220 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 World Health Organization; Licensee MDPI, Basel, Switzerland. This is an open access article distributed under the terms of the Creative Commons Attribution IGO License (http://creativecommons.org/licenses/by/3.0/igo/legalcode), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. In any reproduction of this article there should not be any suggestion that WHO or this article endorse any specific organisation or products. The use of the WHO logo is not permitted.
Share and Cite
Fall, G.; Diallo, D.; Soumaila, H.; Ndiaye, E.H.; Lagare, A.; Sadio, B.D.; Ndione, M.H.D.; Wiley, M.; Dia, M.; Diop, M.; et al. First Detection of the West Nile Virus Koutango Lineage in Sandflies in Niger. Pathogens 2021, 10, 257. https://doi.org/10.3390/pathogens10030257
Fall G, Diallo D, Soumaila H, Ndiaye EH, Lagare A, Sadio BD, Ndione MHD, Wiley M, Dia M, Diop M, et al. First Detection of the West Nile Virus Koutango Lineage in Sandflies in Niger. Pathogens. 2021; 10(3):257. https://doi.org/10.3390/pathogens10030257
Chicago/Turabian StyleFall, Gamou, Diawo Diallo, Hadiza Soumaila, El Hadji Ndiaye, Adamou Lagare, Bacary Djilocalisse Sadio, Marie Henriette Dior Ndione, Michael Wiley, Moussa Dia, Mamadou Diop, and et al. 2021. "First Detection of the West Nile Virus Koutango Lineage in Sandflies in Niger" Pathogens 10, no. 3: 257. https://doi.org/10.3390/pathogens10030257
APA StyleFall, G., Diallo, D., Soumaila, H., Ndiaye, E. H., Lagare, A., Sadio, B. D., Ndione, M. H. D., Wiley, M., Dia, M., Diop, M., Ba, A., Sidikou, F., Ngoy, B. B., Faye, O., Testa, J., Loucoubar, C., Sall, A. A., Diallo, M., & Faye, O. (2021). First Detection of the West Nile Virus Koutango Lineage in Sandflies in Niger. Pathogens, 10(3), 257. https://doi.org/10.3390/pathogens10030257






