Abstract
Burkholderia glumae causes rice (Oryza sativa) bacterial panicle blight, which is an increasingly economically important disease worldwide. As the first B. glumae strain isolated from patients with chronic infections, AU6208 has been reported as an opportunistic clinic pathogen. However, our understanding of the molecular mechanism underlying human pathogenesis by B. glumae remains rudimentary. In this study, we report the complete genome sequence of the human-isolated B. glumae strain AU6208 and compare this to the genome of the rice-pathogenic B. glumae type strain LMG 2196T. Analysis of the average nucleotide identity demonstrated 99.4% similarity between the human- and plant-pathogenic strains. However, the phenotypic results from this study suggest a history of niche adaptation and divergence. In particular, we found 44 genes were predicted to be horizontally transferred into AU6208, and most of these genes were upregulated in conditions that mimic clinical conditions. In these, the gene pair sbnAB encodes key enzymes in antibiotic biosynthesis. These results suggest that horizontal gene transfer in AU6208 may be responsible for selective advantages in its pathogenicity in humans. Our analysis of the AU6208 genome and comparison with that of LMG 2196T reveal the evolutionary signatures of B. glumae in the process of switching niches from plants to humans.
1. Introduction
Burkholderia glumae is a Gram-negative plant pathogen that causes bacterial panicle blight (BPB) in rice (Oryza sativa) and wilting in a wide range of economically important vegetables such as pepper (Capsicum sp.), eggplant (Solanum melongena), and tomato (Solanum lycopersicum) [1,2]. To date, BPB has been reported in more than 21 countries throughout South and Central America, Africa, and Asia [3,4,5,6,7,8], causing costly damage (up to 75% yield reduction) to the rice industry [1].
To date, many rice-pathogenic B. glumae strains with different virulence levels have been identified in different countries [9]. Genome comparisons revealed weak variation in virulence-related genes between the strain BGR1 and the high-virulence strain 336gr-1 [1,10]. Later, it was proposed that the varied strains and broad host ranges of B. glumae may have been generated by the rapid rearrangement, inversion, or deletion of virulence-related genes [11]. Although comparative omics studies of B. glumae will provide evolutionary clues for exploring the unknown host–bacteria interactions, as well as the overall infection process, we lack comprehensive studies on the human-pathogenic B. glumae strain AU6208, which was isolated from patients with chronic granulomatous disease [12].
To understand the differences between B. glumae strains from different host niches, we generated and analyzed the complete genome sequence of the human-pathogenic strain AU6208. Interestingly, AU6208 was reported to be more virulent on rice than the rice-pathogenic strain BGR1 [13]. This result suggested that this human-pathogenic strain may have strong niche adaptation. Therefore, we investigated several interesting features pertaining to its virulence, evolution, and response to selective pressure by comparing AU6208 to the rice-pathogenic strain LMG 2196T. Our results can help to determine the evolutionary changes that enable B. glumae adaptation for growth on different hosts.
2. Results and Discussion
2.1. The Genetic Relationship between AU6208 and LMG 2196T
To better understand the unique features of the human-pathogenic strain AU6208, we first sequenced its entire genome and analyzed general features of the full-length sequenced genome and assembly information (Table 1 and Figure 1). Based on Pacbio-Illumina hybrid sequencing, a total of 78,783 (14 Kb average read length) and 4,779,545 (2X150 bp pair-end) high-quality filtered reads were obtained from Pacbio and Illumina, respectively. The size of the B. glumae AU6208 whole genome is 6.06 Mbp and the genome contains 5008 predicted coding sequences (CDSs), which are located on two chromosome DNA and one plasmid. Strikingly, the average nucleotide identity (ANI) between AU6208 and LMG 2196T was 99.45%, suggesting an extremely high genetic similarity. The synteny analysis between the two strains also suggested the high synteny with some large-scale rearrangement (Figure 2). In total, 257 genes were found to be unique in AU6208 compared to LMG 2196T (Table S1). Most of these genes are annotated as hypothetical protein. Notably, the whole genome of B. glumae AU6208 contained 147 and 4 genes related to virulence and antibiotic resistance, respectively (Tables S2 and S3). Interestingly, the number of virulence- and antibiotic-resistance-related genes in LMG 2196T is virtually identical to that of AU6208, except that AU6208 contains two additional virulence-related genes encoding pmlR/bspR1 and BCAL3235 (Tables S2 and S3), which play important roles in capsule polysaccharide biosynthesis in Burkholderia cecocepacia [14].

Table 1.
Genome statistics on Burkholderia glumae AU6208.
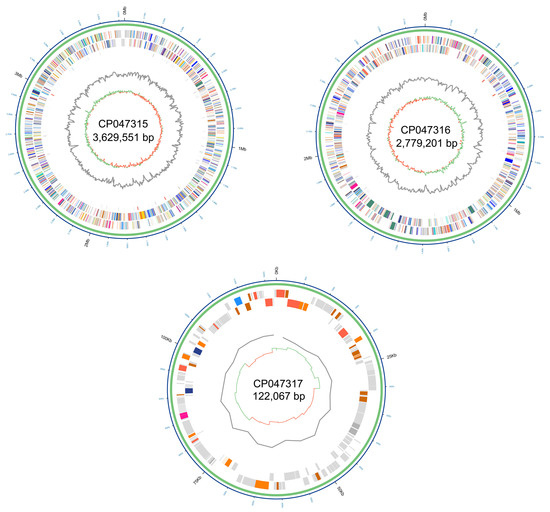
Figure 1.
Circular representation of the complete genome of Burkholderia glumae strain AU6208. Rings from outside to the center: B. glumae AU6208 chromosome or plasmid, genes mapped against Clusters of Orthologous Groups (COG) database, RNA sequences, GC skew, and GC content.
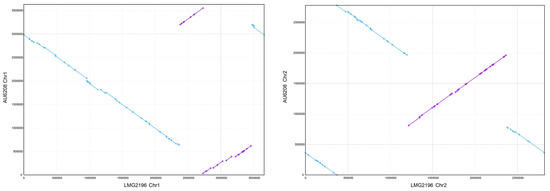
Figure 2.
Genetic synteny visualized by MUMmer alignments. A comparison of the chromosomes of AU6208 and LMG 2196T. The conserved gene synteny and some rearrangement between the chromosomes are observed. The purple dots represent the synteny genes between the two strains, while the blue dots represent the rearrangement genes. The lines connected by dots represent the high synteny regions (purple lines) or rearrangement (reverse arrangement) regions (blue lines).
2.2. AU6208 Showed Increased Biofilm Formation, EPS Secretion, and Virulence
As mentioned above, pmlR/bspR1 and BCAL3235 may give AU6208 some advantages in biofilm formation and exopolysaccharide (EPS) secretion compared to LMG 2196T. To confirm this hypothesis, we measured biofilm formation and EPS secretion in AU6208WT, AU6208ΔpmlR and the AU6208ΔBCAL3235 strains. The morphological appearance and thickness of the biofilm showed obvious differences after 48 h of adhesion at 30 °C without agitation (Figure 3). In particular, the AU6208 strain formed a thicker biofilm compared to the AU6208ΔpmlR and AU6208ΔBCAL3235 strains. When quantified, WT (wild type) yielded readings at least four times as high as AU6208ΔpmlR and AU6208ΔBCAL3235 (Figure 3). The weight of EPS from AU6208WT was at least twice as heavy as that from AU6208ΔpmlR and AU6208ΔBCAL3235 (Figure 3). The quantitative data confirmed that AU6208 exhibited a significantly (p < 0.001) higher biofilm-forming capability and EPS secretion compared to AU6208ΔpmlR and AU6208ΔBCAL3235. The above results strongly suggest that the pmlR and BCAL3235 gene in AU6208 contributes to increased biofilm formation and EPS secretion. In the virulence test in rice, we found that AU6208ΔpmlR and AU6208ΔBCAL3235 showed slightly decreased virulence compared to the WT (Figure 3). The complement strain showed a similar result compared to the WT (Figure 3).
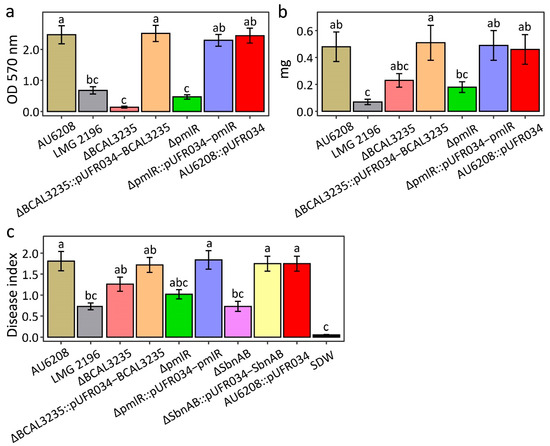
Figure 3.
Biofilm formation, exopolysaccharide (EPS) production, and pathogenicity of B. glumae strains AU6208, LMG 2196T, AU6208ΔBCAL3235, AU6208ΔSbnAB, AU6208ΔpmlR, AU6208- pUFR034 empty vector control and their complementary strains. (a) Biofilm formation after a 48 h incubation; (b) EPS quantification after a 24 h incubation; (c) disease index in rice. All these experiments were repeated three times with four replicates. SDW: distilled water. Different letters denote statistical significance (p < 0.05, identified by analysis of variance comparisons of means, employing a posthoc Tukey-Cramer test for multiple comparisons).
Interestingly, pmlR/bspR1 is a LuxR family transcriptional regulator, which may be involved in quorum sensing (QS). Devescivi et al. (2007) characterized the important role of QS in the strong rice virulence of B. glumae AU6208. They found that, compared with B. glumae LMG 2196T, the active TofR (a LuxR family regulator) present in AU6208 is the reason to cause strong disease symptom in rice [13]. In this study, since we achieved the whole sequences of both strains, the result of comparative genomic analysis presented that the pmlR/bspR1 is unique for AU6208 but not in LMG 2196T. Since the pmlR/bspR1 has been demonstrated as the key regulator responsible for the expression of LuxR and virulence of Burkholderia pseudomallei [15], it is reasonable to suggest that pmlR/bspR1 regulated LuxR system plays a key role in virulence of strain AU6208, which in terms that the weak virulence of LMG 2196 T may be because of the incomplete LuxR system (QS). Moreover, since the QS is responsible for the biofilm formation, the data in this study also suggested that the presence of active pmlR/bspR1 in B. glumae is critical for the formation of biofilm matrix (Figure 3). This is in agreement with reports for many human pathogens in Burkholderia family. For instance, it is well demonstrated that pmlR is critical for full virulence of Burkholderia pseudomallei, the causative agent of melioidosis [15]. In this respect, the presence of the QS system appears to be an indicator of the virulence capacity of Burkholderia family, at least of B. glumae.
Besides pmlR/bspR1, BCAL3235 is another unique gene present in AU6208 compared to LMG 2196T. BCAL3235 encoded UDP-galactopyranose mutase (UGM) is a unique flavin-dependent enzyme, which converts UDP-galactopyranose (UDP-Galp) to UDP-galactofuranose (UDP-Galf), the major component of cell surface glycoproteins and glycolipids [16]. UGM presents in a variety of pathogenic fungi and bacteria, such as Aspergillus, E. coli, Klebsiella, and Mycobacteria [17]. The UGM has been identified in Burkholderia cepacian complex [18], which is a common causal agent of human chronic granulomatous disease. Thus, the finding of BCAL3235 in B. glumae strain AU6208 but not rice pathogen LMG 2196T strongly suggested that the gene may be achieved from bacteria survived in similar niche for adaption.
2.3. Horizontal Gene Transfer May Contribute to Niche Adaptation in AU6208
To further understand the niche adaptation in B. glumae AU6208, we compared the whole-genome expressed proteins of AU6208 under the human mimic [19] and in planta mimic [20] conditions using 2D SDS-PAGE, and the results shown are a representative expression from duplicate experiments (Figure S1). Most of the genes were expressed in both conditions. Notably, 30 proteins were expressed only in one condition (Figure S1). These differentially expressed proteins were digested and further examined by LC–MS/MS and the transcriptomic levels of the corresponding genes were determined by using qRT-PCR. Twenty-two genes showed a dramatic increase in expression under human mimic condition compared to planta mimic condition (Table 2). Of which, two virulence genes (sbnA and sbnB) were uniquely presented in AU6208. These data suggest that these genes may be responsible for bacterial survival and virulence in human host. To investigate whether these genes are achieved from microorganisms for niche adaption, we conducted a comprehensive horizontal gene transfer (HGT) analysis (Method 3.6) and total 44 genes were predicted to be horizontally transferred to AU6208 from other organisms (Table S4). Furthermore, 12 of the 22 (marked yellow at Table 2) significantly expressed genes (54.5%) belonged to the horizontally transferred genes. Although the transmission of B. glumae from rice condition to human condition is still unclear, these data indicated that horizontal gene transfer plays some role in the niche adaption of B. glumae strain AU6208.

Table 2.
List of qPCR results from 30 proteins detected by 2D-MS/MS.
Interestingly, we found that a gene pair, sbnAB, which is involved in staphyloferrin B biosynthesis, is among the 44 horizontally transferred genes [21]. Phylogenetic analysis of both genes strongly suggests their Streptomyces origin with strong bootstrap support (Figure 4). It has been reported that staphyloferrin B is a hydrophilic siderophore, which plays an important role in pathogenicity in both human and plant pathogens [22,23]. Pathogenicity results confirmed that the AU6208ΔSbnAB mutant, which lacks SbnAB, induced no obvious symptoms on the rice plants (Figure 3). The complement strain showed a similar result compared to the WT (Figure 3). This result may be partially explained by the requirement of siderophore biosynthesis in bacteria for plant colonization [24].
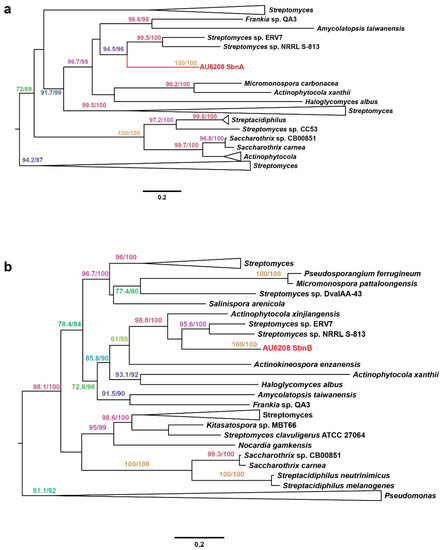
Figure 4.
Phylogeny of (a) SbnA and (b) SbnB. The phylogenetic tree was calculated using the maximum likelihood program PhyML and Bayesian program MrBayes (detailed parameters are described in the main text). Only the values in ML ≥70% and Bayesian posterior probability ≥65% are shown. For key nodes, the actual support values are shown in the order ML bootstraps/Bayesian posterior probability. Bar, 0.1 substitution per site.
Overall in this study, we reported the complete genome of the human-pathogenic B. glumae strain AU6208 and compared it with that of the plant-pathogenic strain LMG 2196T. This study is the first to report the whole genome of a human-pathogenic B. glumae strain, AU6208. Our comparative genomic analysis revealed increased diversity in this pathogen induced by different niche survival needs. We explored the differences in molecular mechanisms of virulence and the difference in the interaction of the human-pathogenic B. glumae strain AU6208 and rice-pathogenic B. glumae strains with rice. Our results provide insight into the different ecological niches of B. glumae strains resulting from differences in virulence and environmental adaptation, as well as their unique interactions with the host during pathogenesis.
3. Materials and Methods
3.1. Bacterial Strains, Plasmids, and Growth Conditions
B. glumae strains and plasmids used in this study are listed in Table 3. All strains were stored at −80 °C in 20% glycerol and grown in nutrient broth (NB) medium (1% peptone, 0.3% yeast extract, and 0.5% NaCl, pH 7.5) at 30 °C. Luria-Bertani (LB) agar without sucrose and LB agar containing 10% (w/v) sucrose were used for deletion mutagenesis. Antibiotics were added at the following concentrations when required: 50 µg/mL kanamycin (Km), 100 µg/mL ampicillin (Amp) and 10 ug/mL neomycin (Nm).

Table 3.
Strains and plasmids used in this study.
3.2. DNA Extraction, Genome Sequencing, and Genomic Analysis
Bacterial genomic DNA was extracted using TIANamp Bacteria DNA kits (Tiangen Biotech, Beijing, China). Whole-genome DNA sequencing of B. glumae AU6208 was conducted using the Pacific Biosciences (PacBio) RSII Single Molecule Real Time (SMRT) system and Illumina with the NEBNext Ultra DNA Library Prep Kit (New England Biolabs, MA, USA). The combination of PacBio and Illumina sequence shotgun data was assembled with the SMRT analysis packages (v2.1) using hierarchical genome assembly [25]. Low-quality sequences were filtered by Sickle (v1.33) with Q30 (https://github.com/najoshi/sickle). Protein-coding regions, tRNAs, and rRNAs were predicted using rapid annotation subsystem technology [26]. Hits were then analyzed for signal peptide characteristics using SignalP [27]. Transmembrane domains and functional protein domains were analyzed using tied-mixture hidden Markov modeling and InterProScan, respectively [28,29]. The secondary metabolite clusters, antibiotic resistance clusters and toxins, and virulence factors were predicted by comparing with DoBISCUIT, CARD, and VFDB databases, respectively [30,31,32]. The category of functional proteins was further annotated by Clusters of Orthologous Groups (COG) and Gene Ontology databases [33]. Genome comparison between AU6208 and LMG 2196T was conducted using average nucleotide identity (ANI) and alignment fraction (AF), which were calculated by an ANIcalculator [34]. Synteny of genome structure between AU6208 and LMG 2196T was performed by Mummer 3.0 with default parameters [35].
3.3. Construction of Deletion Mutants and Complementation
To investigate the role of genes of interest in AU6208, we constructed deletion mutants in AU6208 using homologous recombination. Briefly, two fragments flanking the left and right borders of corresponding genes were amplified from the wild-type genomic DNA with the primer pairs listed in Table S4. The amplified fragments were cloned into pGEM-T (Promega), confirmed by sequence analysis, and then digested and subcloned into vector pKMS1 [36] at BamHI and PstI (or SalI) sites. The resulting recombinant plasmids were introduced into AU6208 by electroporation, and transformants were plated on NA plates supplemented with kanamycin. Colonies resulting from a single homologous crossover (integration of the deletion construct at either the left or right border of the target gene) were then transferred to NBN broth, grown for 12 h at 28 °C, and then plated on NAS plates for sucrose-positive deletion mutant selection. Sucrose-resistant colonies were visible within 3 to 4 days and then transferred to NA plates and NA plus kanamycin plates. Kanamycin-sensitive colonies could be mutants containing a second homologous crossover, which was further confirmed by PCR amplification.
In order to complement the deletion mutants, the full-length of corresponding genes including native promotors were amplified using primer pairs listed in Table S5. After confirmation by sequence analysis, the amplified DNA fragments were cloned into pUFR034 [37] at the BamHI and SalI sites to create the recombinant plasmids, the recombinant plasmids were transferred into corresponding mutants by electroporation, and transformants were screened on NA plates with neomycin. Colonies were visible within 3 to 4 days growing at 28 °C. A putative transformant was shown to contain plasmid by PCR analysis.
3.4. Biofilm Formation and Exopolysaccharide (EPS) Secretion Assay
Biofilm formation was determined using a previously described method [38]. In brief, bacterial cells grown overnight were inoculated 1:100 in fresh NB media in a 96-well microplate (Corning-Costar Corp., Corning, NY, USA). After static culture at 30 °C for 48 h, cells were stained with 1% crystal violet (CV) for 15 min. The planktonic cells were removed by several rinses with H2O. The CV-stained bound cells were air-dried for 1 h and dissolved in 90% ethanol, and the optical density at 590 nm (OD590) of the solution was measured to quantify biofilm formation.
The extraction and quantitation of bacterial EPS were performed as described previously [39]. Briefly, overnight bacterial cultures were inoculated 1:100 in 100 mL fresh NB media to OD600 = 1.0. Cultures were centrifuged at 4 °C, and 100 µl of the supernatant was collected and filtered through 0.22-µm Millipore filters to remove all bacterial cells. A mixture of one volume of supernatant with four volumes of Sevage’s solution (4:1 chloroform:butanol) was incubated with shaking at 180 rpm for 30 min. Subsequently, the supernatants were collected by centrifuging and then mixed with three volumes of 95% ethanol and incubated at 4 °C for 24 h without shaking. The pellet was harvested, dried, and weighed after incubation. The experiments consisted of three biological replicates and were repeated at least three times.
3.5. Pathogenicity Assay
In a greenhouse, rice plants (Oryza sativa cv. kitaake) were inoculated at the flowering stage with approximately 1 × 109 CFU/mL of B. glumae. Disease symptoms in the rice were evaluated on day 10 after inoculation. The disease index was determined as described previously [40]. In detail, the disease score was defined as 0 = healthy panicle, 1 = panicle 0 to 20% discolored, 2 = panicle 20 to 40% discolored, 3 = panicle 40 to 60% discolored, 4 = panicle 60 to 80% discolored, and 5 = panicle 80 to 100% discolored (disease index = (number of samples per score X score)/the total number of panicles). Pathogenicity assays were repeated three times with four replicates.
3.6. Horizontal Gene Transfer (HGT) Analysis
Screening for HGT was first performed by HGTfinder with default parameters [41]. Confirmation of HGT was performed by a combination of BLAST screening and large-scale phylogenetic trees. BLAST screening was performed based on our previous study [42]. In brief, candidate sequences in B. glumae were compared against sequences in the NCBI (National Center for Biotechnology Information) reference genome using BLASTp [43] followed by extracting the sequences with highest similarity for further study. We considered two genes orthologs when the E-value <10−10 and when the alignment similarity was higher than 30% with more than 85% coverage [44]. These sequences were aligned using MAFFT [45], and the conserved region of each alignment was trimmed using TrimAI [46] with stringent settings. Maximum likelihood and Bayesian phylogenies were generated based on our previous study [42].
3.7. Preparation of Bacterial Samples Isolated from Different Niches for 2-Dimensional Electrophoresis
For the human mimic condition, overnight bacterial cultures of B. glumae AU6208 and LMG2196 from one single colony in NB broth were trans-cultured as 1:100 in 100 mL fresh SCFM2 (Human mimic medium) [19] and incubated with shaking at 200 rpm at 30 °C until the OD600 reached 0.8. Cells were then collected by centrifugation at 5000× g for 10 min, washed with 0.5 × phosphate-buffered saline (PBS) three times, and used for protein extraction. For the in planta mimic condition, we collected the bacterial samples using the previous method [20]. In brief, the rice leaves were collected and homogenized by liquid nitrogen with a mortar and pestle. The resultant was aliquoted to 1 g in tubes and stored at −80 °C before using. The 1 g of freezing rice leaf resultant was thaw on ice and added into 40 mL of nutrient broth to ready as the planta mimic medium. Single colony of B. glumae strains was inoculated into the medium and grew to the mid-exponential phase (OD600 = 0.5). The bacterial pellets were harvested from 1.5 mL bacterial suspension by centrifugation at 5000× g for 20 min, washed with 0.5 × PBS three times, and used for protein extraction. Extraction of total proteins and 2-dimensional electrophoresis were performed as previously described [47]. The resulting protein profiles were analyzed with ImageMaster 2D platinum, version 5.0 (Amersham Biosciences, Uppsala, Sweden).
3.8. Protein in-Gel Digestion and Liquid Chromatography–Tandem Mass Spectrometry (LC–MS/MS) Analysis
The tryptic in-gel digestion protocol, optimized for a polyacrylamide gel of 1.5 mm thickness, was performed based on a previous method [48]. Briefly, protein spots were excised from the gel and put into the 96-well polypropylene ZipPlate Micro-SPE plate (Thermo). Then, the gel piece was rinsed for 40 min with 100 μL of 25 mM ammonium bicarbonate/acetonitrile (95:5, v/v) followed by two rinses with 100 μL of 25 mM ammonium bicarbonate/acetonitrile (50:50, v/v). The gel piece was then dehydrated for 12 min with 200 μL of acetonitrile. Digestion was performed with 15 μL of sequencing grade porcine trypsin (10 ng μL−1) at 37 °C for 3 h.
The peptides released from trypsin digestion for LC–MS/MS analysis were prepared in two biological replicates. LC was performed using a Dionex Ultimate 3000 nano-LC system following the method in our previous paper [49]. The MASCOT LC-MS/MS ion search algorithm (Matrix Sciences) was applied to evaluate the LC/MS spectra, and the sequence similarity of resulting peptides to the B. glumae AU6208 strain was compared to other Burkholderia species accessible on NCBI. To further identify the proteins, the cross-correlation (X corr) scores of singly, doubly, and triply charged peptides were fixed greater than 1.8, 2.5, and 3.5, respectively [49]. Peptide sequences with the highest X corr values were then identified.
3.9. Quantitative Real-Time PCR
Total RNAs were extracted from exponentially growing cells, using a RNeasy Mini spin columns Kit (Qiagen) and was treated with a unit of RNase-free DNase I (Qiagen), and cDNA synthesis was performed with a Moloney murine leukemia virus reverse transcriptase first-strand cDNA synthesis kit (QIAGEN). The cDNA was then used directly as the template for qRT-PCR using a SYBER Green master mix (Protech Technology Enterprise Co., Ltd.) on an ABI Prism 7000 sequence detection system (Applied Biosystems). Primers for quantitative real-time PCR (qRT-PCR) of the selected genes were designed by using Primer 3 [50] and gyrB was used as an internal control. Ct values were shown on Table S5.
3.10. Statistical Analysis
Statistically significant differences in the disease index, biofilm formation and EPS production were identified with a one-way ANOVA and Tukey–Kramer post-hoc test in the agricolae package in R.
3.11. Availability of Data and Materials
All data supporting the conclusions of this article are included in this article and its additional files. The complete genome sequences have been deposited at DDBJ/EMBL/GenBank and the bioproject ID is PRJNA595825.
Supplementary Materials
The following are available online at https://www.mdpi.com/2076-0817/10/2/87/s1, Figure S1: 2-DE maps of total proteins from B. glumae strain AU6208 expressed under human mimic and in planta mimic conditions. The labeled spots are the typical results of specific expressed proteins., Table S1: Unique genes in AU6208 compared to LMG 2196T, Table S2: Antibiotic Resistance genes in AU6208 and LMG 2196, Table S3: Virulence genes in AU6208 and LMG 2196, Table S4: Horizontal gene transfer in AU6208, Table S5: List of qPCR results from 30 proteins detected by 2D-MS/MS.
Author Contributions
Z.C. conceived and led the project, prepared bacteria, DNA for sequencing and phenotypic experiments; S.W. assembled the genome, made predictions, annotated genes and performed other bioinformatics analyses; K.U.K., G.X. and B.L. contributed to the project; B.L. acted as a scientific consultant; B.Z. wrote the manuscript. All authors prepared tables and figures, edited and approved the final manuscript. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by National Key R&D Program of China (2018YFD0201202, 2017YFD0201108), Zhejiang National Natural Science Foundation of China (LY17C010006), The Agri-X Interdisciplinary Fund of Shanghai Jiao Tong University (Agri-X2017010), State Key Laboratory for Biology of Plant Diseases and Insect Pests (SKLOF201802), the Open Project Program of State Key Laboratory of Rice Biology (20190109) and Shanghai Committee of Science and Technology (19390743300).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
All data supporting the conclusions of this article are included in this article and its additional files. The complete genome sequences have been deposited at DDBJ/EMBL/GenBank and the bioproject ID is PRJNA160245.
Acknowledgments
We thank John LiPuma at Michigan Medicine Ped Infectious Disease. C. S. Mott Children’s Hospital for providing the AU6208 strain to us.
Conflicts of Interest
The authors declare that they have no competing interests.
References
- Ashburner, M.; Ball, C.A.; Blake, J.A.; Botstein, D.; Butler, H.; Cherry, J.M.; Davis, A.P.; Dolinski, K.; Dwight, S.S.; Eppig, J.T.; et al. Gene ontology: Tool for the unification of biology. Nat. Genet. 2000, 25, 25–29. [Google Scholar] [CrossRef] [PubMed]
- Bernier, S.P.; Sokol, P.A. Use of suppression-subtractive hybridization to identify genes in the Burkholderia cepacia complex that are unique to Burkholderia cenocepacia. J. Bacteriol. 2005, 187, 5278–5291. [Google Scholar] [CrossRef]
- Beverley, S.M.; Owens, K.L.; Showalter, M.; Griffith, C.L.; Doering, T.L.; Jones, V.C.; McNeil, M.R. Eukaryotic UDP-Galactopyranose Mutase (GLF Gene) in microbial and metazoal pathogens. Eukaryot. Cell 2005, 4, 1147–1154. [Google Scholar] [CrossRef]
- Bhatt, G.; Denny, T.P. Ralstonia solanacearum iron scavenging by the siderophore Staphyloferrin B is controlled by PhcA, the global virulence regulator. J. Bacteriol. 2004, 186, 7896–7904. [Google Scholar] [CrossRef] [PubMed]
- Capella-Gutiérrez, S.; Silla-Martínez, J.M.; Gabaldón, T. trimAl: A tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics 2009, 25, 1972–1973. [Google Scholar] [CrossRef] [PubMed]
- Caruso, G.; Cavaliere, C.; Foglia, P.; Gubbiotti, R.; Samperi, R.; Laganà, A. Analysis of drought responsive proteins in wheat (Triticum durum) by 2D-PAGE and MALDI-TOF mass spectrometry. Plant. Sci. 2009, 177, 570–576. [Google Scholar] [CrossRef]
- Chen, L.; Yang, J.; Yu, J.; Yao, Z.; Sun, L.; Shen, Y.; Jin, Q. VFDB: A reference database for bacterial virulence factors. Nucleic Acids Res. 2005, 33, D325–D328. [Google Scholar] [CrossRef]
- Cheung, J.; Beasley, F.C.; Liu, S.; Lajoie, G.A.; Heinrichs, D.E. Molecular characterization of staphyloferrin B bio-synthesis in Staphylococcus aureus. Mol. Microbiol. 2009, 74, 594–608. [Google Scholar] [CrossRef]
- Coenye, T.; Peeters, E.; Nelis, H.J. Biofilm formation by Propionibacterium acnes is associated with increased resistance to antimicrobial agents and increased production of putative virulence factors. Res. Microbiol. 2007, 158, 386–392. [Google Scholar] [CrossRef]
- Dale, S.E.; Doherty-Kirby, A.; Lajoie, G.; Heinrichs, D.E. Role of siderophore biosynthesis in virulence of Staphylo-coccus aureus: Identification and characterization of genes involved in production of a siderophore. Infect. Immun. 2004, 72, 29–37. [Google Scholar] [CrossRef]
- DeFeyter, R.; Kado, C.I.; Gabriel, D.W. Small, stable shuttle vectors for use in Xanthomonas. Gene 1990, 88, 65–72. [Google Scholar] [CrossRef]
- Devescovi, G.; Bigirimana, J.; Degrassi, G.; Cabrio, L.; Lipuma, J.J.; Kim, J.; Hwang, I.; Venturi, V. Involvement of a quorum-sensing-regulated lipase secreted by a clinical isolate of Burkholderia glumae in severe disease symptoms in rice. Appl. Environ. Microbiol. 2007, 73, 4950–4958. [Google Scholar] [CrossRef] [PubMed]
- Francis, F.; Kim, J.; Ramaraj, T.; Farmer, A.; Rush, M.C.; Ham, J.H. Comparative genomic analysis of two Burkholderia glumae strains from different geographic origins reveals a high degree of plasticity in genome structure associated with genomic islands. Mol. Genet. Genom. 2013, 288, 195–203. [Google Scholar] [CrossRef] [PubMed]
- Goo, E.; Kang, Y.; Kim, H.; Hwang, I. Proteomic analysis of quorum sensing-dependent proteins in Burkholderia glumae. J. Proteome Res. 2010, 9, 3184–3199. [Google Scholar] [CrossRef] [PubMed]
- Goo, E.; Kang, Y.; Lim, J.Y.; Ham, H.; Hwang, I. Lethal consequences of overcoming metabolic restrictions imposed on a cooperative bacterial population. mBio 2017, 8, e00042-17. [Google Scholar] [CrossRef]
- Guo, W.; Cai, L.-L.; Zou, H.-S.; Ma, W.-X.; Liu, X.-L.; Zou, L.-F.; Li, Y.-R.; Chen, X.-B.; Chen, G.-Y. Ketoglutarate transport protein KgtP is secreted through the type III secretion system and contributes to virulence in Xanthomonas oryzae pv. oryzae. Appl. Environ. Microbiol. 2012, 78, 5672–5681. [Google Scholar] [CrossRef]
- Ham, J.H.; Melanson, R.A.; Rush, M.C. Burkholderia glumae: Next major pathogen of rice? Mol. Plant. Pathol. 2011, 12, 329–339. [Google Scholar] [CrossRef]
- Hao, L.-Y.; Willis, D.K.; Andrews-Polymenis, H.; McClelland, M.; Barak, J.D. Requirement of siderophore biosyn-thesis for plant colonization by Salmonella enterica. Appl. Environ. Microbiol. 2012, 78, 4561–4570. [Google Scholar] [CrossRef]
- Ichikawa, N.; Sasagawa, M.; Yamamoto, M.; Komaki, H.; Yoshida, Y.; Yamazaki, S.; Fujita, N. DoBISCUIT: A database of secondary metabolite biosynthetic gene clusters. Nucleic Acids Res. 2013, 41, D408–D414. [Google Scholar] [CrossRef]
- Jeong, Y.; Kim, J.; Kim, S.; Kang, Y.; Nagamatsu, T.; Hwang, I. Toxoflavin produced by Burkholderia glumae causing rice grain rot is responsible for inducing bacterial wilt in many field crops. Plant. Dis. 2003, 87, 890–895. [Google Scholar] [CrossRef]
- Jia, B.; Raphenya, A.R.; Alcock, B.; Waglechner, N.; Guo, P.; Tsang, K.K.; Lago, B.A.; Dave, B.M.; Pereira, S.; Sharma, A.N.; et al. CARD 2017: Expansion and model-centric curation of the comprehensive antibiotic resistance database. Nucleic Acids Res. 2016, 45, D566–D573. [Google Scholar] [CrossRef] [PubMed]
- Katoh, K.; Standley, D.M. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef]
- Kim, J.-W.; Kang, Y.-S.; Kim, J.-G.; Choi, O.-H.; Hwang, I.-G. Occurrence of Burkholderia glumae on rice and field crops in Korea. Plant. Pathol. J. 2010, 26, 271–272. [Google Scholar] [CrossRef][Green Version]
- Kim, S.; Cho, Y.-J.; Song, E.-S.; Lee, S.H.; Kim, J.-G.; Kang, L.-W. Time-resolved pathogenic gene expression analysis of the plant pathogen Xanthomonas oryzae pv. oryzae. BMC Genom. 2016, 17, 345. [Google Scholar] [CrossRef] [PubMed]
- Tanner, J.J.; Sobrado, P. Targeting UDP-Galactopyranose mutases from eukaryotic human pathogens. Curr. Pharm. Des. 2013, 19, 2561–2573. [Google Scholar] [CrossRef]
- Krogh, A.; Larsson, B.; Von Heijne, G.; Sonnhammer, E.L. Predicting transmembrane protein topology with a hidden markov model: Application to complete genomes. J. Mol. Biol. 2001, 305, 567–580. [Google Scholar] [CrossRef] [PubMed]
- Kurtz, S.; Phillippy, A.M.; Delcher, A.L.; Smoot, M.; Shumway, M.; Antonescu, C.; Salzberg, S.L. Versatile and open software for comparing large genomes. Genome Biol. 2004, 5, R12. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.-H.; Park, J.; Kim, J.; Park, I.; Seo, Y.-S. Understanding the direction of evolution in Burkholderia glumae through comparative genomics. Curr. Genet. 2016, 62, 115–123. [Google Scholar] [CrossRef]
- Liu, H.; Yang, C.L.; Ge, M.Y.; Ibrahim, M.; Li, B.; Zhao, W.J.; Chen, G.Y.; Zhu, B.; Xie, G.L. Regulatory role of tetR gene in a novel gene cluster of Acidovorax avenae subsp. avenae RS-1 under oxidative stress. Front. Microbiol. 2014, 5, 547. [Google Scholar] [CrossRef]
- Mondal, K.K.; Mani, C.; Verma, G. Emergence of bacterial panicle blight caused by Burkholderia glumae in North India. Plant. Dis. 2015, 99, 1268. [Google Scholar] [CrossRef]
- Nandakumar, R.; Rush, M.; Shahjahan, A.; O’Reilly, K.; Groth, D. Bacterial panicle blight of rice in the southern United States caused by Burkholderia glumae and B-gladioli. Phytopathology 2005, 95, S73. [Google Scholar]
- Nandakumar, R.; Rush, M.C.; Correa, F. Association of Burkholderia glumae and B. gladioli with panicle blight symptoms on rice in panama. Plant. Dis. 2007, 91, 767. [Google Scholar] [CrossRef] [PubMed]
- Nandakumar, R.; Shahjahan, A.K.M.; Yuan, X.L.; Dickstein, E.R.; Groth, D.; Clark, C.A.; Cartwright, R.D.; Rush, M.C. Burkholderia glumae and B. gladioli cause bacterial panicle blight in rice in the Southern United States. Plant. Dis. 2009, 93, 896–905. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, M.; Ekstrom, A.; Li, X.; Yin, Y. HGT-Finder: A new tool for horizontal gene transfer finding and application to aspergillus genomes. Toxins 2015, 7, 4035–4053. [Google Scholar] [CrossRef] [PubMed]
- Nogueira, T.; Rankin, D.J.; Touchon, M.; Taddei, F.; Brown, S.P.; Rocha, E.P. Horizontal gene transfer of the se-cretome drives the evolution of bacterial cooperation and virulence. Curr. Biol. 2009, 19, 1683–1691. [Google Scholar] [CrossRef]
- Overbeek, R.; Olson, R.; Pusch, G.D.; Olsen, G.J.; Davis, J.J.; Disz, T.; Edwards, R.A.; Gerdes, S.; Parrello, B.; Shukla, M.; et al. The SEED and the Rapid Annotation of microbial genomes using Subsystems Technology (RAST). Nucleic Acids Res. 2014, 42, D206–D214. [Google Scholar] [CrossRef]
- Patel, A.K.; Laroche, C.; Marcati, A.; Ursu, A.V.; Jubeau, S.; Marchal, L.; Petit, E.; Djelveh, G.; Michaud, P. Separation and fractionation of exopolysaccharides from Porphyridium cruentum. Bioresour. Technol. 2013, 145, 345–350. [Google Scholar] [CrossRef]
- Petersen, T.N.; Brunak, S.; Von Heijne, G.; Nielsen, H. SignalP 4.0: Discriminating signal peptides from trans-membrane regions. Nat. Methods 2011, 8, 785–786. [Google Scholar] [CrossRef]
- Pruitt, K.D.; Tatusova, T.; Maglott, D.R. NCBI reference sequences (RefSeq): A curated non-redundant sequence database of genomes, transcripts and proteins. Nucleic Acids Res. 2006, 35, D61–D65. [Google Scholar] [CrossRef]
- Quevillon, E.; Silventoinen, V.; Pillai, S.; Harte, N.; Mulder, N.; Apweiler, R.; Lopez, R. InterProScan: Protein do-mains identifier. Nucleic Acids Res. 2005, 33, W116–W120. [Google Scholar] [CrossRef]
- Riera-Ruiz, C.; Vargas, J.; Cedeño, C.; Quirola, P.; Escobar, M.; Cevallos-Cevallos, J.M.; Ratti, M.; Peralta, E.L. First report of Burkholderia glumae causing bacterial panicle blight on rice in Ecuador. Plant. Dis. 2014, 98, 988. [Google Scholar] [CrossRef] [PubMed]
- Salloum, T.; Nassour, E.; Araj, G.F.; Abboud, E.; Tokajian, S. Insights into the genome diversity and virulence of two clinical isolates of Burkholderia cenocepacia. J. Med. Microbiol. 2018, 67, 1157–1167. [Google Scholar] [CrossRef] [PubMed]
- Seo, Y.-S.; Lim, J.Y.; Park, J.; Kim, S.; Lee, H.-H.; Cheong, H.; Kim, S.-M.; Moon, J.S.; Hwang, I. Comparative genome analysis of rice-pathogenic Burkholderia provides insight into capacity to adapt to different environments and hosts. BMC Genom. 2015, 16, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Turner, K.H.; Wessel, A.K.; Palmer, G.C.; Murray, J.L.; Whiteley, M. Essential genome of Pseudomonas aeruginosa in cystic fibrosis sputum. Proc. Natl. Acad. Sci. USA 2015, 112, 4110–4115. [Google Scholar] [CrossRef]
- Untergasser, A.; Cutcutache, I.; Koressaar, T.; Ye, J.; Faircloth, B.C.; Remm, M.; Rozen, S.G. Primer3—New capabilities and interfaces. Nucleic Acids Res. 2012, 40, e115. [Google Scholar] [CrossRef]
- Valade, E.; Thibault, F.; Gauthier, Y.; Palencia, M.; Popoff, M.; Vidal, D. The PmlI-PmlR quorum-sensing system in Burkholderia pseudomallei plays a key role in virulence and modulates production of the MprA protease. J. Bacteriol. 2004, 186, 2288–2294. [Google Scholar] [CrossRef]
- Varghese, N.J.; Mukherjee, S.; Ivanova, N.; Konstantinidis, K.T.; Mavrommatis, K.; Kyrpides, N.C.; Pati, A. Microbial species delineation using whole genome sequences. Nucleic Acids Res. 2015, 43, 6761–6771. [Google Scholar] [CrossRef]
- Weinberg, J.B.; Alexander, B.D.; Majure, J.M.; Williams, L.W.; Kim, J.Y.; Vandamme, P.; Lipuma, J.J. Burkholderia glumae infection in an infant with chronic granulomatous disease. J. Clin. Microbiol. 2007, 45, 662–665. [Google Scholar] [CrossRef]
- Zhou, X.G. First report of bacterial panicle blight of rice caused by Burkholderia glumae in South Africa. Plant. Dis. 2014, 98, 566. [Google Scholar] [CrossRef]
- Zhu, B.; Lou, M.-M.; Xie, G.-L.; Zhang, X.; Zhou, X.; Li, B.; Jin, G. Horizontal gene transfer in silkworm, Bombyx mori. BMC Genom. 2011, 12, 248–249. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).