Discrepancies in Serology-Based and Nucleic Acid-Based Detection and Quantitation of Tomato Spotted Wilt Orthotospovirus in Leaf and Root Tissues from Symptomatic and Asymptomatic Peanut Plants
Abstract
1. Introduction
2. Results
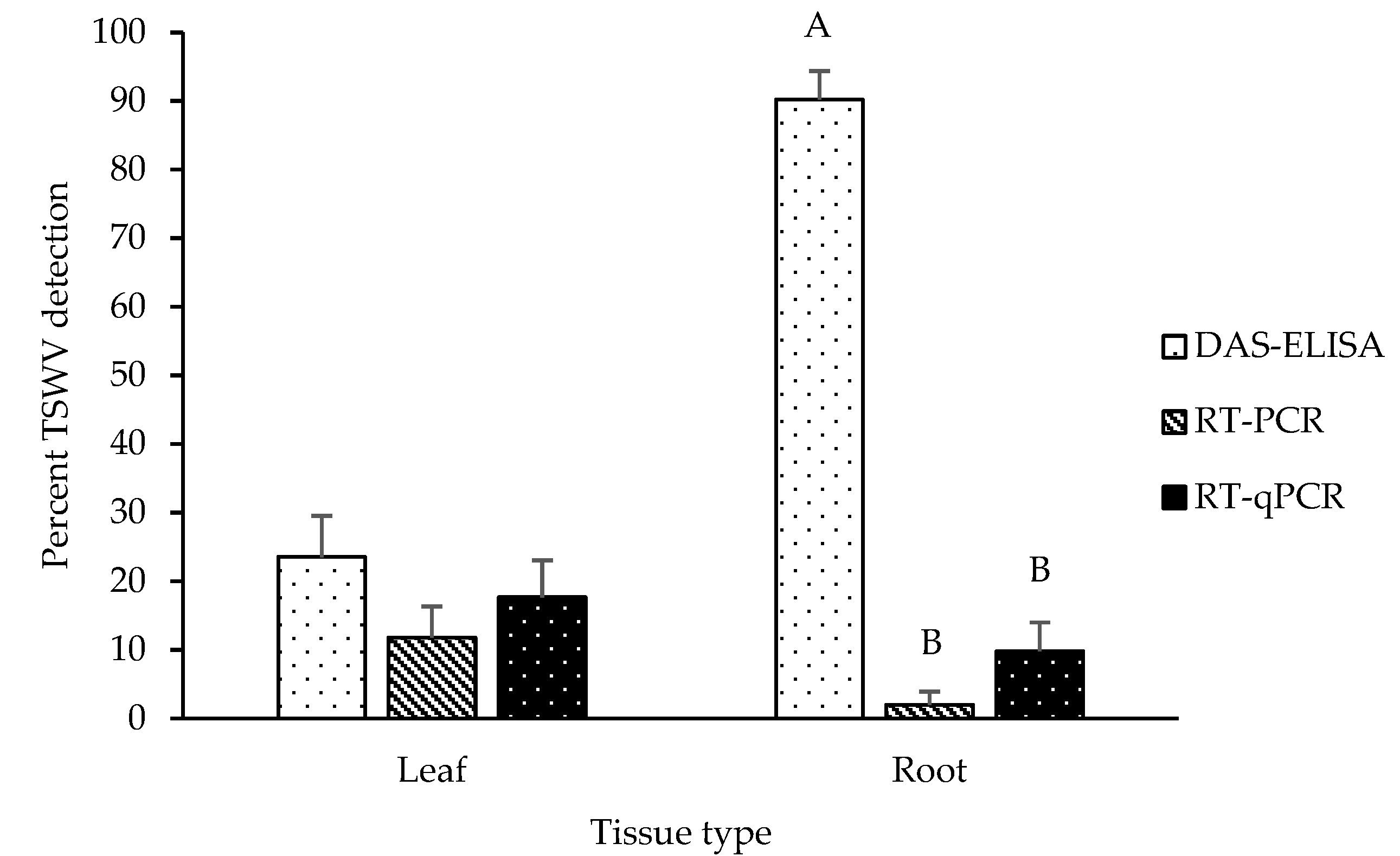
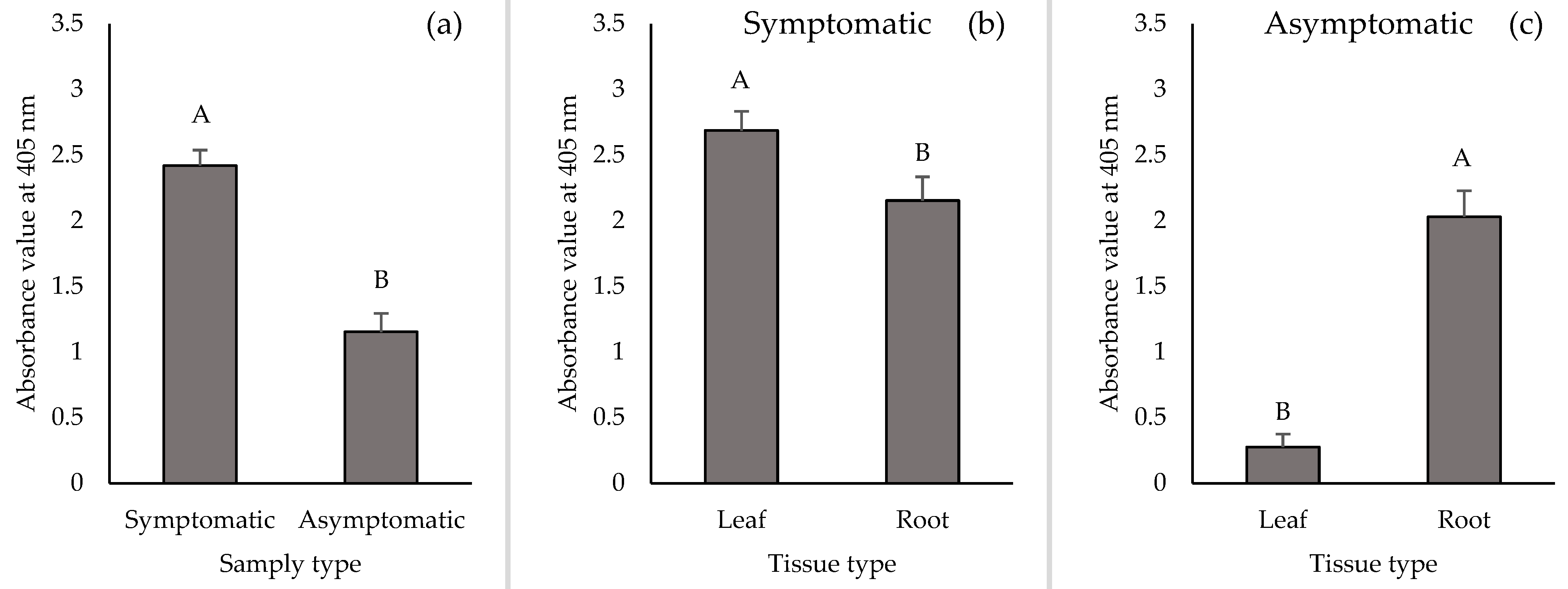
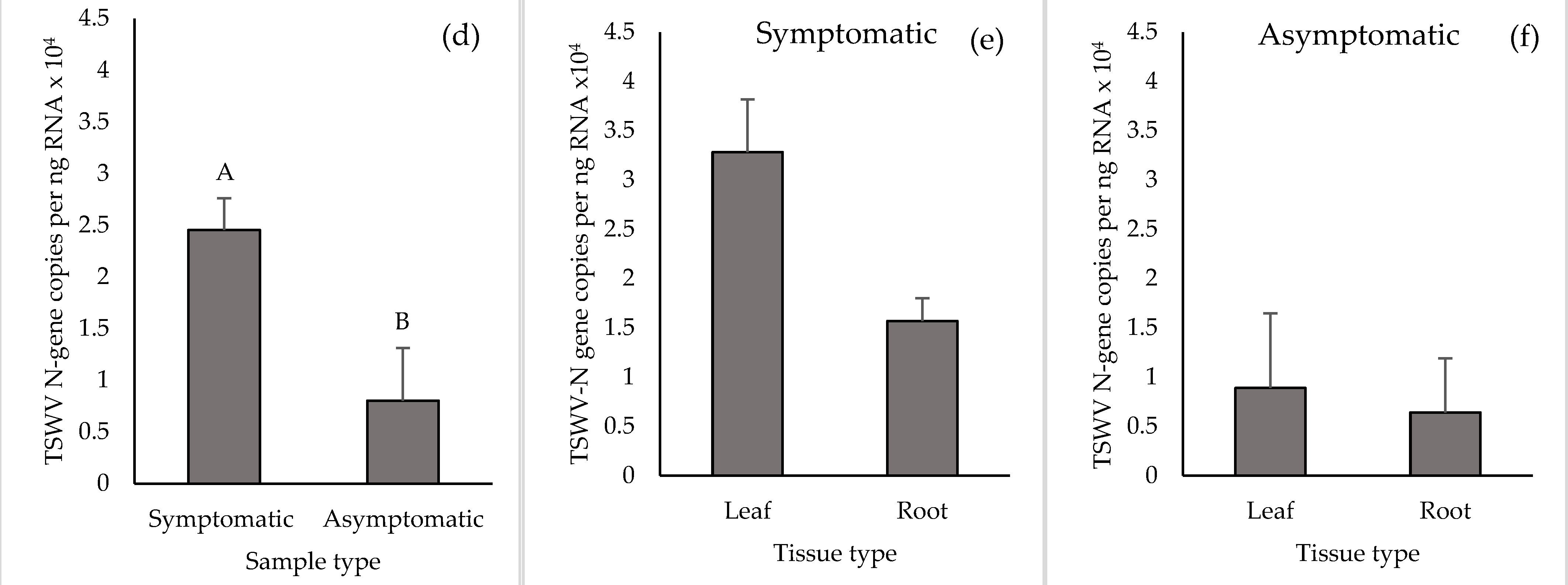
2.1. Detection of TSWV in Leaf and Root Tissues of Symptomatic Plants
2.2. Detection of TSWV in Leaf and Root Tissues of Asymptomatic Plants
2.3. TSWV Accumulation in Leaf and Root Tissues of Symptomatic and Asymptomatic Plants by DAS-ELISA and RT-qPCR
3. Discussion
4. Materials and Methods
4.1. Sample Collection
4.2. Detection and Quantitation of TSWV in Leaf and Root Tissue Samples by DAS-ELISA
4.3. Detection and Quantitation of TSWV in Leaf and Root Tissue Samples by RT-PCR and RT-qPCR
4.4. Statistical Analyses
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Culbreath, A.K.; Todd, J.W.; Brown, S.L. Epidemiology and Management of Tomato Spotted Wilt in Peanut. Annu. Rev. Phytopathol. 2003, 41, 53–75. [Google Scholar] [CrossRef] [PubMed]
- Riley, D.G.; Joseph, S.V.; Srinivasan, R.; Diffie, S. Thrips Vectors of Tospoviruses. J. Integr. Pest Manag. 2011, 1, 1–10. [Google Scholar] [CrossRef]
- Rotenberg, D.; Jacobson, A.L.; Schneweis, D.J.; Whitfield, A.E. Thrips Transmission of Tospoviruses. Curr. Opin. Virol. 2015, 15, 80–89. [Google Scholar] [CrossRef]
- Parrella, G.; Gognalons, P.; Gebre-Selassiè, K.; Vovlas, C.; Marchoux, G. An update of the host range of tomato spotted wilt virus. J. Plant Pathol. 2003, 85, 227–264. Available online: http://www.jstor.org/stable/41998156 (accessed on 28 October 2021).
- Black, M.C.; Lummus, P.F.; Smith, D.H.; Demski, J.W. An Epidemic of Spotted Wilt Disease in South Texas Peanuts in 1985. Proc. Am. Peanut Res. Ed. Soc. 1986, 18, 66. [Google Scholar]
- Culbreath, A.K.; Todd, J.W.; Demski, J.W.; Chamberlin, J.R. Disease Progress of Spotted Wilt in Peanut Cultivars Florunner and Southern Runner. Phytopathology 1992, 82, 766–771. [Google Scholar] [CrossRef]
- Garcia, L.E.; Brandenburg, R.L.; Bailey, J.E. Incidence of Tomato Spotted Wilt Virus (Bunyaviridae) and Tobacco Thrips in Virginia-Type Peanuts in North Carolina. Plant Dis. 2000, 84, 459–464. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Hagan, A.K.; Weeks, J.R.; French, J.C.; Gudauskas, R.T.; Mullen, J.M.; Gazaway, W.S.; Shelby, R. Tomato Spotted Wilt Virus in Peanut in Alabama. Plant Dis. 1990, 74, 615. [Google Scholar] [CrossRef]
- Halliwell, R.S.; Philley, G. Spotted Wilt of Peanut in Texas. Plant Dis. Rep. 1974, 58, 23–25. [Google Scholar]
- Reed, J.T.; Sukamto, S. Thrips and Tomato Spotted Wilt Virus in a Mississippi Peanut Field. In Thrips Biology and Management; Parker, B.L., Skinner, M., Lewis, T., Eds.; Springer: Boston, MA, USA, 1995; pp. 171–173. [Google Scholar] [CrossRef]
- Little, E.L. 2018 Georgia Plant Disease Loss Estimates. 2020. Available online: https://extension.uga.edu/publications/detail.html?number=AP102-11 (accessed on 13 August 2021).
- Little, E.L. 2017 Georgia Plant Disease Loss Estimates. 2019. Available online: https://extension.uga.edu/publications/detail.html?number=AP102-10 (accessed on 13 August 2021).
- Little, E.L. 2016 Georgia Plant Disease Loss Estimates. 2019. Available online: https://extension.uga.edu/publications/detail.html?number=AP102-9 (accessed on 13 August 2021).
- Little, E.L. 2015 Georgia Plant Disease Loss Estimates. Athens, GA. 2017. Available online: https://extension.uga.edu/publications/detail.html?number=AP102-8 (accessed on 13 August 2021).
- USDA-NASS Crop Production 2019 Summary. 2020. Available online: https://www.nass.usda.gov/Publications/Todays_Reports/reports/cropan20.pdf (accessed on 13 August 2021).
- Culbreath, A.K.; Todd, J.W.; Demski, J.W. Comparison of Hidden and Apparent Spotted Wilt Epidemics in Peanut. Proc. Am. Peanut Res. Ed. Soc. 1992, 24, 39. [Google Scholar]
- Culbreath, A.K.; Todd, J.W.; Demski, J.W. Productivity of Florunner Peanut Infected with Tomato Spotted Wilt Virus. Peanut Sci. 1992, 19, 11–14. [Google Scholar] [CrossRef]
- Cantonwine, E.G.; Culbreath, A.K.; Stevenson, K.L.; Kemerait, R.C.; Brenneman, T.B.; Smith, N.B.; Mullinix, B.G., Jr. Integrated Disease Management of Leaf Spot and Spotted Wilt of Peanut. Plant Dis. 2006, 90, 493–500. [Google Scholar] [CrossRef]
- Culbreath, A.K.; Tillman, B.L.; Tubbs, R.S.; Beasley, J.P.; Kemerait, R.C.; Brenneman, T.B. Interactive Effects of Planting Date and Cultivar on Tomato Spotted Wilt of Peanut. Plant Dis. 2010, 94, 898–904. [Google Scholar] [CrossRef]
- Wells, M.L.; Culbreath, A.K.; Todd, J.W.; Brown, S.L.; Gorbet, D.W. A Regression Approach for Comparing Field Resistance of Peanut Cultivars to Tomato Spotted Wilt Tospovirus. Crop Prot. 2002, 21, 467–474. [Google Scholar] [CrossRef]
- Culbreath, A.K.; Csinos, A.S.; Brenneman, T.B.; Demski, J.W.; Todd, J.W. Association of Tomato Spotted Wilt Virus with Foliar Chlorosis of Peanut in Georgia. Plant Dis. 1991, 75, 863C. [Google Scholar] [CrossRef]
- Dang, P.M.; Rowland, D.L.; Faircloth, W.H. Comparison of ELISA and RT-PCR Assays for the Detection of Tomato Spotted Wilt Virus in Peanut. Peanut Sci. 2009, 36, 133–137. [Google Scholar] [CrossRef][Green Version]
- Jain, R.K.; Pappu, S.S.; Pappu, H.R.; Culbreath, A.K.; Todd, J.W. Molecular Diagnosis of Tomato Spotted Wilt Tospovirus Infection of Peanut and Other Field and Greenhouse Crops. Plant Dis. 1998, 82, 900–904. [Google Scholar] [CrossRef]
- Kresta, K.K.; Mitchell, F.L.; Smith, J.W. Survey by ELISA of Thrips (Thysanoptera: Thripidae) Vectored Tomato Spotted Wilt Virus Distribution in Foliage and Flowers of Field-Infected Peanut. Peanut Sci. 1995, 22, 141–149. [Google Scholar] [CrossRef]
- Murakami, M.; Gallo-Meagher, M.; Gorbet, D.W.; Meagher, R.L. Utilizing Immunoassays to Determine Systemic Tomato Spotted Wilt Virus Infection for Elucidating Field Resistance in Peanut. Crop Prot. 2006, 25, 235–243. [Google Scholar] [CrossRef]
- Pappu, S.S.; Pappu, H.R.; Culbreath, A.K.; Todd, J.W. Localization of Tomato Spotted Wilt Virus (Genus Tospovirus, Family Bunyaviridae) in Peanut Pods. Peanut Sci. 1999, 26, 98–100. [Google Scholar] [CrossRef]
- Rowland, D.; Dorner, J.; Sorensen, R.; Beasley, J.P.; Todd, J. Tomato Spotted Wilt Virus in Peanut Tissue Types and Physiological Effects Related to Disease Incidence and Severity. Plant Pathol. 2005, 54, 431–440. [Google Scholar] [CrossRef]
- Wang, M.L.; Pinnow, D.L.; Pittman, R.N. Preliminary Screening of Peanut Germplasm in the US Collection for TSWV Resistance and High Flavonoid Content. Plant Pathol. J. 2007, 6, 219–226. [Google Scholar] [CrossRef]
- Shrestha, A.; Sundaraj, S.; Culbreath, A.K.; Riley, D.G.; Abney, M.R.; Srinivasan, R. Effects of Thrips Density, Mode of Inoculation, and Plant Age on Tomato Spotted Wilt Virus Transmission in Peanut Plants. Environ. Entomol. 2015, 44, 136–143. [Google Scholar] [CrossRef] [PubMed]
- Shrestha, A.; Srinivasan, R.; Sundaraj, S.; Culbreath, A.K.; Riley, D.G. Second Generation Peanut Genotypes Resistant to Thrips-Transmitted Tomato Spotted Wilt Virus Exhibit Tolerance Rather than True Resistance and Differentially Affect Thrips Fitness. J. Econ. Entomol. 2013, 106, 587–596. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Sundaraj, S.; Srinivasan, R.; Culbreath, A.K.; Riley, D.G.; Pappu, H.R. Host Plant Resistance against Tomato Spotted Wilt Virus in Peanut (Arachis hypogaea) and Its Impact on Susceptibility to the Virus, Virus Population Genetics, and Vector Feeding Behavior and Survival. Phytopathology 2014, 104, 202–210. [Google Scholar] [CrossRef] [PubMed]
- Culbreath, A.K.; Tubbs, R.S.; Tillman, B.L.; Beasley, J.P.; Branch, W.D.; Holbrook, C.C.; Smith, A.R.; Smith, N.B. Effects of Seeding Rate and Cultivar on Tomato Spotted Wilt of Peanut. Crop Prot. 2013, 53, 118–124. [Google Scholar] [CrossRef]
- Culbreath, A.K.; Tillman, B.L.; Gorbet, D.W.; Holbrook, C.C.; Nischwitz, C. Response of New Field-Resistant Peanut Cultivars to Twin-Row Pattern or in-Furrow Applications of Phorate for Management of Spotted Wilt. Plant Dis. 2008, 92, 1307–1312. [Google Scholar] [CrossRef]
- Culbreath, A.K.; Todd, J.W.; Gorbet, D.W.; Brown, S.L.; Baldwin, J.; Pappu, H.R.; Shokes, F.M. Reaction of Peanut Cultivars to Spotted Wilt. Peanut Sci. 2000, 27, 35–39. [Google Scholar] [CrossRef]
- Clark, M.F.; Adams, A.N. Characteristics of the Microplate Method of Enzyme-linked Immunosorbent Assay for the Detection of Plant Viruses. J. Gen. Virol. 1977, 34, 475–483. [Google Scholar] [CrossRef]
- Sherwood, J.L.; Sanborn, M.R.; Keyser, G.C.; Myers, L.D. Use of Monoclonal Antibodies in Detection of Tomato Spotted Wilt Virus. Phytopathology 1989, 79, 61–64. [Google Scholar] [CrossRef]
- Boonham, N.; Kreuze, J.; Winter, S.; van der Vlugt, R.; Bergervoet, J.; Tomlinson, J.; Mumford, R. Methods in Virus Diagnostics: From ELISA to Next Generation Sequencing. Virus Res. 2014, 186, 20–31. [Google Scholar] [CrossRef] [PubMed]
- Torrance, L.; Jones, R.A.C. Recent Developments in Serological Methods Suited for Use in Routine Testing for Plant Viruses. Plant Pathol. 1981, 30, 1–24. [Google Scholar] [CrossRef]
- Ward, E.; Foster, S.J.; Fraaije, B.A.; McCartney, H.A. Plant Pathogen Diagnostics: Immunological and Nucleic Acid-Based Approaches. Ann. Appl. Biol. 2004, 145, 1–16. [Google Scholar] [CrossRef]
- Schochetman, G.; Ou, C.-Y.; Jones, W.K. Polymerase Chain Reaction. J. Infect. Dis. 1988, 158, 1154–1157. Available online: https://www.jstor.org/stable/30137034 (accessed on 13 August 2021). [CrossRef]
- Putnam, M.L. Evaluation of Selected Methods of Plant Disease Diagnosis. Crop Prot. 1995, 14, 517–525. [Google Scholar] [CrossRef]
- Mackay, I.M.; Arden, K.E.; Nitsche, A. Real-Time PCR in Virology. Nucleic Acids Res. 2002, 30, 1292–1305. [Google Scholar] [CrossRef]
- Boonham, N.; Smith, P.; Walsh, K.; Tame, J.; Morris, J.; Spence, N.; Bennison, J.; Barker, I. The Detection of Tomato Spotted Wilt Virus (TSWV) in Individual Thrips Using Real Time Fluorescent RT-PCR (TaqMan). J. Virol. Methods 2002, 101, 37–48. [Google Scholar] [CrossRef]
- Rotenberg, D.; Kumar, N.K.K.; Ullman, D.E.; Montero-Astúa, M.; Willis, D.K.; German, T.L.; Whitfield, A.E. Variation in Tomato Spotted Wilt Virus Titer in Frankliniella occidentalis and Its Association with Frequency of Transmission. Phytopathology 2009, 99, 404–410. [Google Scholar] [CrossRef]
- Culbreath, A.K.; Srinivasan, R. Epidemiology of Spotted Wilt Disease of Peanut Caused by Tomato Spotted Wilt Virus in the Southeastern U.S. Virus Res. 2011, 159, 101–109. [Google Scholar] [CrossRef] [PubMed]
- Lyerly, J.H.; Stalker, H.T.; Moyer, J.W.; Hoffman, K. Evaluation of Arachis Species for Resistance to Tomato Spotted Wilt Virus. Peanut Sci. 2002, 29, 79–84. [Google Scholar] [CrossRef]
- Lacroix, C.; Renner, K.; Cole, E.; Seabloom, E.W.; Borer, E.T.; Malmstrom, C.M. Methodological Guidelines for Accurate Detection of Viruses in Wild Plant Species. Appl. Environ. Microbiol. 2016, 82, 1966–1975. [Google Scholar] [CrossRef] [PubMed]
- Mahmoud, S.Y.M. Detection of Cucumber Mosaic Virus in Some Ornamental Plants and Elimination of Nonspecific ELISA Reactions. Arch. Phytopathol. Plant Prot. 2011, 44, 1410–1424. [Google Scholar] [CrossRef]
- Sikora, E.J.; Andrianifahanana, M.; Murphy, J.F. Detection of Cucumber Mosaic Cucumovirus in Weed Species: A Cautionary Report on Nonspecific Reactions in ELISA. Can. J. Plant Pathol. 1999, 21, 338–344. [Google Scholar] [CrossRef]
- Van Schadewijk, A.R.; Meekes, E.T.M.; Verbeek, M.; Verhoeven, J.T.J. Validation of Plant Virus Detection. Acta Hortic. 2011, 901, 81–86. [Google Scholar] [CrossRef]
- Hoffmann, K.; Geske, S.M.; Moyer, J.W. Pathogenesis of Tomato Spotted Wilt Virus in Peanut Plants Dually Infected with Peanut Mottle Virus. Plant Dis. 1998, 82, 610–614. [Google Scholar] [CrossRef]
- Pappu, S.S.; Black, M.C.; Pappu, H.R.; Brenneman, T.B.; Culbreath, A.K.; Todd, J.W. First Report of Natural Infection of Peanut (Groundnut) by Impatiens Necrotic Spot Tospovirus (Family Bunyaviridae). Plant Dis. 1999, 83, 966. [Google Scholar] [CrossRef]
- Sreenivasulu, P.; Demski, J.W.; Reddy, D.V.R.; Misari, S.M.; Olorunju, P.E.; Kuhn, C.W. Tomato Spotted Wilt Virus (TSWV) and Strains of Peanut Mottle Virus That Mimic TSWV Symptoms in Peanut in Georgia. Plant Dis. 1988, 72, 546. [Google Scholar] [CrossRef]
- Brecke, B.J.; Funderburk, J.E.; Teare, I.D.; Gorbet, D.W. Interaction of Early-Season Herbicide Injury, Tobacco Thrips Injury, and Cultivar on Peanut. Agron. J. 1996, 88, 14–18. [Google Scholar] [CrossRef]
- Mandal, B.; Pappu, H.R.; Culbreath, A.K.; Holbrook, C.C.; Gorbet, D.W.; Todd, J.W. Differential Response of Selected Peanut (Arachis hypogaea) Genotypes to Mechanical Inoculation by Tomato Spotted Wilt Virus. Plant Dis. 2002, 86, 939–944. [Google Scholar] [CrossRef][Green Version]
- Morichetti, S.; Ferrell, J.; Devotka, P. Diagnosing Herbicide Injury in Peanut. EDIS 2010, 4. Available online: https://journals.flvc.org/edis/article/view/118669 (accessed on 13 August 2021).
- López, M.M.; Bertolini, E.; Olmos, A.; Caruso, P.; Gorris, M.T.; Llop, P.; Penyalver, R.; Cambra, M. Innovative Tools for Detection of Plant Pathogenic Viruses and Bacteria. Int. Microbiol. 2003, 6, 233–243. [Google Scholar] [CrossRef]
- Schaad, N.W.; Frederick, R.D.; Shaw, J.; Schneider, W.L.; Hickson, R.; Petrillo, M.D.; Luster, D.G. Advances in Molecular-Based Diagnostics in Meeting Crop Biosecurity and Phytosanitary Issues. Annu. Rev. Phytopathol. 2003, 41, 305–324. [Google Scholar] [CrossRef]
- Adams, A.N.; Guise, C.M.; Crossley, S.J. Plum Pox Virus Detection in Dormant Plum Trees by PCR and ELISA. Plant Pathol. 1999, 48, 240–244. [Google Scholar] [CrossRef]
- Hu, J.S.; Li, H.P.; Barry, K.; Wang, M.; Jordan, R. Comparison of Dot Blot, ELISA, and RT-PCR Assays for Detection of Two Cucumber Mosaic Virus Isolates Infecting Banana in Hawaii. Plant Dis. 1995, 79, 902–906. [Google Scholar] [CrossRef]
- Mekuria, G.; Ramesh, S.A.; Alberts, E.; Bertozzi, T.; Wirthensohn, M.; Collins, G.; Sedgley, M. Comparison of ELISA and RT-PCR for the Detection of Prunus Necrotic Ring Spot Virus and Prune Dwarf Virus in Almond (Prunus dulcis). J. Virol. Methods 2003, 114, 65–69. [Google Scholar] [CrossRef]
- Mumford, R.A.; Barker, I.; Wood, K.R. The Detection of Tomato Spotted Wilt Virus Using the Polymerase Chain Reaction. J. Virol. Methods 1994, 46, 303–311. [Google Scholar] [CrossRef]
- Sanchez-Navarro, J.A.; Aparicio, F.; Rowhani, A.; Pallás, V. Comparative Analysis of ELISA, Nonradioactive Molecular Hybridization and PCR for the Detection of Prunus Necrotic Ringspot Virus in Herbaceous and Prunus Hosts. Plant Pathol. 1998, 47, 780–786. [Google Scholar] [CrossRef]
- Webster, C.G.; Turechek, W.W.; Li, W.; Kousik, C.S.; Adkins, S. Development and Evaluation of ELISA and QRT-PCR for Identification of Squash Vein Yellowing Virus in Cucurbits. Plant Dis. 2017, 101, 178–185. [Google Scholar] [CrossRef] [PubMed]
- Gunn, L.V.; Pares, R.D. Effect of Potato Physiology on the Interpretation of ELISA Results for Potato Leafroll Virus. Plant Pathol. 1988, 37, 516–521. [Google Scholar] [CrossRef]
- Naidu, R.A.; Hughes, J.D.A. Methods for the Detection of Plant Virus Diseases. In Proceedings of the Plant Virology in sub-Saharan Africa: Proceedings of a Conference Organized by IITA, Ibadan, Nigeria, 4–8 June 2001; pp. 233–260. [Google Scholar]
- Etzler, M.E. Plant Lectins: Molecular and Biological Aspects. Ann. Rev. Plant Physiol. 1985, 36, 209–234. [Google Scholar] [CrossRef]
- Redolfi, P. Occurrence of Pathogenesis-Related (b) and Similar Proteins in Different Plant Species. Neth. J. Plant Pathol. 1983, 89, 245–254. [Google Scholar] [CrossRef]
- Baetz, U.; Martinoia, E. Root Exudates: The Hidden Part of Plant Defense. Trends Plant Sci. 2014, 19, 90–98. [Google Scholar] [CrossRef] [PubMed]
- Kijne, J.W.; Bauchrowitz, M.A.; Diaz, C.L. Root Lectins and Rhizobia. Plant Physiol. 1997, 115, 869–873. [Google Scholar] [CrossRef][Green Version]
- Peumans, W.J.; Van Damme, E.J.M. Lectins as Plant Defense Proteins. Plant Physiol. 1995, 109, 347–352. [Google Scholar] [CrossRef] [PubMed]
- Sutula, C.L.; Gillett, J.M.; Morrissey, S.M.; Ramsdell, D.C. Interpreting ELISA Data and Establishing the Positive-Negative Threshold. Plant Dis. 1986, 70, 722. [Google Scholar] [CrossRef]
- Rådström, P.; Löfström, C.; Lövenklev, M.; Knutsson, R.; Wolffs, P. Strategies for Overcoming PCR Inhibition. Cold Spring Harb. Protoc. 2008, 3, 1–10. [Google Scholar] [CrossRef]
- Wilson, I.G. Inhibition and Facilitation of Nucleic Acid Amplification. Appl. Environ. Microbiol. 1997, 63, 3741–3751. [Google Scholar] [CrossRef]
- Rådström, P.; Knutsson, R.; Wolffs, P.; Lövenklev, M.; Löfström, C. Pre-PCR Processing: Strategies to Generate PCR-Compatible Samples. Mol. Biotechnol. 2004, 26, 133–146. [Google Scholar] [CrossRef]
- Schaad, N.W.; Frederick, R.D. Real-Time PCR and Its Application for Rapid Plant Disease Diagnostics. Can. J. Plant Pathol. 2002, 24, 250–258. [Google Scholar] [CrossRef]
- Zhu, Y.J.; Lim, S.T.S.; Schenck, S.; Arcinas, A.; Komor, E. RT-PCR and Quantitative Real-Time RT-PCR Detection of Sugarcane Yellow Leaf Virus (SCYLV) in Symptomatic and Asymptomatic Plants of Hawaiian Sugarcane Cultivars and the Correlation of SCYLV Titre to Yield. Eur. J. Plant Pathol. 2010, 127, 263–273. [Google Scholar] [CrossRef]
- Branch, W.D. Registration of ‘Georgia-16HO’ Peanut. J. Plant Regist. 2017, 11, 231–234. [Google Scholar] [CrossRef]
- Branch, W.D. Registration of ‘Georgia-06G’ Peanut. J. Plant Regist. 2007, 1, 120. [Google Scholar] [CrossRef]
- Branch, W.D. Registration of ‘Georgia Green’ Peanut. Crop Sci. 1996, 36, 806. [Google Scholar] [CrossRef]
- Tillman, B.L.; Gorbet, D.W. Registration of ‘TUFRunner “511”’ Peanut. J. Plant Regist. 2017, 11, 235–239. [Google Scholar] [CrossRef]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lai, P.-C.; Abney, M.R.; Chen, Y.-J.; Bag, S.; Srinivasan, R. Discrepancies in Serology-Based and Nucleic Acid-Based Detection and Quantitation of Tomato Spotted Wilt Orthotospovirus in Leaf and Root Tissues from Symptomatic and Asymptomatic Peanut Plants. Pathogens 2021, 10, 1476. https://doi.org/10.3390/pathogens10111476
Lai P-C, Abney MR, Chen Y-J, Bag S, Srinivasan R. Discrepancies in Serology-Based and Nucleic Acid-Based Detection and Quantitation of Tomato Spotted Wilt Orthotospovirus in Leaf and Root Tissues from Symptomatic and Asymptomatic Peanut Plants. Pathogens. 2021; 10(11):1476. https://doi.org/10.3390/pathogens10111476
Chicago/Turabian StyleLai, Pin-Chu, Mark R. Abney, Yi-Ju Chen, Sudeep Bag, and Rajagopalbabu Srinivasan. 2021. "Discrepancies in Serology-Based and Nucleic Acid-Based Detection and Quantitation of Tomato Spotted Wilt Orthotospovirus in Leaf and Root Tissues from Symptomatic and Asymptomatic Peanut Plants" Pathogens 10, no. 11: 1476. https://doi.org/10.3390/pathogens10111476
APA StyleLai, P.-C., Abney, M. R., Chen, Y.-J., Bag, S., & Srinivasan, R. (2021). Discrepancies in Serology-Based and Nucleic Acid-Based Detection and Quantitation of Tomato Spotted Wilt Orthotospovirus in Leaf and Root Tissues from Symptomatic and Asymptomatic Peanut Plants. Pathogens, 10(11), 1476. https://doi.org/10.3390/pathogens10111476







