Efficacy of a Modified Live Virus Vaccine against Porcine Reproductive and Respiratory Syndrome Virus 1 (PRRSV-1) Administered to 1-Day-Old Piglets in Front of Heterologous PRRSV-1 Challenge
Abstract
:1. Introduction
2. Results
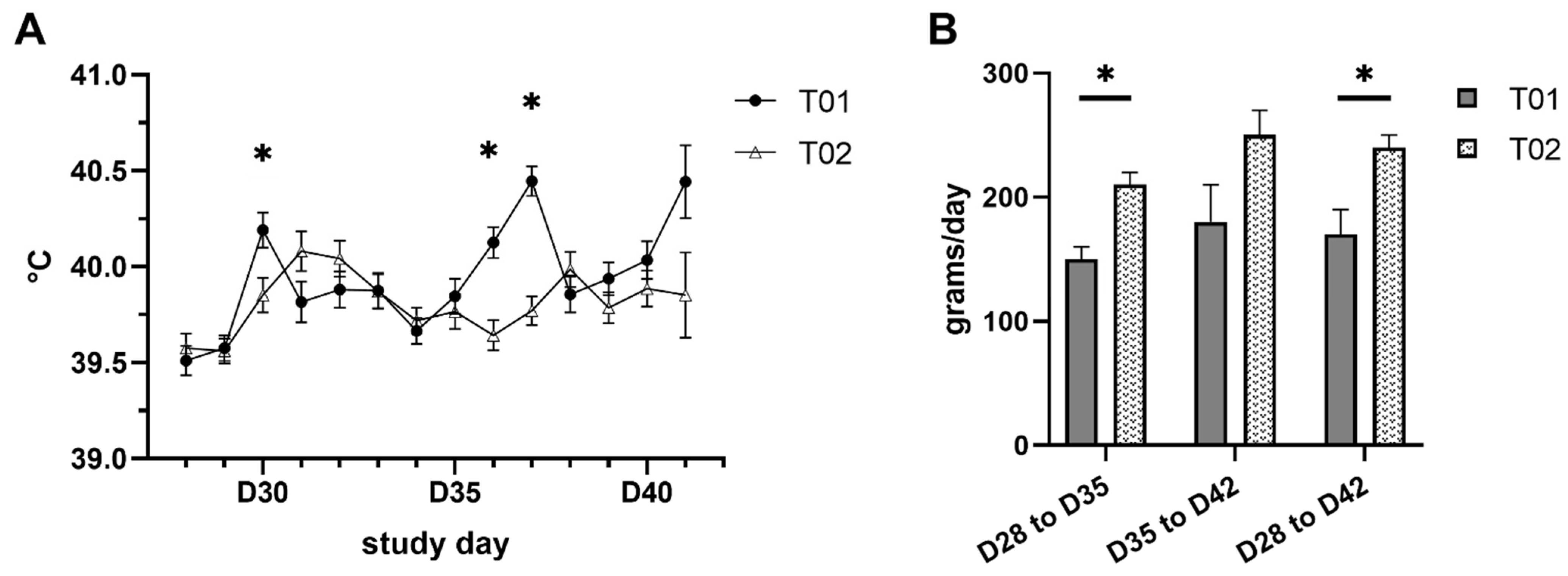
2.1. Clinical Signs and Body Weight
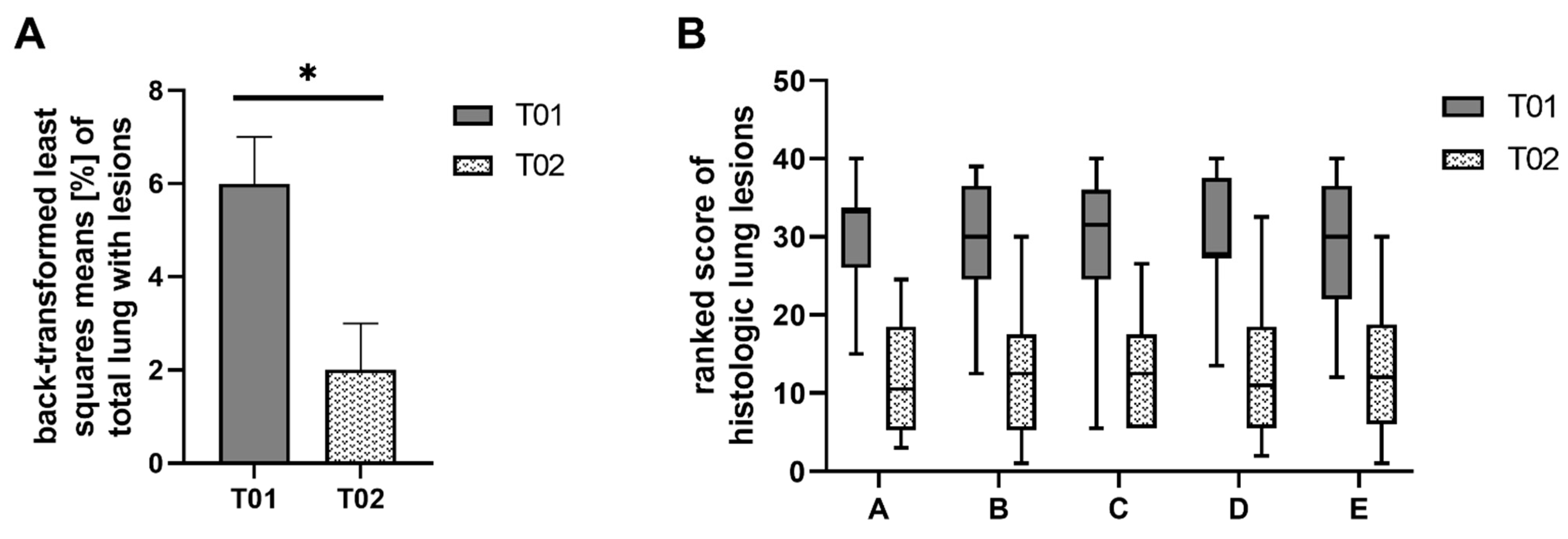
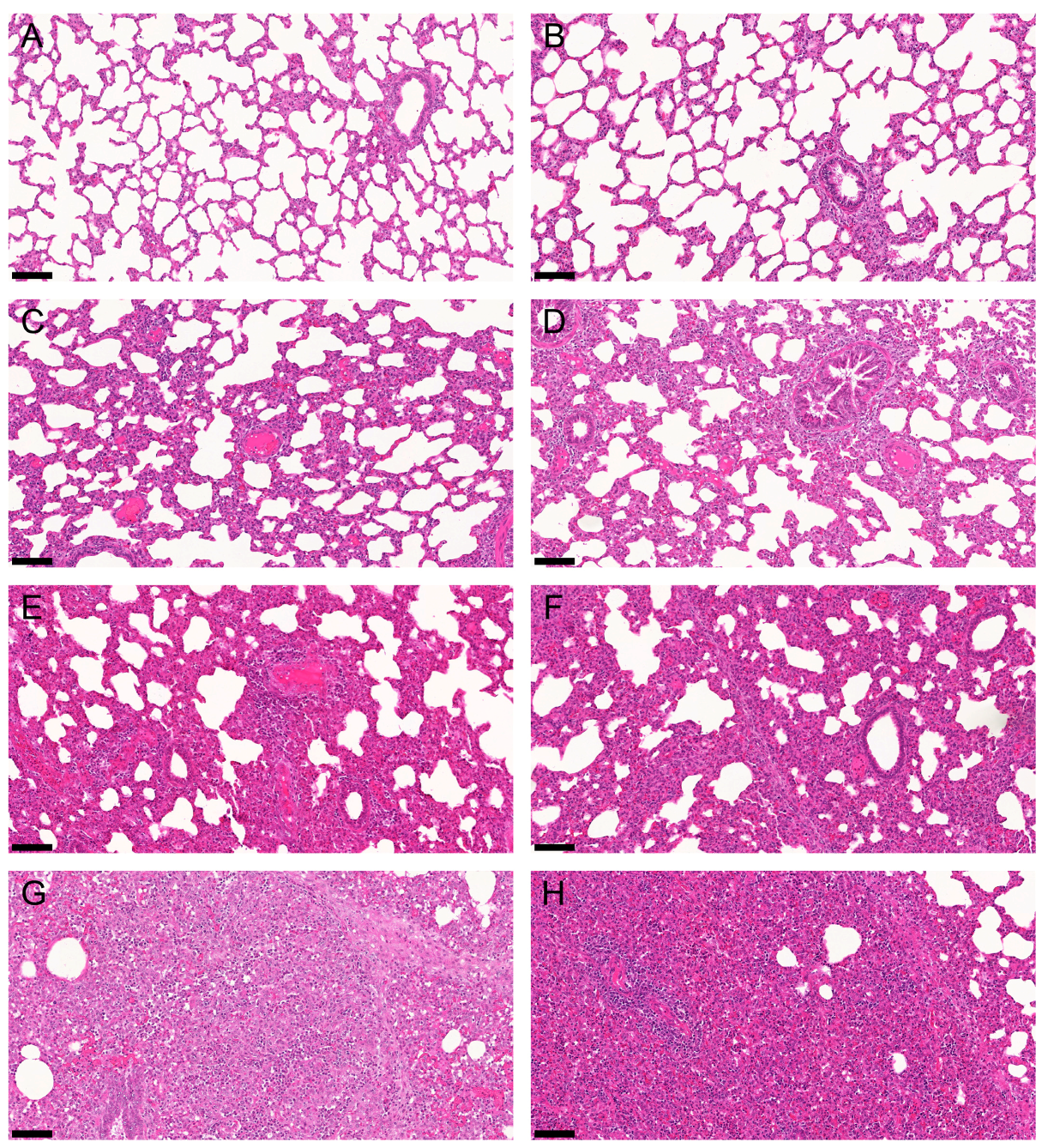
2.2. Gross and Histologic Lung Lesions
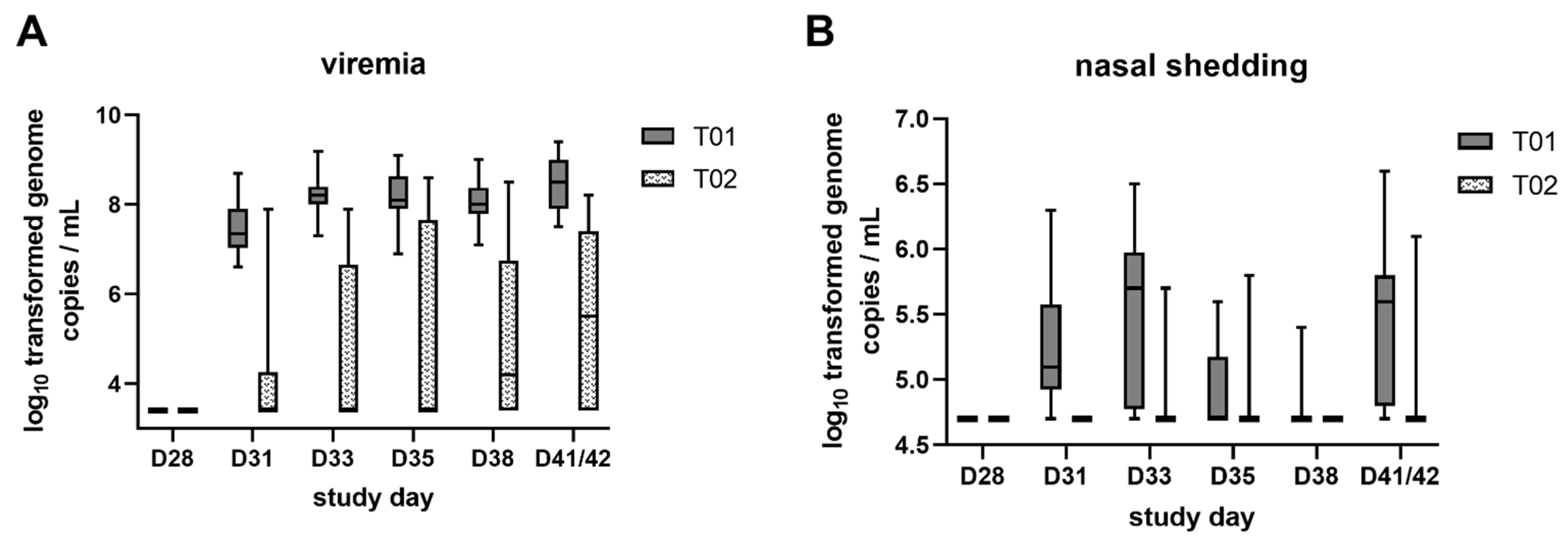
2.3. Viremia and Viral Shedding
2.4. IFN-γ Producing Peripheral Blood Lymphocytes and PRRSV Nucleoprotein Antibodies
2.5. Correlation of Viremia and IFN-γ Producing Peripheral Blood Lymphocytes
3. Discussion
4. Materials and Methods
4.1. Animals and Study Design
4.2. Vaccine and Challenge Virus
4.3. Clinical Examination and Necropsy
4.4. RTq-PCR Analysis
4.5. Serology
4.6. PRRSV-Specific IFN-γ ELISpot
4.7. Statistical Analysis
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Holtkamp, D.J.; Kliebenstein, J.B.; Neumann, E.J.; Zimmerman, J.J.; Rotto, H.F.; Yoder, T.K.; Wang, C.; Yeske, P.E.; Mowrer, C.L.; Haley, C.A. Assessment of the economic impact of porcine reproductive and respiratory syndrome virus on United States pork producers. J. Swine Health Prod. 2013, 21, 72–84. [Google Scholar]
- Oppeneder, A.; Griessler, A.; Voglmayr, T.; Reitböck, R.; Renzhammer, R.; Ritzmann, M.; Stadler, J.; Ladinig, A. Economic impact of a PRRS virus introduction via semen into farms with different PRRSV status. Berl. Münchener Tierärztliche Wochenschr. 2020, 133, 49–58. [Google Scholar] [CrossRef]
- Kuhn, J.H.; Lauck, M.; Bailey, A.L.; Shchetinin, A.M.; Vishnevskaya, T.V.; Bao, Y.M.; Ng, T.F.F.; LeBreton, M.; Schneider, B.S.; Gillis, A.; et al. Reorganization and expansion of the nidoviral family Arteriviridae. Arch. Virol. 2016, 161, 755–768. [Google Scholar] [CrossRef]
- Shi, M.; Lam, T.T.; Hon, C.C.; Murtaugh, M.P.; Davies, P.R.; Hui, R.K.; Li, J.; Wong, L.T.; Yip, C.W.; Jiang, J.W.; et al. Phylogeny-based evolutionary, demographical, and geographical dissection of North American type 2 porcine reproductive and respiratory syndrome viruses. J. Virol. 2010, 84, 8700–8711. [Google Scholar] [CrossRef] [Green Version]
- Stadejek, T.; Stankevicius, A.; Murtaugh, M.P.; Oleksiewicz, M.B. Molecular evolution of PRRSV in Europe: Current state of play. Vet. Microbiol. 2013, 165, 21–28. [Google Scholar] [CrossRef]
- Balka, G.; Podgorska, K.; Brar, M.S.; Balint, A.; Cadar, D.; Celer, V.; Denes, L.; Dirbakova, Z.; Jedryczko, A.; Marton, L.; et al. Genetic diversity of PRRSV 1 in Central Eastern Europe in 1994-2014: Origin and evolution of the virus in the region. Sci. Rep. 2018, 8, 7811. [Google Scholar] [CrossRef]
- Balint, A.; Balka, G.; Horvath, P.; Kecskemeti, S.; Dan, A.; Farsang, A.; Szeredi, L.; Banyai, K.; Bartha, D.; Olasz, F.; et al. Full-length genome sequence analysis of a Hungarian porcine reproductive and respiratory syndrome virus isolated from a pig with severe respiratory disease. Arch. Virol. 2015, 160, 417–422. [Google Scholar] [CrossRef] [PubMed]
- Frydas, I.S.; Trus, I.; Kvisgaard, L.K.; Bonckaert, C.; Reddy, V.R.; Li, Y.; Larsen, L.E.; Nauwynck, H.J. Different clinical, virological, serological and tissue tropism outcomes of two new and one old Belgian type 1 subtype 1 porcine reproductive and respiratory virus (PRRSV) isolates. Vet. Res. 2015, 46, 37. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sinn, L.J.; Klingler, E.; Lamp, B.; Brunthaler, R.; Weissenböck, H.; Rümenapf, T.; Ladinig, A. Emergence of a virulent porcine reproductive and respiratory syndrome virus (PRRSV) 1 strain in Lower Austria. Porcine Health Manag. 2016, 2, 28. [Google Scholar] [CrossRef] [Green Version]
- Canelli, E.; Catella, A.; Borghetti, P.; Ferrari, L.; Ogno, G.; De Angelis, E.; Corradi, A.; Passeri, B.; Bertani, V.; Sandri, G.; et al. Phenotypic characterization of a highly pathogenic Italian porcine reproductive and respiratory syndrome virus (PRRSV) type 1 subtype 1 isolate in experimentally infected pigs. Vet. Microbiol. 2017, 210, 124–133. [Google Scholar] [CrossRef]
- Dürlinger, S.; Balka, G.; Rathkjen, P.H.; Kraft, C.; Morgenstern, R.; Rümenapf, T.; Ladinig, A. Efficacy of Ingelvac PRRSFLEX® EU against experimental challenge with PRRSV AUT15-33 (“ACRO” PRRSV). In Proceedings of the 11th European Symposium of Porcine Health Management (ESPHM), Utrecht, The Netherlands, 22–24 May 2019. [Google Scholar]
- Nathues, H.; Alarcon, P.; Rushton, J.; Jolie, R.; Fiebig, K.; Jimenez, M.; Geurts, V.; Nathues, C. Cost of porcine reproductive and respiratory syndrome virus at individual farm level—An economic disease model. Prev. Vet. Med. 2017, 142, 16–29. [Google Scholar] [CrossRef] [PubMed]
- Bonckaert, C.; van der Meulen, K.; Rodriguez-Ballara, I.; Pedrazuela Sanz, R.; Martinez, M.F.; Nauwynck, H.J. Modified-live PRRSV subtype 1 vaccine UNISTRAIN((R)) PRRS provides a partial clinical and virological protection upon challenge with East European subtype 3 PRRSV strain Lena. Porcine Health Manag. 2016, 2, 12. [Google Scholar] [CrossRef] [Green Version]
- Canelli, E.; Catella, A.; Borghetti, P.; Ferrari, L.; Ogno, G.; De Angelis, E.; Bonilauri, P.; Guazzetti, S.; Nardini, R.; Martelli, P. Efficacy of a modified-live virus vaccine in pigs experimentally infected with a highly pathogenic porcine reproductive and respiratory syndrome virus type 1 (HP-PRRSV-1). Vet. Microbiol. 2018, 226, 89–96. [Google Scholar] [CrossRef]
- Thomann, B.; Rushton, J.; Schuepbach-Regula, G.; Nathues, H. Modeling Economic Effects of Vaccination Against Porcine Reproductive and Respiratory Syndrome: Impact of Vaccination Effectiveness, Vaccine Price, and Vaccination Coverage. Front. Vet. Sci. 2020, 7, 500. [Google Scholar] [CrossRef] [PubMed]
- Charerntantanakul, W. Porcine reproductive and respiratory syndrome virus vaccines: Immunogenicity, efficacy and safety aspects. World J. Virol. 2012, 1, 23–30. [Google Scholar] [CrossRef] [PubMed]
- Balasch, M.; Fort, M.; Taylor, L.P.; Calvert, J.G. Vaccination of 1-day-old pigs with a porcine reproductive and respiratory syndrome virus (PRRSV) modified live attenuated virus vaccine is able to overcome maternal immunity. Porcine Health Manag. 2018, 4, 25. [Google Scholar] [CrossRef]
- Jeong, J.; Kim, S.; Park, K.H.; Kang, I.; Park, S.J.; Yang, S.; Oh, T.; Chae, C. Vaccination with a porcine reproductive and respiratory syndrome virus vaccine at 1-day-old improved growth performance of piglets under field conditions. Vet. Microbiol. 2018, 214, 113–124. [Google Scholar] [CrossRef]
- Balasch, M.; Fort, M.; Taylor, L.P.; Diaz, I.; Mateu, E.; Calvert, J.G. Immune response development after vaccination of 1-day-old naive pigs with a Porcine Reproductive and Respiratory Syndrome 1-based modified live virus vaccine. Porcine Health Manag. 2019, 5, 2. [Google Scholar] [CrossRef]
- Mateu, E.; Diaz, I. The challenge of PRRS immunology. Vet. J. 2008, 177, 345–351. [Google Scholar] [CrossRef]
- Loving, C.L.; Osorio, F.A.; Murtaugh, M.P.; Zuckermann, F.A. Innate and adaptive immunity against Porcine Reproductive and Respiratory Syndrome Virus. Vet. Immunol. Immunopathol. 2015, 167, 1–14. [Google Scholar] [CrossRef]
- Holtkamp, D.J.; Polson, D.D.; Torremorell, M.; Morrison, B.; Classen, D.M.; Becton, L.; Henry, S.; Rodibaugh, M.T.; Rowland, R.R.; Snelson, H.; et al. Terminology for classifying swine herds by porcine reproductive and respiratory syndrome virus status. J. Swine Health Prod. 2011, 19, 44–56. [Google Scholar]
- Stevenson, G.W.; Van Alstine, W.G.; Kanitz, C.L.; Keffaber, K.K. Endemic porcine reproductive and respiratory syndrome virus infection of nursery pigs in two swine herds without current reproductive failure. J. Vet. Diagn. Investig. 1993, 5, 432–434. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- De Regge, N.; Cay, B. Comparison of PRRSV Nucleic Acid and Antibody Detection in Pen-Based Oral Fluid and Individual Serum Samples in Three Different Age Categories of Post-Weaning Pigs from Endemically Infected Farms. PLoS ONE 2016, 11, e0166300. [Google Scholar] [CrossRef]
- Lopez, W.A.; Angulo, J.; Zimmerman, J.J.; Linhares, D.C.L. Porcine reproductive and respiratory syndrome monitoring in breeding herds using processing fluids. J. Swine Health Prod. 2018, 26, 146–150. [Google Scholar]
- Corzo, C.A.; Mondaca, E.; Wayne, S.; Torremorell, M.; Dee, S.; Davies, P.; Morrison, R.B. Control and elimination of porcine reproductive and respiratory syndrome virus. Virus Res. 2010, 154, 185–192. [Google Scholar] [CrossRef] [PubMed]
- Johnson, W.; Roof, M.; Vaughn, E.; Christopher-Hennings, J.; Johnson, C.R.; Murtaugh, M.P. Pathogenic and humoral immune responses to porcine reproductive and respiratory syndrome virus (PRRSV) are related to viral load in acute infection. Vet. Immunol. Immunopathol. 2004, 102, 233–247. [Google Scholar] [CrossRef]
- Martelli, P.; Gozio, S.; Ferrari, L.; Rosina, S.; De Angelis, E.; Quintavalla, C.; Bottarelli, E.; Borghetti, P. Efficacy of a modified live porcine reproductive and respiratory syndrome virus (PRRSV) vaccine in pigs naturally exposed to a heterologous European (Italian cluster) field strain: Clinical protection and cell-mediated immunity. Vaccine 2009, 27, 3788–3799. [Google Scholar] [CrossRef]
- Murtaugh, M.P.; Genzow, M. Immunological solutions for treatment and prevention of porcine reproductive and respiratory syndrome (PRRS). Vaccine 2011, 29, 8192–8204. [Google Scholar] [CrossRef]
- Li, X.; Galliher-Beckley, A.; Pappan, L.; Trible, B.; Kerrigan, M.; Beck, A.; Hesse, R.; Blecha, F.; Nietfeld, J.C.; Rowland, R.R.; et al. Comparison of host immune responses to homologous and heterologous type II porcine reproductive and respiratory syndrome virus (PRRSV) challenge in vaccinated and unvaccinated pigs. Biomed. Res. Int 2014, 2014, 416727. [Google Scholar] [CrossRef]
- Trus, I.; Bonckaert, C.; van der Meulen, K.; Nauwynck, H.J. Efficacy of an attenuated European subtype 1 porcine reproductive and respiratory syndrome virus (PRRSV) vaccine in pigs upon challenge with the East European subtype 3 PRRSV strain Lena. Vaccine 2014, 32, 2995–3003. [Google Scholar] [CrossRef]
- Wensvoort, G.; Terpstra, C.; Pol, J.M.; ter Laak, E.A.; Bloemraad, M.; de Kluyver, E.P.; Kragten, C.; van Buiten, L.; den Besten, A.; Wagenaar, F.; et al. Mystery swine disease in The Netherlands: The isolation of Lelystad virus. Vet. Q. 1991, 13, 121–130. [Google Scholar] [CrossRef] [PubMed]
- Rossow, K.D.; Bautista, E.M.; Goyal, S.M.; Molitor, T.W.; Murtaugh, M.P.; Morrison, R.B.; Benfield, D.A.; Collins, J.E. Experimental porcine reproductive and respiratory syndrome virus infection in one-, four-, and 10-week-old pigs. J. Vet. Diagn. Investig. 1994, 6, 3–12. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lunney, J.K.; Fang, Y.; Ladinig, A.; Chen, N.; Li, Y.; Rowland, B.; Renukaradhya, G.J. Porcine Reproductive and Respiratory Syndrome Virus (PRRSV): Pathogenesis and Interaction with the Immune System. Annu. Rev. Anim. Biosci. 2016, 4, 129–154. [Google Scholar] [CrossRef] [PubMed]
- Balka, G.; Ladinig, A.; Ritzmann, M.; Saalmüller, A.; Gerner, W.; Käser, T.; Jakab, C.; Rusvai, M.; Weissenböck, H. Immunohistochemical characterization of type II pneumocyte proliferation after challenge with type I porcine reproductive and respiratory syndrome virus. J. Comp. Pathol. 2013, 149, 322–330. [Google Scholar] [CrossRef] [PubMed]
- Duan, X.; Nauwynck, H.J.; Pensaert, M.B. Virus quantification and identification of cellular targets in the lungs and lymphoid tissues of pigs at different time intervals after inoculation with porcine reproductive and respiratory syndrome virus (PRRSV). Vet. Microbiol. 1997, 56, 9–19. [Google Scholar] [CrossRef]
- Labarque, G.; Van Gucht, S.; Nauwynck, H.; Van Reeth, K.; Pensaert, M. Apoptosis in the lungs of pigs infected with porcine reproductive and respiratory syndrome virus and associations with the production of apoptogenic cytokines. Vet. Res. 2003, 34, 249–260. [Google Scholar] [CrossRef] [Green Version]
- Lee, S.M.; Kleiboeker, S.B. Porcine reproductive and respiratory syndrome virus induces apoptosis through a mitochondria-mediated pathway. Virology 2007, 365, 419–434. [Google Scholar] [CrossRef] [Green Version]
- Zimmerman, J.J.; Dee, S.A.; Holtkamp, D.J.; Murtaugh, M.P.; Stadejek, T.; Stevenson, G.W.; Torremorell, M.; Yang, H.; Zhang, J. Porcine Reproductive and Respiratory Syndrome Viruses (Porcine Arteriviruses). In Diseases of Swine; Zimmerman, J.J., Karriker, L.A., Ramirez, A., Schwartz, K.J., Stevenson, G.W., Zhang, J., Eds.; Wiley-Blackwell: Hoboken, NJ, USA, 2019; Volume 11, pp. 685–708. [Google Scholar]
- Drigo, M.; Franzo, G.; Tucciarone, C.M.; Manfredda, A.; Zanardelli, P.; Basano, F.S.; Nazzari, R. Effect of early PRRSV (Porcine Reproductive and Respiratory Syndrome Virus) vaccination on pig health and performance: The earlier the better? In Proceedings of the 11th European Symposium of Porcine Health Management (ESPHM), Utrecht, The Netherlands, 22–24 May 2019. [Google Scholar]
- Wills, R.W.; Zimmerman, J.J.; Yoon, K.J.; Swenson, S.L.; Hoffman, L.J.; McGinley, M.J.; Hill, H.T.; Platt, K.B. Porcine reproductive and respiratory syndrome virus: Routes of excretion. Vet. Microbiol. 1997, 57, 69–81. [Google Scholar] [CrossRef]
- Cho, J.G.; Dee, S.A.; Deen, J.; Trincado, C.; Fano, E.; Jiang, Y.; Faaberg, K.; Murtaugh, M.P.; Guedes, A.; Collins, J.E.; et al. The impact of animal age, bacterial coinfection, and isolate pathogenicity on the shedding of porcine reproductive and respiratory syndrome virus in aerosols from experimentally infected pigs. Can. J. Vet. Res. 2006, 70, 297–301. [Google Scholar]
- Pileri, E.; Gibert, E.; Soldevila, F.; Garcia-Saenz, A.; Pujols, J.; Diaz, I.; Darwich, L.; Casal, J.; Martin, M.; Mateu, E. Vaccination with a genotype 1 modified live vaccine against porcine reproductive and respiratory syndrome virus significantly reduces viremia, viral shedding and transmission of the virus in a quasi-natural experimental model. Vet. Microbiol. 2015, 175, 7–16. [Google Scholar] [CrossRef]
- Yoon, I.J.; Joo, H.S.; Goyal, S.M.; Molitor, T.W. A modified serum neutralization test for the detection of antibody to porcine reproductive and respiratory syndrome virus in swine sera. J. Vet. Diagn. Investig. 1994, 6, 289–292. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lopez, O.J.; Osorio, F.A. Role of neutralizing antibodies in PRRSV protective immunity. Vet. Immunol. Immunopathol. 2004, 102, 155–163. [Google Scholar] [CrossRef] [PubMed]
- Cancel-Tirado, S.M.; Evans, R.B.; Yoon, K.J. Monoclonal antibody analysis of porcine reproductive and respiratory syndrome virus epitopes associated with antibody-dependent enhancement and neutralization of virus infection. Vet. Immunol. Immunopathol. 2004, 102, 249–262. [Google Scholar] [CrossRef] [PubMed]
- Fablet, C.; Renson, P.; Eono, F.; Mahe, S.; Eveno, E.; Le Dimna, M.; Normand, V.; Lebret, A.; Rose, N.; Bourry, O. Maternally-derived antibodies (MDAs) impair piglets’ humoral and cellular immune responses to vaccination against porcine reproductive and respiratory syndrome (PRRS). Vet. Microbiol. 2016, 192, 175–180. [Google Scholar] [CrossRef]
- Renson, P.; Fablet, C.; Andraud, M.; Normand, V.; Lebret, A.; Paboeuf, F.; Rose, N.; Bourry, O. Maternally-derived neutralizing antibodies reduce vaccine efficacy against porcine reproductive and respiratory syndrome virus infection. Vaccine 2019, 37, 4318–4324. [Google Scholar] [CrossRef]
- Talker, S.C.; Käser, T.; Reutner, K.; Sedlak, C.; Mair, K.H.; Koinig, H.; Graage, R.; Viehmann, M.; Klingler, E.; Ladinig, A.; et al. Phenotypic maturation of porcine NK- and T-cell subsets. Dev. Comp. Immunol. 2013, 40, 51–68. [Google Scholar] [CrossRef]
- Chase, C.; Lunney, J.K. Immune System. In Diseases of Swine; Zimmerman, J.J., Karriker, L.A., Ramirez, A., Schwartz, K.J., Stevenson, G.W., Zhang, J., Eds.; Wiley-Blackwell: Hoboken, NJ, USA, 2019; Volume 11, pp. 264–291. [Google Scholar]
- Diaz, I.; Darwich, L.; Pappaterra, G.; Pujols, J.; Mateu, E. Immune responses of pigs after experimental infection with a European strain of Porcine reproductive and respiratory syndrome virus. J. Gen. Virol. 2005, 86, 1943–1951. [Google Scholar] [CrossRef]
- Mair, K.H.; Koinig, H.; Gerner, W.; Hohne, A.; Bretthauer, J.; Kroll, J.J.; Roof, M.B.; Saalmüller, A.; Stadler, K.; Libanova, R. Carbopol improves the early cellular immune responses induced by the modified-life vaccine Ingelvac PRRS(R) MLV. Vet. Microbiol. 2015, 176, 352–357. [Google Scholar] [CrossRef] [Green Version]
- Rowland, R.R.; Robinson, B.; Stefanick, J.; Kim, T.S.; Guanghua, L.; Lawson, S.R.; Benfield, D.A. Inhibition of porcine reproductive and respiratory syndrome virus by interferon-gamma and recovery of virus replication with 2-aminopurine. Arch. Virol. 2001, 146, 539–555. [Google Scholar] [CrossRef]
- Zuckermann, F.A.; Garcia, E.A.; Luque, I.D.; Christopher-Hennings, J.; Doster, A.; Brito, M.; Osorio, F. Assessment of the efficacy of commercial porcine reproductive and respiratory syndrome virus (PRRSV) vaccines based on measurement of serologic response, frequency of gamma-IFN-producing cells and virological parameters of protection upon challenge. Vet. Microbiol. 2007, 123, 69–85. [Google Scholar] [CrossRef]
- Straw, B.E.; Backstrom, L.; Leman, A.D. Examination of Swine at Slaughter.2. Findings at Slaughter and Their Significance. Comp. Cont. Educ. Pract. 1986, 8, S106–S110. [Google Scholar]


| Histologic Lesion | LSM Difference between T01 and T02 | Degrees of Freedom | T-Value | p-Value |
|---|---|---|---|---|
| Intra-alveolar accumulation of inflammatory cells | 18.4 | 38 | 8.23 | <0.0001 |
| Perivascular accumulation of inflammatory cells | 17.4 | 36 | 7.35 | <0.0001 |
| Necrotic debris | 17.5 | 38 | 7.29 | <0.0001 |
| Pneumocytic hypertrophy and hyperplasia | 17.2 | 38 | 6.98 | <0.0001 |
| Septal infiltration with mononuclear cells | 16.0 | 38 | 5.98 | <0.0001 |
| Study Day | Sample | LSM Difference between T01 and T02 | Standard Error | Degrees of Freedom | p-Value |
|---|---|---|---|---|---|
| D31 | Serum | 3.40 | 0.40 | 56 | <0.0001 |
| Nasal swab | 0.54 | 0.10 | 42 | <0.0001 | |
| D33 | Serum | 3.47 | 0.40 | 56 | <0.0001 |
| Nasal swab | 0.72 | 0.13 | 41 | <0.0001 | |
| D35 | Serum | 2.88 | 0.40 | 56 | <0.0001 |
| Nasal swab | 0.08 | 0.10 | 37 | 0.405 | |
| D38 | Serum | 2.91 | 0.40 | 56 | <0.0001 |
| Nasal swab | 0.05 | 0.04 | 41 | 0.157 | |
| D41/42 | Serum | 3.04 | 0.40 | 56 | <0.0001 |
| Nasal swab | 0.59 | 0.17 | 36 | 0.002 |
| Study Day | LSM Difference between T01 and T02 | Standard Error | Degrees of Freedom | p-Value |
|---|---|---|---|---|
| D28 | −3.5 | 0.2 | 24 | <0.0001 |
| D35 | −2.7 | 0.3 | 34 | <0.0001 |
| D41/42 | −2.0 | 0.4 | 36 | <0.0001 |
| Study Day | LSM Difference between T01 and T02 | Standard Error | Degrees of Freedom | p-Value |
|---|---|---|---|---|
| D28 | −2.4 | 0.2 | 74 | <0.0001 |
| D41/42 | −0.7 | 0.2 | 74 | 0.002 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kreutzmann, H.; Dürlinger, S.; Knecht, C.; Koch, M.; Cabana, M.; Torrent, G.; Balasch, M.; Taylor, L.P.; Balka, G.; Gerner, W.; et al. Efficacy of a Modified Live Virus Vaccine against Porcine Reproductive and Respiratory Syndrome Virus 1 (PRRSV-1) Administered to 1-Day-Old Piglets in Front of Heterologous PRRSV-1 Challenge. Pathogens 2021, 10, 1342. https://doi.org/10.3390/pathogens10101342
Kreutzmann H, Dürlinger S, Knecht C, Koch M, Cabana M, Torrent G, Balasch M, Taylor LP, Balka G, Gerner W, et al. Efficacy of a Modified Live Virus Vaccine against Porcine Reproductive and Respiratory Syndrome Virus 1 (PRRSV-1) Administered to 1-Day-Old Piglets in Front of Heterologous PRRSV-1 Challenge. Pathogens. 2021; 10(10):1342. https://doi.org/10.3390/pathogens10101342
Chicago/Turabian StyleKreutzmann, Heinrich, Sophie Dürlinger, Christian Knecht, Michaela Koch, Marta Cabana, Gerard Torrent, Mònica Balasch, Lucas P. Taylor, Gyula Balka, Wilhelm Gerner, and et al. 2021. "Efficacy of a Modified Live Virus Vaccine against Porcine Reproductive and Respiratory Syndrome Virus 1 (PRRSV-1) Administered to 1-Day-Old Piglets in Front of Heterologous PRRSV-1 Challenge" Pathogens 10, no. 10: 1342. https://doi.org/10.3390/pathogens10101342
APA StyleKreutzmann, H., Dürlinger, S., Knecht, C., Koch, M., Cabana, M., Torrent, G., Balasch, M., Taylor, L. P., Balka, G., Gerner, W., & Ladinig, A. (2021). Efficacy of a Modified Live Virus Vaccine against Porcine Reproductive and Respiratory Syndrome Virus 1 (PRRSV-1) Administered to 1-Day-Old Piglets in Front of Heterologous PRRSV-1 Challenge. Pathogens, 10(10), 1342. https://doi.org/10.3390/pathogens10101342






