The Effect of Impaired Polyamine Transport on Pneumococcal Transcriptome
Abstract
:1. Introduction
2. Results
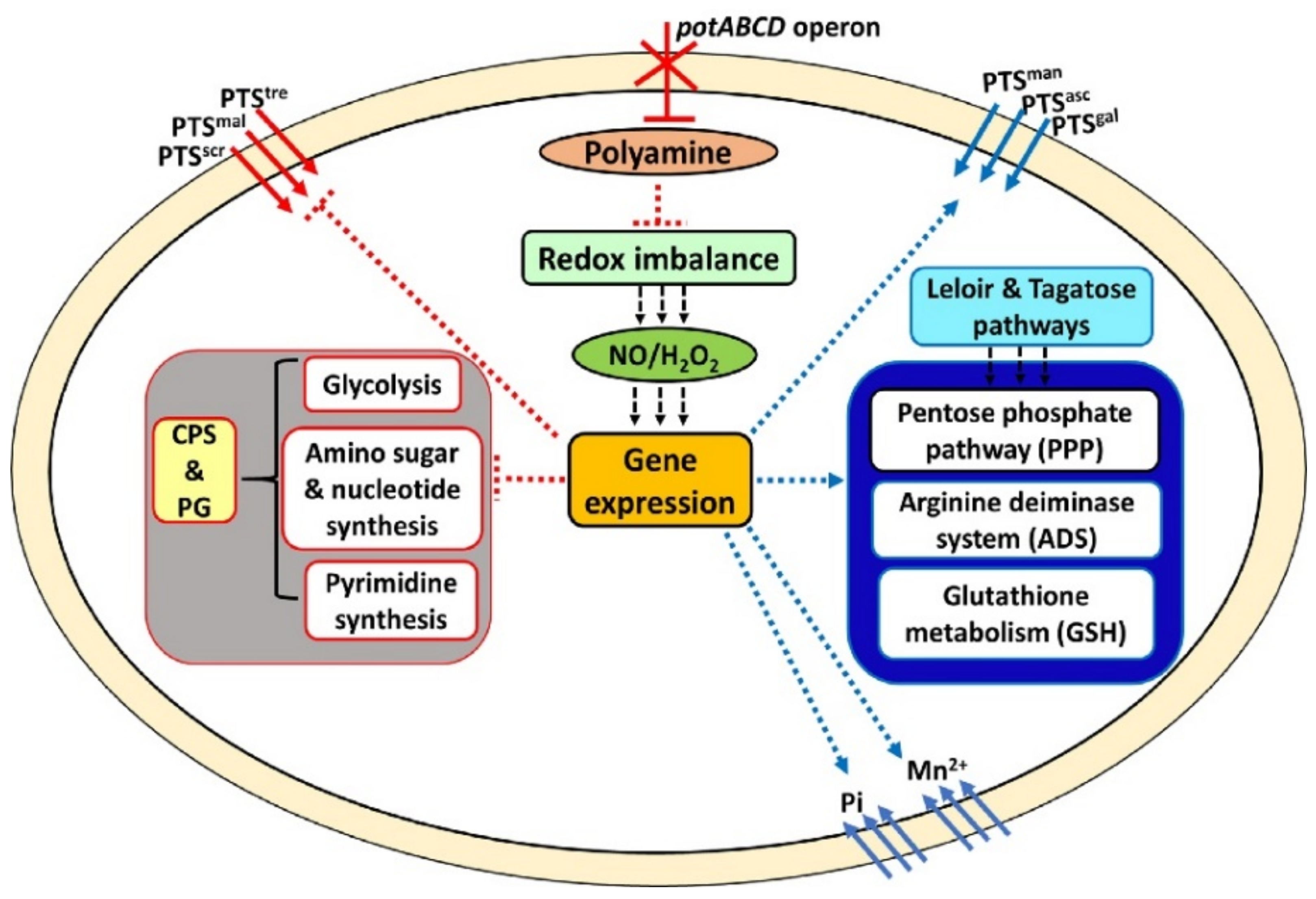
2.1. Polyamine Transport Modulates Pneumococcal Gene Expression
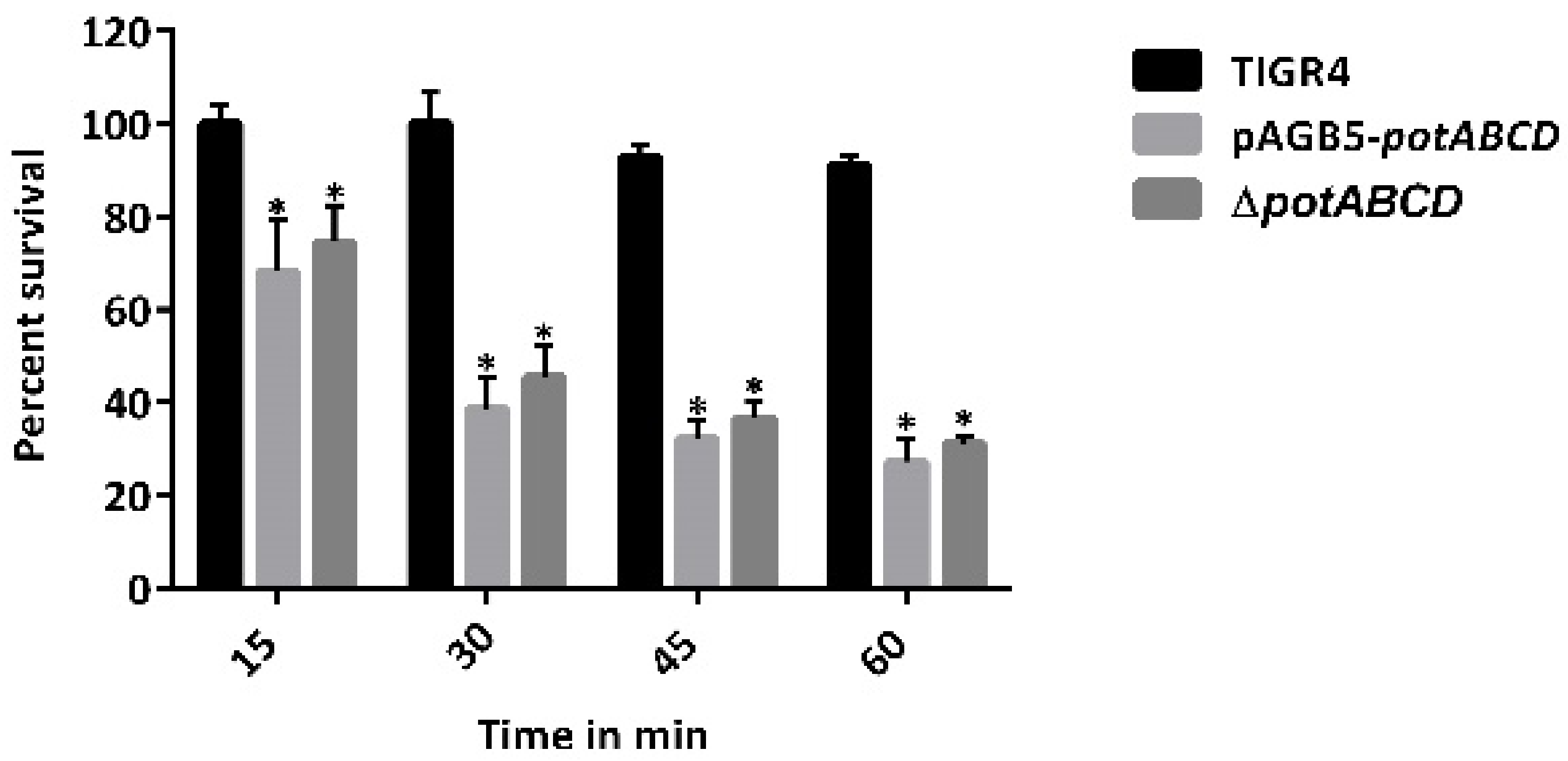
2.2. Polyamine Transporter and Pneumococcal Stress Responses
2.3. Galactose Utilization and the Pentose Phosphate Pathway (PPP)
2.4. Glycolysis and Production of Precursors for the Pneumococcal Capsule
2.5. Redox State and Regulation of Intracellular pH in S. pneumoniae
2.6. Metabolic Profile of Polyamine Transport-Deficient S. pneumoniae
2.7. PotABCD Is Required for Pneumococcal Hydrogen Peroxide and Nitrosative Stress Response
3. Discussion
4. Materials and Methods
4.1. Bacterial Strains and Growth Conditions
4.2. RNA Sequencing
4.3. Quantitative Real Time PCR
4.4. Measurement of Intracellular pH
4.5. Measurement of Intracellular Nicotinamide Adenine Dinucleotide Phosphate (NADPH)
4.6. Measurement of Intracellular Glutathione
4.7. UPLC-HRMS Untargeted Metabolomics
4.8. Hydrogen Peroxide Production
4.9. Hydrogen Peroxide Susceptibility
4.10. S-Nitrosoglutathione (GSNO) Susceptibility
4.11. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- O’Brien, K.L.; Wolfson, L.J.; Watt, J.P.; Henkle, E.; Deloria-Knoll, M.; McCall, N.; Lee, E.; Mulholland, K.; Levine, O.S.; Cherian, T. Burden of disease caused by Streptococcus pneumoniae in children younger than 5 years: Global estimates. Lancet 2009, 374, 893–902. [Google Scholar] [CrossRef]
- Dion, C.F.; Ashurst, J.V. Streptococcus pneumoniae. In StatPearls [Internet]; StatPearls: Treasure Island, FL, USA, 2020. [Google Scholar]
- Henriques-Normark, B.; Tuomanen, E.I. The Pneumococcus: Epidemiology, Microbiology, and Pathogenesis. Cold Spring Harb. Perspect. Med. 2013, 3, a010215. [Google Scholar] [CrossRef]
- Berical, A.C.; Harris, D.; Dela Cruz, C.S.; Possick, J.D. Pneumococcal Vaccination Strategies. An Update and Perspective. Ann. Am. Thorac. Soc. 2016, 13, 933–944. [Google Scholar] [CrossRef]
- Balsells, E.; Guillot, L.; Nair, H.; Kyaw, M.H. Serotype distribution of Streptococcus pneumoniae causing invasive disease in children in the post-PCV era: A systematic review and meta-analysis. PLoS ONE 2017, 12, e0177113. [Google Scholar] [CrossRef]
- Cherazard, R.; Epstein, M.; Doan, T.-L.; Salim, T.; Bharti, S.; Smith, M.A. Antimicrobial Resistant Streptococcus pneumoniae: Prevalence, Mechanisms, and Clinical Implications. Am. J. Ther. 2017, 24, e361–e369. [Google Scholar] [CrossRef]
- Igarashi, K.; Kashiwagi, K. Characteristics of cellular polyamine transport in prokaryotes and eukaryotes. Plant Physiol. Biochem. 2010, 48, 506–512. [Google Scholar] [CrossRef] [PubMed]
- Shah, P.; Swiatlo, E. A multifaceted role for polyamines in bacterial pathogens. Mol. Microbiol. 2008, 68, 4–16. [Google Scholar] [CrossRef] [PubMed]
- Shah, P.; Romero, D.G.; Swiatlo, E. Role of polyamine transport in Streptococcus pneumoniae response to physiological stress and murine septicemia. Microb. Pathog. 2008, 45, 167–172. [Google Scholar] [CrossRef] [PubMed]
- Shah, P.; Nanduri, B.; Swiatlo, E.; Ma, Y.; Pendarvis, K. Polyamine biosynthesis and transport mechanisms are crucial for fitness and pathogenesis of Streptococcus pneumoniae. Microbiology 2011, 157, 504–515. [Google Scholar] [CrossRef] [PubMed]
- Converso, T.R.; Goulart, C.; Rodriguez, D.; Darrieux, M.; Leite, L. Systemic immunization with rPotD reduces Streptococcus pneumoniae nasopharyngeal colonization in mice. Vaccine 2017, 35, 149–155. [Google Scholar] [CrossRef] [PubMed]
- Shah, P.; Swiatlo, E. Immunization with Polyamine Transport Protein PotD Protects Mice against Systemic Infection with Streptococcus pneumoniae. Infect. Immun. 2006, 74, 5888–5892. [Google Scholar] [CrossRef]
- Shah, P.; Briles, D.E.; King, J.; Hale, Y.; Swiatlo, E. Mucosal Immunization with Polyamine Transport Protein D (PotD) Protects Mice against Nasopharyngeal Colonization with Streptococcus pneumoniae. Exp. Biol. Med. 2009, 234, 403–409. [Google Scholar] [CrossRef]
- Rai, A.N.; Thornton, J.A.; Stokes, J.; Sunesara, I.; Swiatlo, E.; Nanduri, B. Polyamine transporter in Streptococcus pneumoniae is essential for evading early innate immune responses in pneumococcal pneumonia. Sci. Rep. 2016, 6, 26964. [Google Scholar] [CrossRef] [PubMed]
- Ayoola, M.B.; Shack, L.A.; Nakamya, M.F.; Thornton, J.A.; Swiatlo, E.; Nanduri, B. Polyamine Synthesis Effects Capsule Expression by Reduction of Precursors in Streptococcus pneumoniae. Front. Microbiol. 2019, 10, 1996. [Google Scholar] [CrossRef] [PubMed]
- Ayoola, M.B.; Nakamya, M.F.; Shack, L.A.; Park, S.; Lim, J.; Lee, J.H.; Ross, M.K.; Eoh, H.; Nanduri, B. SP_0916 Is an Arginine Decarboxylase That Catalyzes the Synthesis of Agmatine, Which Is Critical for Capsule Biosynthesis in Streptococcus pneumoniae. Front. Microbiol. 2020, 11, 578533. [Google Scholar] [CrossRef]
- Rhee, H.J.; Kim, E.-J.; Lee, J.K. Physiological polyamines: Simple primordial stress molecules. J. Cell. Mol. Med. 2007, 11, 685–703. [Google Scholar] [CrossRef] [PubMed]
- Gómez-Mejia, A.; Gámez, G.; Hirschmann, S.; Kluger, V.; Rath, H.; Böhm, S.; Voss, F.; Kakar, N.; Petruschka, L.; Völker, U.; et al. Pneumococcal metabolic adaptation and colonization is regulated by the two-component regulatory system 08. mSphere 2018, 3, e00165-18. [Google Scholar] [CrossRef] [PubMed]
- Kwon, H.-Y.; Kim, E.-H.; Tran, T.D.H.; Pyo, S.-N.; Rhee, D.-K. Reduction-sensitive and cysteine residue-mediated Streptococcus pneumoniae HrcA oligomerization in vitro. Mol. Cells 2009, 27, 149–157. [Google Scholar] [CrossRef]
- Torrent, M.; Chalancon, G.; de Groot, N.S.; Wuster, A.; Babu, M.M. Cells alter their tRNA abundance to selectively regulate protein synthesis during stress conditions. Sci. Signal. 2018, 11, eaat6409. [Google Scholar] [CrossRef] [PubMed]
- Carvalho, S.M.; Kloosterman, T.G.; Manzoor, I.; Caldas, J.; Vinga, S.; Martinussen, J.; Saraiva, L.; Kuipers, O.P.; Neves, A.R. Interplay between Capsule Expression and Uracil Metabolism in Streptococcus pneumoniae D39. Front. Microbiol. 2018, 9, 321. [Google Scholar] [CrossRef]
- Crawford, M.A.; Henard, C.A.; Tapscott, T.; Porwollik, S.; McClelland, M.; Vázquez-Torres, A. DksA-Dependent Transcriptional Regulation in Salmonella Experiencing Nitrosative Stress. Front. Microbiol. 2016, 7, 444. [Google Scholar] [CrossRef]
- Ogunniyi, A.D.; Mahdi, L.K.; Jennings, M.P.; McEwan, A.G.; McDevitt, C.; Van der Hoek, M.B.; Bagley, C.J.; Hoffmann, P.; Gould, K.; Paton, J.C. Central Role of Manganese in Regulation of Stress Responses, Physiology, and Metabolism in Streptococcus pneumoniae. J. Bacteriol. 2010, 192, 4489–4497. [Google Scholar] [CrossRef] [PubMed]
- Christodoulou, D.; Link, H.; Fuhrer, T.; Kochanowski, K.; Gerosa, L.; Sauer, U. Reserve Flux Capacity in the Pentose Phosphate Pathway Enables Escherichia coli’s Rapid Response to Oxidative Stress. Cell Syst. 2018, 6, 569–578.e7. [Google Scholar] [CrossRef] [PubMed]
- Stincone, A.; Prigione, A.; Cramer, T.; Wamelink, M.M.C.; Campbell, K.; Cheung, E.; Olin-Sandoval, V.; Grüning, N.-M.; Krüger, A.; Alam, M.T.; et al. The return of metabolism: Biochemistry and physiology of the pentose phosphate pathway. Biol. Rev. 2015, 90, 927–963. [Google Scholar] [CrossRef]
- Frey, P.A. The Leloir pathway: A mechanistic imperative for three enzymes to change the stereochemical configuration of a single carbon in galactose. FASEB J. 1996, 10, 461–470. [Google Scholar] [CrossRef] [PubMed]
- Afzal, M.; Shafeeq, S.; Kuipers, O.P. LacR Is a Repressor of lacABCD and LacT Is an Activator of lacTFEG, Constituting the lac Gene Cluster in Streptococcus pneumoniae. Appl. Environ. Microbiol. 2014, 80, 5349–5358. [Google Scholar] [CrossRef]
- Bidossi, A.; Mulas, L.; Decorosi, F.; Colomba, L.; Ricci, S.; Pozzi, G.; Deutscher, J.; Viti, C.; Oggioni, M.R. A Functional Genomics Approach to Establish the Complement of Carbohydrate Transporters in Streptococcus pneumoniae. PLoS ONE 2012, 7, e33320. [Google Scholar] [CrossRef]
- Zeng, L.; Das, S.; Burne, R.A. Utilization of Lactose and Galactose by Streptococcus mutans: Transport, Toxicity, and Carbon Catabolite Repression. J. Bacteriol. 2010, 192, 2434–2444. [Google Scholar] [CrossRef]
- Shaw, J.A.; Henard, C.A.; Liu, L.; Dieckman, L.M.; Vázquez-Torres, A.; Bourret, T.J. Salmonella enterica serovar Typhimurium has three transketolase enzymes contributing to the pentose phosphate pathway. J. Biol. Chem. 2018, 293, 11271–11282. [Google Scholar] [CrossRef]
- Laass, S.; Kleist, S.; Bill, N.; Drüppel, K.; Kossmehl, S.; Wöhlbrand, L.; Rabus, R.; Klein, J.; Rohde, M.; Bartsch, A.; et al. Gene Regulatory and Metabolic Adaptation Processes of Dinoroseobacter shibae DFL12T during Oxygen Depletion. J. Biol. Chem. 2014, 289, 13219–13231. [Google Scholar] [CrossRef]
- Grüning, N.-M.; Lehrach, H.; Ralser, M. Regulatory crosstalk of the metabolic network. Trends Biochem. Sci. 2010, 35, 220–227. [Google Scholar] [CrossRef]
- Jones, C.; Currie, F.; Forster, M.J. N.m.r. and conformational analysis of the capsular polysaccharide from Streptococcus pneumoniae type 4. Carbohydr. Res. 1991, 221, 95–121. [Google Scholar] [CrossRef]
- Geno, K.A.; Gilbert, G.L.; Song, J.Y.; Skovsted, I.C.; Klugman, K.P.; Jones, C.; Konradsen, H.B.; Nahm, M.H. Pneumococcal Capsules and Their Types: Past, Present, and Future. Clin. Microbiol. Rev. 2015, 28, 871–899. [Google Scholar] [CrossRef] [PubMed]
- Rodríguez-Díaz, J.; Rubio-Del-Campo, A.; Yebra, M.J. Regulatory insights into the production of UDP-N-acetylglucosamine by Lactobacillus casei. Bioengineered 2012, 3, 339–342. [Google Scholar] [CrossRef] [PubMed]
- Bonnet, J.; Durmort, C.; Mortier-Barrière, I.; Campo, N.; Jacq, M.; Moriscot, C.; Straume, D.; Berg, K.; Håvarstein, L.; Wong, Y.-S.; et al. Nascent teichoic acids insertion into the cell wall directs the localization and activity of the major pneumococcal autolysin LytA. Cell Surf. 2018, 2, 24–37. [Google Scholar] [CrossRef] [PubMed]
- Kilstrup, M.; Hammer, K.; Jensen, P.R.; Martinussen, J. Nucleotide metabolism and its control in lactic acid bacteria. FEMS Microbiol. Rev. 2005, 29, 555–590. [Google Scholar] [CrossRef] [PubMed]
- Grunenwald, C.M.; Choby, J.E.; Juttukonda, L.J.; Beavers, W.N.; Weiss, A.; Torres, V.J.; Skaar, E.P. Manganese Detoxification by MntE Is Critical for Resistance to Oxidative Stress and Virulence of Staphylococcus aureus. mBio 2019, 10, e02915-18. [Google Scholar] [CrossRef] [PubMed]
- Patil, M.D.; Rathod, V.P.; Bihade, U.R.; Banerjee, U.C. Purification and characterization of arginine deiminase from Pseudomonas putida: Structural insights of the differential affinities of l-arginine analogues. J. Biosci. Bioeng. 2019, 127, 129–137. [Google Scholar] [CrossRef]
- Jakubovics, N.S.; Robinson, J.C.; Samarian, D.S.; Kolderman, E.; Yassin, S.A.; Bettampadi, D.; Bashton, M.; Rickard, A.H. Critical roles of arginine in growth and biofilm development by Streptococcus gordonii. Mol. Microbiol. 2015, 97, 281–300. [Google Scholar] [CrossRef]
- Potter, A.J.; Trappetti, C.; Paton, J.C. Streptococcus pneumoniae Uses Glutathione to Defend against Oxidative Stress and Metal Ion Toxicity. J. Bacteriol. 2012, 194, 6248–6254. [Google Scholar] [CrossRef] [PubMed]
- Lisher, J.P.; Tsui, H.C.T.; Ramos-Montañez, S.; Hentchel, K.L.; Martin, J.E.; Trinidad, J.C.; Winkler, M.E.; Giedroc, D.P. Biological and Chemical Adaptation to Endogenous Hydrogen Peroxide Production in Streptococcus pneumoniae D39. mSphere 2017, 2, e00291-16. [Google Scholar] [CrossRef]
- Li, Y.; Hugenholtz, J.; Abee, T.; Molenaar, D. Glutathione Protects Lactococcus lactis against Oxidative Stress. Appl. Environ. Microbiol. 2003, 69, 5739–5745. [Google Scholar] [CrossRef]
- Carmel-Harel, O.; Storz, G. Roles of the Glutathione- and Thioredoxin-Dependent Reduction Systems in the Escherichia coli and Saccharomyces cerevisiae Responses to Oxidative Stress. Annu. Rev. Microbiol. 2000, 54, 439–461. [Google Scholar] [CrossRef] [PubMed]
- Igarashi, K.; Kashiwagi, K. Effects of polyamines on protein synthesis and growth of Escherichia coli. J. Biol. Chem. 2018, 293, 18702–18709. [Google Scholar] [CrossRef] [PubMed]
- Paluscio, E.; Watson, M.E., Jr.; Caparon, M.G. CcpA Coordinates Growth/Damage Balance for Streptococcus pyogenes Pathogenesis. Sci. Rep. 2018, 8, 14254. [Google Scholar] [CrossRef] [PubMed]
- Antunes, A.; Camiade, E.; Monot, M.; Courtois, E.; Barbut, F.; Sernova, N.V.; Rodionov, D.; Martin-Verstraete, I.; Dupuy, B. Global transcriptional control by glucose and carbon regulator CcpA in Clostridium difficile. Nucleic Acids Res. 2012, 40, 10701–10718. [Google Scholar] [CrossRef]
- Kay, E.J.; Yates, L.E.; Terra, V.S.; Cuccui, J.; Wren, B.W. Recombinant expression of Streptococcus pneumoniae capsular polysaccharides in Escherichia coli. Open Biol. 2016, 13, 150243. [Google Scholar] [CrossRef]
- Chen, L.L.; Han, D.L.; Zhai, Y.F.; Wang, J.H.; Wang, Y.F.; Chen, M. Characterization and mutational analysis of two UDP-galactose 4-epimerases in Streptococcus pneumoniae TIGR4. Biochemistry 2018, 83, 37–44. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.; Tan, M.; Zhang, C.; Xu, Z.; Li, L.; Zhou, R. Functional characterization of murB-potABCD operon for polyamine uptake and peptidoglycan synthesis in Streptococcus suis. Microbiol. Res. 2018, 207, 177–187. [Google Scholar] [CrossRef]
- Ware, D.; Watt, J.; Swiatlo, E. Utilization of putrescine by Streptococcus pneumoniae during growth in choline-limited medium. J. Microbiol. 2005, 43, 398–405. [Google Scholar] [PubMed]
- Rai, P.; Parrish, M.; Tay, I.J.J.; Li, N.; Ackerman, S.; He, F.; Kwang, J.; Chow, V.; Engelward, B.P. Streptococcus pneumoniae secretes hydrogen peroxide leading to DNA damage and apoptosis in lung cells. Proc. Natl. Acad. Sci. USA 2015, 112, E3421–E3430. [Google Scholar] [CrossRef] [PubMed]
- Spellerberg, B.; Cundell, D.R.; Sandros, J.; Pearce, B.J.; Idänpään-Heikkilä, I.; Rosenow, C.; Masure, H.R. Pyruvate oxidase, as a determinant of virulence in Streptococcus pneumoniae. Mol. Microbiol. 1996, 19, 803–813. [Google Scholar] [CrossRef] [PubMed]
- Pericone, C.D.; Overweg, K.; Hermans, P.W.M.; Weiser, J.N. Inhibitory and Bactericidal Effects of Hydrogen Peroxide Production by Streptococcus pneumoniae on Other Inhabitants of the Upper Respiratory Tract. Infect. Immun. 2000, 68, 3990–3997. [Google Scholar] [CrossRef] [PubMed]
- Tkachenko, A.G.; Akhova, A.V.; Shumkov, M.S.; Nesterova, L.Y. Polyamines reduce oxidative stress in Escherichia coli cells exposed to bactericidal antibiotics. Res. Microbiol. 2012, 163, 83–91. [Google Scholar] [CrossRef] [PubMed]
- Cabiscol Català, E.; Tamarit Sumalla, J.; Ros Salvador, J. Oxidative stress in bacteria and protein damage by reactive oxygen species. Int. Microbiol. 2000, 3, 3–8. [Google Scholar]
- Chattopadhyay, M.K.; Tabor, C.W.; Tabor, H. Polyamines protect Escherichia coli cells from the toxic effect of oxygen. Proc. Natl. Acad. Sci. USA 2003, 100, 2261–2265. [Google Scholar] [CrossRef]
- Nakamya, M.F.; Ayoola, M.; Shack, L.; Mohamed, M.; Swiatlo, E.; Nanduri, B. Arginine Decarboxylase Is Essential for Pneumococcal Stress Responses. Pathogens 2021, 10, 286. [Google Scholar] [CrossRef]
- Espinel, I.C.; Guerra, P.R.; Jelsbak, L. Multiple roles of putrescine and spermidine in stress resistance and virulence of Salmonella enterica serovar Typhimurium. Microb. Pathog. 2016, 95, 117–123. [Google Scholar] [CrossRef]
- Bower, J.M.; Mulvey, M.A. Polyamine-Mediated Resistance of Uropathogenic Escherichia coli to Nitrosative Stress. J. Bacteriol. 2006, 188, 928–933. [Google Scholar] [CrossRef]
- Winterhoff, N.; Goethe, R.; Gruening, P.; Rohde, M.; Kalisz, H.; Smith, H.E.; Valentin-Weigand, P. Identification, and characterization of two temperature-induced surface-associated proteins of Streptococcus suis with high homologies to members of the Arginine Deiminase system of Streptococcus pyogenes. J. Bacteriol. 2002, 184, 6768–6776. [Google Scholar] [CrossRef]
- Imlay, J.A. Iron-sulphur clusters and the problem with oxygen. Mol. Microbiol. 2006, 59, 1073–1082. [Google Scholar] [CrossRef] [PubMed]
- Belenky, P.; Ye, J.D.; Porter, C.B.; Cohen, N.R.; Lobritz, M.A.; Ferrante, T.; Jain, S.; Korry, B.J.; Schwarz, E.G.; Walker, G.C.; et al. Bactericidal Antibiotics Induce Toxic Metabolic Perturbations that Lead to Cellular Damage. Cell Rep. 2015, 13, 968–980. [Google Scholar] [CrossRef] [PubMed]
- Brynildsen, M.P.; Winkler, J.A.; Spina, C.; MacDonald, I.C.; Collins, J.J. Potentiating antibacterial activity by predictably enhancing endogenous microbial ROS production. Nat. Biotechnol. 2013, 31, 160–165. [Google Scholar] [CrossRef] [PubMed]
- Tettelin, H.; Nelson, K.E.; Durkin, A.S.; Gwinn, M.; Kolonay, J.F.; Nelson, W.C.; Peterson, J.D.; Umayam, L.A.; White, O.; Salzberg, S.L.; et al. Complete Genome Sequence of a Virulent Isolate of Streptococcus pneumoniae. Science 2001, 293, 498–506. [Google Scholar] [CrossRef] [PubMed]
- Texeira, E.; Checa, J.; Ríal, A.; Chabalgoity, J.A.; Suárez, N. A new chemically defined medium for cultivation of Streptococcus pneumoniae Serotype 1. J. Biotech Res. 2015, 6, 54–62. [Google Scholar]
- Martens, L.; Chambers, M.; Sturm, M.; Kessner, D.; Levander, F.; Shofstahl, J.; Tang, W.H.; Römpp, A.; Neumann, S.; Pizarro, A.D.; et al. mzML—A Community Standard for Mass Spectrometry Data. Mol. Cell. Proteom. 2011, 10, R110.000133. [Google Scholar] [CrossRef]
- Keseler, I.M.; Mackie, A.; Santos-Zavaleta, A.; Billington, R.; Bonavides-Martínez, C.; Caspi, R.; Fulcher, C.; Gama-Castro, S.; Kothari, A.; Krummenacker, M.; et al. The EcoCyc database: Reflecting new knowledge about Escherichia coli K-12. Nucleic Acids Res. 2017, 45, D543–D550. [Google Scholar] [CrossRef]
- The Gene Ontology Consortium. Expansion of the Gene Ontology knowledgebase and resources. Nucleic Acids Res. 2017, 45, D331–D338. [Google Scholar] [CrossRef]
- Kanehisa, M.; Goto, S.; Sato, Y.; Furumichi, M.; Tanabe, M. KEGG for integration and interpretation of large-scale molecular data sets. Nucleic Acids Res. 2012, 40, D109–D114. [Google Scholar] [CrossRef]
- The UniProt Consortium. UniProt: The universal protein knowledgebase. Nucleic Acids Res. 2018, 46, 2699. [Google Scholar] [CrossRef]
- Szklarczyk, D.; Gable, A.L.; Lyon, D.; Junge, A.; Wyder, S.; Huerta-Cepas, J.; Simonovic, M.; Doncheva, N.T.; Morris, J.H.; Bork, P.; et al. STRING v11: Protein–protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019, 47, D607–D613. [Google Scholar] [CrossRef] [PubMed]
- Qian, W.-J.; Jacobs, J.M.; Davis, R.W.; Tompkins, R.G.; Smith, R.D.; Camp, D.G.; Monroe, M.E.; Moore, R.J.; Gritsenko, M.A.; Calvano, S.E.; et al. Comparative proteome analyses of human plasma following in vivo lipopolysaccharide administration using multidimensional separations coupled with tandem mass spectrometry. Proteomics 2005, 5, 572–584. [Google Scholar] [CrossRef] [PubMed]
- Clementi, E.A.; Marks, L.; Roche-Håkansson, H.; Håkansson, A.P. Monitoring Changes in Membrane Polarity, Membrane Integrity, and Intracellular Ion Concentrations in Streptococcus pneumoniae Using Fluorescent Dyes. J. Vis. Exp. 2014, e51008. [Google Scholar] [CrossRef] [PubMed]
- Lu, W.; Clasquin, M.F.; Melamud, E.; Amador-Noguez, D.; Caudy, A.; Rabinowitz, J.D. Metabolomic Analysis via Reversed-Phase Ion-Pairing Liquid Chromatography Coupled to a Stand Alone Orbitrap Mass Spectrometer. Anal. Chem. 2010, 82, 3212–3221. [Google Scholar] [CrossRef]
- Clasquin, M.F.; Melamud, E.; Rabinowitz, J.D. LC-MS Data Processing with MAVEN: A Metabolomic Analysis and Visualization Engine. Curr. Protoc. Bioinform. 2012, 37, 9818–9826. [Google Scholar] [CrossRef]
- Melamud, E.; Vastag, L.; Rabinowitz, J.D. Metabolomic Analysis and Visualization Engine for LC-MS Data. Anal. Chem. 2010, 82, 9818–9826. [Google Scholar] [CrossRef]
- Chong, J.; Soufan, O.; Li, C.; Caraus, I.; Li, S.; Bourque, G.; Wishart, D.S.; Xia, J. MetaboAnalyst 4.0: Towards more transparent and integrative metabolomics analysis. Nucleic Acids Res. 2018, 46, W486–W494. [Google Scholar] [CrossRef]
- Lee, J.; Park, J.; Lim, M.-S.; Seong, S.J.; Seo, J.J.; Park, S.M.; Lee, H.W.; Yoon, Y.-R. Quantile Normalization Approach for Liquid Chromatography–Mass Spectrometry-based Metabolomic Data from Healthy Human Volunteers. Anal. Sci. 2012, 28, 801–805. [Google Scholar] [CrossRef]


| Locus Tag | Gene | Fold Change ΔpotABCD/TIGR4 | FDR p Value | Description |
|---|---|---|---|---|
| SP_1884 | SP_1884 | −3.6 | <0.0001 | PTS trehalose transporter subunit IIBC |
| SP_1883 | SP_1883 | −3.2 | <0.0001 | Trehalose 6-phosphate hydrolase |
| SP_1907 | groES | −3.9 | <0.0001 | Co-chaperone |
| SP_0519 | dnaJ | −3.8 | <0.0001 | Molecular chaperone |
| SP_0517 | dnaK | −4.2 | <0.0001 | Molecular chaperone |
| SP_1906 | groEL | −3.3 | <0.0001 | Molecular chaperone |
| SP_1870 | SP_1870 | 2.7 | <0.0001 | Iron-compound ABC transporter |
| SP_1871 | SP_1871 | 2.9 | <0.0001 | Iron ABC transporter ATP-binding protein |
| SP_1241 | SP_1241 | 3.1 | <0.0001 | Glutamine transport system substrate-binding |
| SP_1242 | SP_1242 | 3.2 | <0.0001 | Glutamine transport system ATP-binding protein |
| SP_2087 | pstB | 41.8 | <0.0001 | Phosphate ABC transporter ATP-binding protein |
| SP_2085 | pstC | 37.2 | <0.0001 | Phosphate ABC transporter permease subunit |
| SP_2084 | pstS | 27.2 | <0.0001 | Phosphate ABC transporter substrate-binding |
| SP_2086 | pstA | 40.6 | <0.0001 | Phosphate ABC transporter permease protein |
| SP_1650 | psaA | 3.2 | <0.0001 | Manganese ABC transporter substrate-binding |
| SP_1648 | psaB | 3.4 | <0.0001 | Metal ABC transporter ATP-binding protein |
| SP_1649 | psaC | 3.5 | <0.0001 | Metal ABC transporter permease |
| SP_0502 | glnA | 4.2 | <0.0001 | Glutamine synthetase |
| SP_2148 | arcA | 4.6 | <0.0001 | Arginine deiminase |
| SP_2151 | arcC | 3.7 | <0.0001 | Carbamate kinase |
| SP_0798 | ciaR | 3.0 | <0.0001 | DNA-binding response regulator |
| SP_0799 | ciaH | 3.0 | <0.0001 | Two-component sensor histidine kinase |
| SP_0501 | merR | 4.0 | <0.0001 | MerR family transcriptional regulator |
| SP_0515 | hrcA | −4.3 | <0.0001 | Transcriptional regulator |
| SP_2088 | phoU | 47.2 | <0.0001 | Phosphate uptake regulator |
| Locus Tag | Gene | Fold Change ΔpotABCD/TIGR4 | FDR p Value | Description |
|---|---|---|---|---|
| SP_2127 | tktN | 222.7 | <0.0001 | Transketolase |
| SP_2128 | tktC | 225.5 | <0.0001 | Transketolase |
| SP_2130 | SP_2130 | 211.5 | <0.0001 | Ascorbate PTS system EIIB |
| SP_2129 | SP_2129 | 214.7 | <0.0001 | PTS ascorbate transporter subunit IIC |
| SP_1192 | lacB | 2.8 | <0.0001 | Galactose 6-phosphate isomerase subunit |
| SP_1193 | lacA | 2.8 | <0.0001 | Galactose 6-phosphate isomerase subunit |
| SP_1190 | lacD | 2.8 | <0.0001 | Tagatose 1,6-diphosphate aldolase |
| SP_1191 | lacC | 2.8 | <0.0001 | Tagatose 6-phosphate kinase |
| SP_1186 | lacF-2 | 2.0 | <0.0001 | Lactose PTS system EIIA component |
| SP_2165 | fucU | 4.5 | <0.0001 | Fucose isomerase |
| SP_2166 | fucA | 3.8 | <0.0001 | L-fuculose phosphate aldolase |
| SP_1853 | galK | 6.1 | <0.0001 | Galactokinase |
| SP_0066 | galM | 2.7 | <0.0001 | Galactose mutarotase |
| SP_0064 | SP_0064 | 3.4 | <0.0001 | PTS mannose PTS system EIIA |
| SP_0645 | SP_0645 | 8.8 | <0.0001 | PTS galactose PTS system EIIA |
| SP_0646 | SP_0646 | 9.0 | <0.0001 | PTS galactose PTS system EIIB |
| SP_2164 | SP_2164 | 3.7 | <0.0001 | PTS mannose transporter subunit IIA |
| SP_2161 | SP_2161 | 3.3 | <0.0001 | PTS mannose transporter subunit IID |
| SP_2162 | SP_2162 | 3.7 | <0.0001 | PTS mannose PTS system EIIC |
| Locus Tag | Gene | Fold Change ΔpotABCD/TIGR4 | FDR p Value | Description |
|---|---|---|---|---|
| SP_2131 | bglG | 247.3 | <0.0001 | Transcriptional regulator |
| SP_0100 | padR | 6.8 | <0.0001 | Transcriptional regulator |
| SP_1854 | galR | 3.9 | <0.0001 | LacI family transcriptional regulator |
| SP_2109 | malC | −2.5 | <0.0001 | Maltodextrin ABC transporter permease |
| SP_2110 | malD | −2.4 | <0.0001 | Maltodextrin ABC transporter permease |
| SP_2108 | malX | −2.4 | <0.0001 | Maltose/maltodextrin-binding protein |
| SP_1894 | gtfA | 2.3 | <0.0001 | Sucrose phosphorylase |
| SP_1722 | SP_1722 | −51.0 | <0.0001 | PTS sucrose system EIIBCA or EIIBC |
| SP_0648 | bgaA | 7.0 | <0.0001 | Beta galactosidase |
| SP_1898 | aga | 2.2 | <0.0001 | Alpha galactosidase |
| SP_1721 | scrK | −10.0 | <0.0001 | Fructokinase |
| SP_1725 | scrR | −9.2 | <0.0001 | LacI family transcriptional regulator, |
| SP_1278 | pyrR | −2.7 | <0.0001 | Bifunctional pyrimidine operon transcriptional regulator |
| SP_1724 | scrB | −10.5 | <0.0001 | Sucrose 6-phosphate hydrolase |
| SP_1415 | nagB | 3.2 | <0.0001 | Glucosamine 6-phosphate deaminase |
| SP_2056 | nagA | 2.3 | <0.0001 | N-acetylglucosamine 6-phosphate deacetylase |
| SP_0321 | SP_0321 | −2.3 | <0.0001 | PTS N-acetylgalactosamine transporter subunit IIA |
| SP_0323 | SP_0323 | −2.6 | <0.0001 | PTS N-acetylgalactosamine PTS system EIIB |
| SP_1277 | pyrB | −2.4 | <0.0001 | Aspartate carbamoyltransferase |
| SP_1014 | dapA | −4.3 | <0.0001 | 4-hydroxy-tetrahydrodipicolinate synthase |
| SP_1013 | asd | −5.5 | <0.0001 | Aspartate-semialdehyde dehydrogenase |
| Locus Tag | Gene | Fold Change ΔpotABCD/TIGR4 | p Value | Description |
|---|---|---|---|---|
| SP_0916 | speA | 16.9 | <0.0001 | Arginine decarboxylase |
| SP_0918 | speE | 14.4 | <0.0001 | Spermidine synthase |
| SP_2127 | SP_2127 | 20.3 | <0.0001 | Transketolase C-terminal subunit |
| SP_2128 | SP_2128 | 27.7 | <0.0001 | Transketolase N-terminal subunit |
| SP_2131 | BglG | 5.5 | <0.0001 | Transcriptional regulator |
| SP_2136 | pcpA | 7.2 | <0.0001 | Choline-binding protein |
| SP_1650 | psaA | 5.40 | <0.0001 | Manganese ABC transporter |
| Locus Tag | Gene | Fold Change ΔpotABCD/TIGR4 | p Value | Description |
|---|---|---|---|---|
| SP_0916 | speA | 16.9 | <0.0001 | Arginine decarboxylase |
| SP_0918 | speE | 14.4 | <0.0001 | Spermidine synthase |
| SP_2127 | SP_2127 | 20.3 | <0.0001 | Transketolase, C-terminal subunit |
| SP_2128 | SP_2128 | 27.7 | <0.0001 | Transketolase, N-terminal subunit |
| SP_2131 | BglG | 5.5 | <0.0001 | Transcriptional regulator |
| SP_2136 | pcpA | 7.2 | <0.0001 | Choline-binding protein |
| SP_1650 | psaA | 5.40 | <0.0001 | Manganese ABC transporter |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nakamya, M.F.; Ayoola, M.B.; Shack, L.A.; Swiatlo, E.; Nanduri, B. The Effect of Impaired Polyamine Transport on Pneumococcal Transcriptome. Pathogens 2021, 10, 1322. https://doi.org/10.3390/pathogens10101322
Nakamya MF, Ayoola MB, Shack LA, Swiatlo E, Nanduri B. The Effect of Impaired Polyamine Transport on Pneumococcal Transcriptome. Pathogens. 2021; 10(10):1322. https://doi.org/10.3390/pathogens10101322
Chicago/Turabian StyleNakamya, Mary F., Moses B. Ayoola, Leslie A. Shack, Edwin Swiatlo, and Bindu Nanduri. 2021. "The Effect of Impaired Polyamine Transport on Pneumococcal Transcriptome" Pathogens 10, no. 10: 1322. https://doi.org/10.3390/pathogens10101322
APA StyleNakamya, M. F., Ayoola, M. B., Shack, L. A., Swiatlo, E., & Nanduri, B. (2021). The Effect of Impaired Polyamine Transport on Pneumococcal Transcriptome. Pathogens, 10(10), 1322. https://doi.org/10.3390/pathogens10101322





