Emergence and Characterization of a Novel Reassortant Canine Influenza Virus Isolated from Cats
Abstract
:1. Introduction
2. Material and Method
2.1. Virus Isolation
2.2. RT-PCR, Sequencing and Analysis of Sequences
3. Results
3.1. Virus Isolation
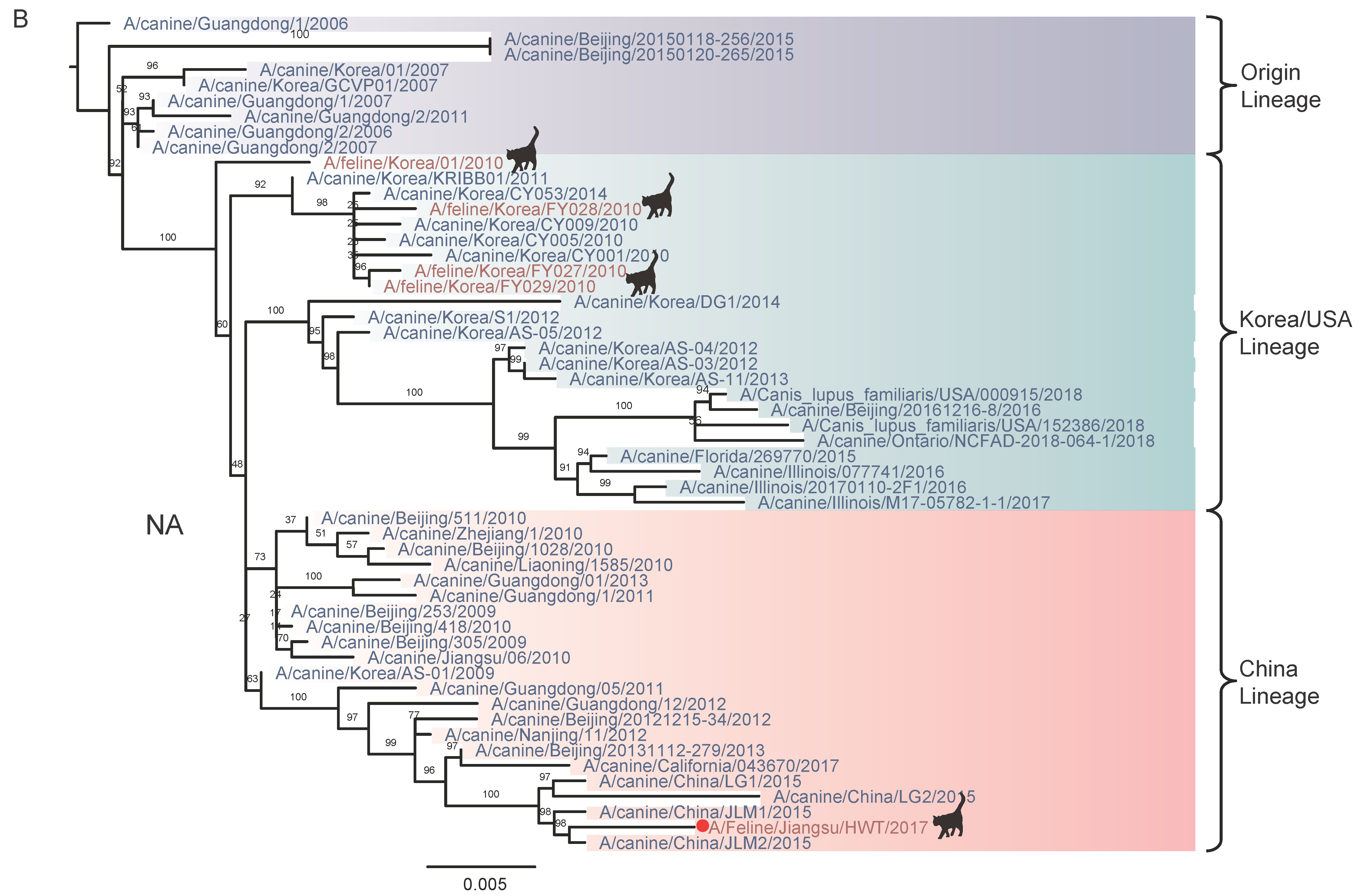
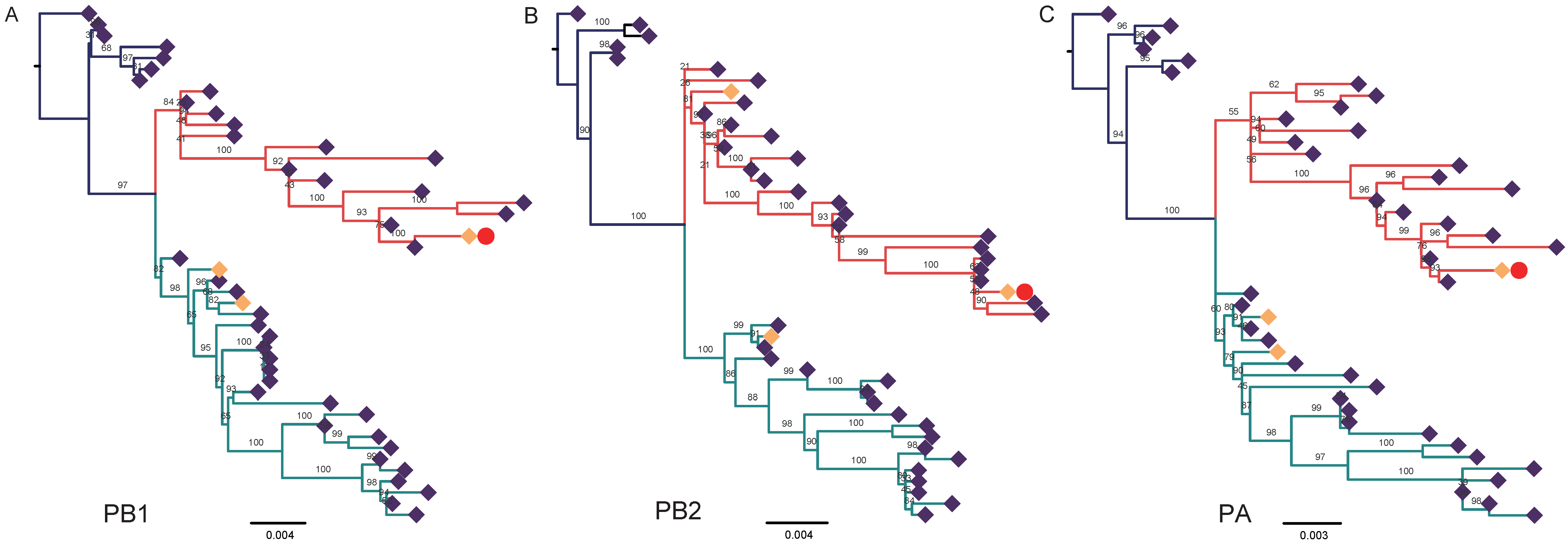
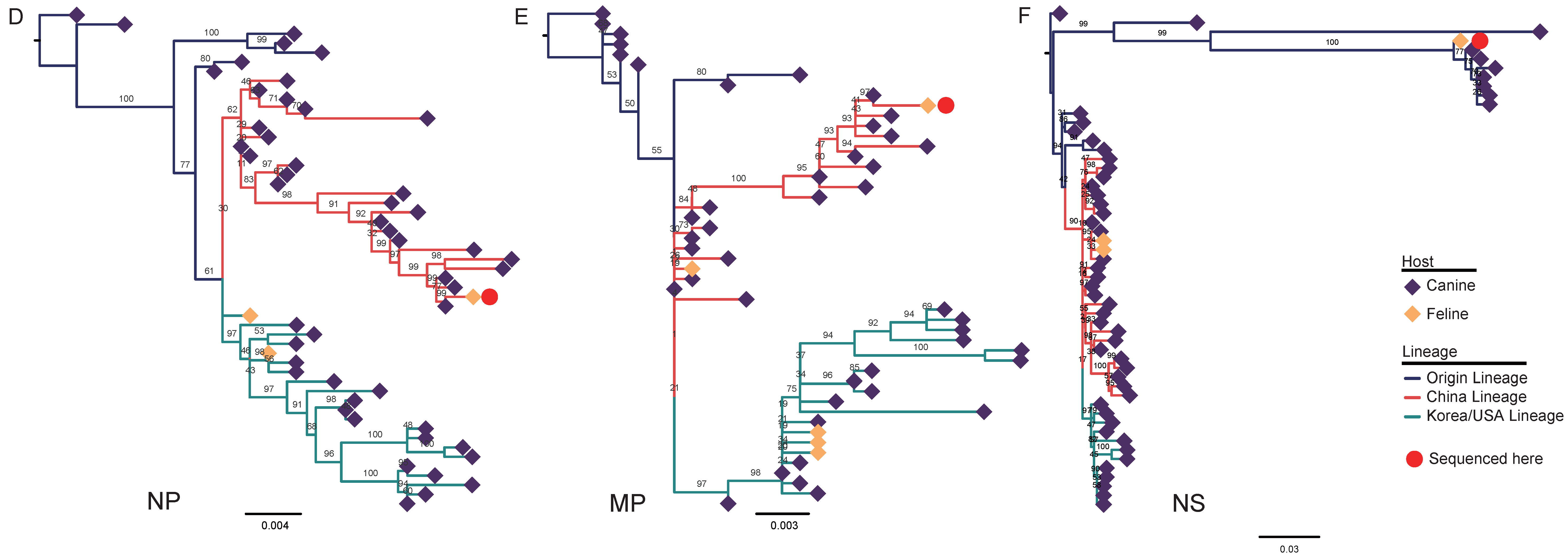
3.2. Nucleotide Sequencing and Phylogenetic Analysis
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Su, S.; Chen, J.; Jia, K.; Khan, S.U.; He, S.; Fu, X.; Hong, M.; Sun, L.; Qi, W.; Gray, G.C.; et al. Evidence for subclinical influenza A(H1N1) pdm09 virus infection among dogs in Guangdong Province, China. J. Clin. Microbiol. 2014, 52, 1762–1765. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Su, S.; Li, H.T.; Zhao, F.R.; Chen, J.D.; Xie, J.X.; Chen, Z.M.; Huang, Z.; Hu, Y.M.; Zhang, M.Z.; Tan, L.K.; et al. Avian-origin H3N2 canine influenza virus circulating in farmed dogs in Guangdong, China. Infect. Genet. Evol. 2013, 14, 444–449. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.I.; Lee, I.; Park, S.; Lee, S.; Hwang, M.W.; Bae, J.Y.; Heo, J.; Kim, D.; Jang, S.I.; Kim, K.; et al. Phylogenetic analysis of a swine influenza A(H3N2) virus isolated in Korea in 2012. PLoS ONE 2014, 9, e88782. [Google Scholar] [CrossRef] [PubMed]
- Song, D.S.; An, D.J.; Moon, H.J.; Yeom, M.J.; Jeong, H.Y.; Jeong, W.S.; Park, S.J.; Kim, H.K.; Han, S.Y.; Oh, J.S.; et al. Interspecies transmission of the canine influenza H3N2 virus to domestic cats in South Korea, 2010. J. Gen. Virol. 2011, 92, 2350–2355. [Google Scholar] [CrossRef]
- Kim, H.M.; Park, E.H.; Yum, J.; Kim, H.S.; Seo, S.H. Greater virulence of highly pathogenic H5N1 influenza virus in cats than in dogs. Arch. Virol. 2015, 160, 305–313. [Google Scholar] [CrossRef] [PubMed]
- Feng, H.; Liu, Y.; Song, Q.; Ling, Z.; Zhang, F.; Zhu, Y.; Jiang, S.; Xie, Z. Interspecies transmission of canine influenza virus H5N2 to cats and chickens by close contact with experimentally infected dogs. Vet. Microbiol. 2014, 170, 414–417. [Google Scholar]
- Jeoung, H.Y.; Lim, S.I.; Shin, B.H.; Lim, J.A.; Song, J.Y.; Song, D.S.; Kang, B.K.; Moon, H.J.; An, D.J. A novel canine influenza H3N2 virus isolated from cats in an animal shelter. Vet. Microbiol. 2013, 165, 281–286. [Google Scholar] [CrossRef]
- Belser, J.A.; Pulit-Penaloza, J.A.; Sun, X.; Brock, N.; Pappas, C.; Creager, H.M.; Zeng, H.; Tumpey, T.M.; Maines, T.R. A Novel A(H7N2) Influenza Virus Isolated from a Veterinarian Caring for Cats in a New York City Animal Shelter Causes Mild Disease and Transmits Poorly in the Ferret Model. J. Virol. 2017, 91, e00672-17. [Google Scholar] [CrossRef] [Green Version]
- Zhang, K.; Zhang, Z.; Yu, Z.; Li, L.; Cheng, K.; Wang, T.; Huang, G.; Yang, S.; Zhao, Y.; Feng, N.; et al. Domestic cats and dogs are susceptible to H9N2 avian influenza virus. Virus Res. 2013, 175, 52–57. [Google Scholar] [CrossRef]
- Harder, T.C.; Vahlenkamp, T.W. Influenza virus infections in dogs and cats. Vet. Immunol. Immunopathol. 2010, 134, 54–60. [Google Scholar] [CrossRef]
- Wang, H.; Wu, X.; Cheng, Y.; An, Y.; Ning, Z. Tissue distribution of human and avian type sialic acid influenza virus receptors in domestic cat. Acta. Vet. Hung. 2013, 61, 537–546. [Google Scholar] [CrossRef]
- Thongratsakul, S.; Suzuki, Y.; Hiramatsu, H.; Sakpuaram, T.; Sirinarumitr, T.; Poolkhet, C.; Moonjit, P.; Yodsheewan, R.; Songserm, T. Avian and human influenza A virus receptors in trachea and lung of animals. Asian Pac. J. Allergy. Immunol. 2010, 28, 294–301. [Google Scholar]
- Lei, N.; Yuan, Z.G.; Huang, S.F.; Zhang, D.W.; Zhang, A.G.; Huang, B.H.; Zhang, G.H.; Li, S.J. Transmission of avian-origin canine influenza viruses A (H3N2) in cats. Vet. Microbiol. 2012, 160, 481–483. [Google Scholar] [CrossRef]
- Tian, J.; Zhang, X.; Wu, H.; Liu, C.; Liu, J.; Hu, X.; Qu, L. Assessment of the IFN-β response to four feline caliciviruses: Infection in CRFK cells. Infect. Genet. Evol. 2015, 34, 352–360. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Liu, Y.; Liu, D.; Guo, D.; Liu, M.; Li, Y.; Qu, L. Complete Genome Sequence of Feline Calicivirus Strain HRB-SS from a Cat in Heilongjiang Province, Northeastern China. Genome Announc. 2014, 2, e00698-14. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, C.; Liu, Y.; Liu, D.; Qiu, Z.; Tian, J.; Guo, D.; Li, Z.; Liu, M.; Li, Y.; Qu, L. Complete Genome Sequence of Feline Panleukopenia Virus Strain HRB-CS1, Isolated from a Domestic Cat in Northeastern China. Genome Announc. 2015, 3, e01556-14. [Google Scholar] [CrossRef] [Green Version]
- Radford, A.D.; Addie, D.; Belak, S.; Boucraut-Baralon, C.; Egberink, H.; Frymus, T.; Gruffydd-Jones, T.; Hartmann, K.; Hosie, M.J.; Lloret, A.; et al. Feline calicivirus infection. ABCD guidelines on prevention and management. J. Feline Med. Surg. 2009, 11, 556–564. [Google Scholar] [CrossRef] [PubMed]
- Thiry, E.; Addie, D.; Belak, S.; Boucraut-Baralon, C.; Egberink, H.; Frymus, T.; Gruffydd-Jones, T.; Hartmann, K.; Hosie, M.J.; Lloret, A.; et al. Feline herpesvirus infection. ABCD guidelines on prevention and management. J. Feline Med. Surg. 2009, 11, 547–555. [Google Scholar] [CrossRef] [PubMed]
- Lee, C.; Song, D.; Kang, B.; Kang, D.; Yoo, J.; Jung, K.; Na, G.; Lee, K.; Park, B.; Oh, J. A serological survey of avian origin canine H3N2 influenza virus in dogs in Korea. Vet. Microbiol. 2009, 137, 359–362. [Google Scholar] [CrossRef]
- Hoffmann, E.; Stech, J.; Guan, Y.; Webster, R.G.; Perez, D.R. Universal primer set for the full-length amplification of all influenza A viruses. Arch. Virol. 2001, 146, 2275–2289. [Google Scholar] [CrossRef]
- Nguyen, L.T.; Schmidt, H.A.; von Haeseler, A.; Minh, B.Q. IQ-TREE: A fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 2015, 32, 268–274. [Google Scholar] [CrossRef] [PubMed]
- Kalyaanamoorthy, S.; Minh, B.Q.; Wong, T.K.F.; von Haeseler, A.; Jermiin, L.S. ModelFinder: Fast model selection for accurate phylogenetic estimates. Nat. Methods 2017, 14, 587–589. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Matrosovich, M.; Zhou, N.; Kawaoka, Y.; Webster, R. The surface glycoproteins of H5 influenza viruses isolated from humans, chickens, and wild aquatic birds have distinguishable properties. J. Virol. 1999, 73, 1146–1155. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Belshe, R.B.; Smith, M.H.; Hall, C.B.; Betts, R.; Hay, A.J. Genetic basis of resistance to rimantadine emerging during treatment of influenza virus infection. J. Virol. 1988, 62, 1508–1512. [Google Scholar] [CrossRef] [Green Version]
- Zhai, X.; Sun, J.; Yan, Z.; Zhang, J.; Zhao, J.; Zhao, Z.; Gao, Q.; He, W.T.; Veit, M.; Su, S. Comparison of severe acute respiratory syndrome coronavirus 2 spike protein binding to ACE2 receptors from human, pets, farm animals, and putative intermediate hosts. J. Virol. 2020, 94, e00831. [Google Scholar] [CrossRef] [PubMed]
- Sun, J.; He, W.T.; Wang, L.; Lai, A.; Ji, X.; Zhai, X.; Li, G.; Suchard, M.A.; Tian, J.; Zhou, J.; et al. COVID-19: Epidemiology, Evolution, and Cross-Disciplinary Perspectives. Trends Mol. Med. 2020, 26, 483–495. [Google Scholar] [CrossRef] [Green Version]
- Berg, S.H.; O′Hara, J.K.; Shortt, M.T.; Thune, H.; Bronnick, K.K.; Lungu, D.A.; Roislien, J.; Wiig, S. Health authorities’ health risk communication with the public during pandemics: A rapid scoping review. BMC Public Health 2021, 21, 1401. [Google Scholar] [CrossRef]
- He, W.T.; Ji, X.; He, W.; Dellicour, S.; Wang, S.; Li, G.; Zhang, L.; Gilbert, M.; Zhu, H.; Xing, G.; et al. Genomic Epidemiology, Evolution, and Transmission Dynamics of Porcine Deltacoronavirus. Mol. Biol. Evol. 2020, 37, 2641–2654. [Google Scholar] [CrossRef]
- Li, G.; He, W.; Zhu, H.; Bi, Y.; Wang, R.; Xing, G.; Zhang, C.; Zhou, J.; Yuen, K.Y.; Gao, G.F.; et al. Origin, Genetic Diversity, and Evolutionary Dynamics of Novel Porcine Circovirus 3. Adv. Sci. 2018, 5, 1800275. [Google Scholar] [CrossRef]
- Keawcharoen, J.; Oraveerakul, K.; Kuiken, T.; Fouchier, R.A.; Amonsin, A.; Payungporn, S.; Noppornpanth, S.; Wattanodorn, S.; Theambooniers, A.; Tantilertcharoen, R.; et al. Avian influenza H5N1 in tigers and leopards. Emerg. Infect. Dis. 2004, 10, 2189–2191. [Google Scholar] [CrossRef]
- Cao, X.; Yang, F.; Wu, H.; Xu, L. Genetic characterization of novel reassortant H5N6-subtype influenza viruses isolated from cats in eastern China. Arch. Virol. 2017, 162, 3501–3505. [Google Scholar] [CrossRef] [PubMed]
- Park, S.J.; Kang, B.K.; Jeoung, H.Y.; Moon, H.J.; Hong, M.; Na, W.; Park, B.K.; Poo, H.; Kim, J.K.; An, D.J.; et al. Complete Genome Sequence of a Canine-Origin H3N2 Feline Influenza Virus Isolated from Domestic Cats in South Korea. Genome Announc. 2013, 1, e0025312. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Songserm, T.; Amonsin, A.; Jam-on, R.; Sae-Heng, N.; Meemak, N.; Pariyothorn, N.; Payungporn, S.; Theamboonlers, A.; Poovorawan, Y. Avian influenza H5N1 in naturally infected domestic cat. Emerg. Infect. Dis. 2006, 12, 681–683. [Google Scholar] [CrossRef] [PubMed]
- Sponseller, B.A.; Strait, E.; Jergens, A.; Trujillo, J.; Harmon, K.; Koster, L.; Jenkins-Moore, M.; Killian, M.; Swenson, S.; Bender, H.; et al. Influenza A pandemic (H1N1) 2009 virus infection in domestic cat. Emerg. Infect. Dis. 2010, 16, 534–537. [Google Scholar] [CrossRef]
- Yu, Z.; Gao, X.; Wang, T.; Li, Y.; Xu, Y.; Chu, D.; Sun, H.; Wu, C.; Li, S.; Wang, H.; et al. Fatal H5N6 Avian Influenza Virus Infection in a Domestic Cat and Wild Birds in China. Sci. Rep. 2015, 5, 10704–10718. [Google Scholar] [CrossRef] [Green Version]
- Horimoto, T.; Kawaoka, Y. Reverse genetics provides direct evidence for a correlation of hemagglutinin cleavability and virulence of an avian influenza A virus. J. Virol. 1994, 68, 3120–3128. [Google Scholar] [CrossRef] [Green Version]
- Li, Y.; Chen, S.; Zhang, X.; Fu, Q.; Zhang, Z.; Shi, S.; Zhu, Y.; Gu, M.; Peng, D.; Liu, X. A 20-amino-acid deletion in the neuraminidase stalk and a five-amino-acid deletion in the NS1 protein both contribute to the pathogenicity of H5N1 avian influenza viruses in mallard ducks. PLoS ONE 2014, 9, e95539. [Google Scholar] [CrossRef]
- Chen, S.; Miao, X.; Huangfu, D.; Zhao, X.; Zhang, M.; Qin, T.; Peng, D.; Liu, X. H5N1 avian influenza virus without 80–84 amino acid deletion at the NS1 protein hijacks the innate immune system of dendritic cells for an enhanced mammalian pathogenicity. Transbound Emerg. Dis. 2021, 68, 2401–2413. [Google Scholar] [CrossRef] [PubMed]
- Nogales, A.; Aydillo, T.; Avila-Perez, G.; Escalera, A.; Chiem, K.; Cadagan, R.; DeDiego, M.L.; Li, F.; Garcia-Sastre, A.; Martinez-Sobrido, L. Functional Characterization and Direct Comparison of Influenza A, B, C, and D NS1 Proteins in vitro and in vivo. Front. Microbiol. 2019, 10, 2862. [Google Scholar] [CrossRef] [PubMed]

| Gene | Nucleotide Sequence Compared | Identity (%) | Virus Designation | GenBank Accession Number |
|---|---|---|---|---|
| PB2 | 2280 | 98.6 | A/canine/Guangdong/12/2012 (H3N2) | KF826944 |
| PB1 | 2274 | 98.1 | A/canine/Guangdong/12/2012 (H3N2) | KF826945 |
| PA | 2151 | 98.7 | A/canine/Guangdong/12/2012 (H3N2) A/canine/Heilongjiang/L1/2013(H3N2) | KF826946 KF042275 |
| HA | 1701 | 98.2 | A/canine/Nanjing/11/2012 (H3N2) | KF322105 |
| NP | 1497 | 99.8 | A/canine/Guangdong/05/2011 (H3N2) | JX414247 |
| NA | 1417 | 98.7 | A/canine/Nanjing/11/2012(H3N2) | KF322106 |
| M | 982 | 99.0 | A/canine/Guangdong/12/2012(H3N2) | KF826950 |
| NS | 838 | 99.42 | A/Victoria/55/2015(H3N2) | CY253950 |
| HA | NA | M2 | PB2 | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Strains | Cleavage Site | Receptor Binding Sites | Insertion | Oseltamivir-Resistant Amino Acids | Amantadine Resistant Amino Acids | Virulence Determinant | |||||||
| 324–332 | 226 | 228 | 75–77 | 119 | 222 | 274 | 292 | 26 | 27 | 30 | 31 | 627 | |
| F-JS-17 | PERQTRGLL | Q | G | KEI | E | I | H | R | L | V | A | S | E |
| F-Kor-10 | PERQTRGLF | Q | G | - - - | E | I | H | R | L | V | A | S | E |
| F-Kor-FY-10 | PERQTRGLF | Q | G | - - - | E | I | H | R | L | V | A | S | E |
| F-HLJ-14 | PERQTRGLF | Q | G | KEI | E | I | H | R | L | V | A | S | E |
| F-GD-11 | PEKQTRGLF | Q | G | - - - | E | I | H | R | L | V | A | S | E |
| F-GD-12 | PEKQTRGLF | Q | G | - - - | E | I | H | R | L | V | A | N | E |
| C-GX-12 | PEKQTRGLF | Q | G | - - - | E | I | H | R | L | V | A | S | E |
| D-JS-04 | PEKQTRGLF | Q | G | - - - | E | I | H | R | L | V | A | S | E |
| Can-NJ-12 | PERQTRGLF | Q | G | KEI | E | I | H | R | no | no | no | no | no |
| Can-GD-12 | PERQTRGLF | Q | G | - - - | E | I | H | R | L | V | A | S | E |
| Can-HLJ-13 | PERQTRGLF | Q | G | KEI | E | I | H | R | L | V | A | S | E |
| Can-Kor-15 | PERQTRGLF | Q | G | - - - | E | I | H | R | L | I | A | S | E |
| Can-USA-16 | PERQTRGLF | Q | G | - - - | E | I | H | R | L | I | A | S | E |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhao, J.; He, W.; Lu, M.; He, H.; Lai, A. Emergence and Characterization of a Novel Reassortant Canine Influenza Virus Isolated from Cats. Pathogens 2021, 10, 1320. https://doi.org/10.3390/pathogens10101320
Zhao J, He W, Lu M, He H, Lai A. Emergence and Characterization of a Novel Reassortant Canine Influenza Virus Isolated from Cats. Pathogens. 2021; 10(10):1320. https://doi.org/10.3390/pathogens10101320
Chicago/Turabian StyleZhao, Jin, Wanting He, Meng Lu, Haijian He, and Alexander Lai. 2021. "Emergence and Characterization of a Novel Reassortant Canine Influenza Virus Isolated from Cats" Pathogens 10, no. 10: 1320. https://doi.org/10.3390/pathogens10101320
APA StyleZhao, J., He, W., Lu, M., He, H., & Lai, A. (2021). Emergence and Characterization of a Novel Reassortant Canine Influenza Virus Isolated from Cats. Pathogens, 10(10), 1320. https://doi.org/10.3390/pathogens10101320






