Current Prevalence of Oral Helicobacter pylori among Japanese Adults Determined Using a Nested Polymerase Chain Reaction Assay
Abstract
:1. Introduction
2. Results
2.1. Relative Frequencies by Sex, Age and Medical History
2.2. Relative Frequencies by Sampling Site and after Eradication Therapy
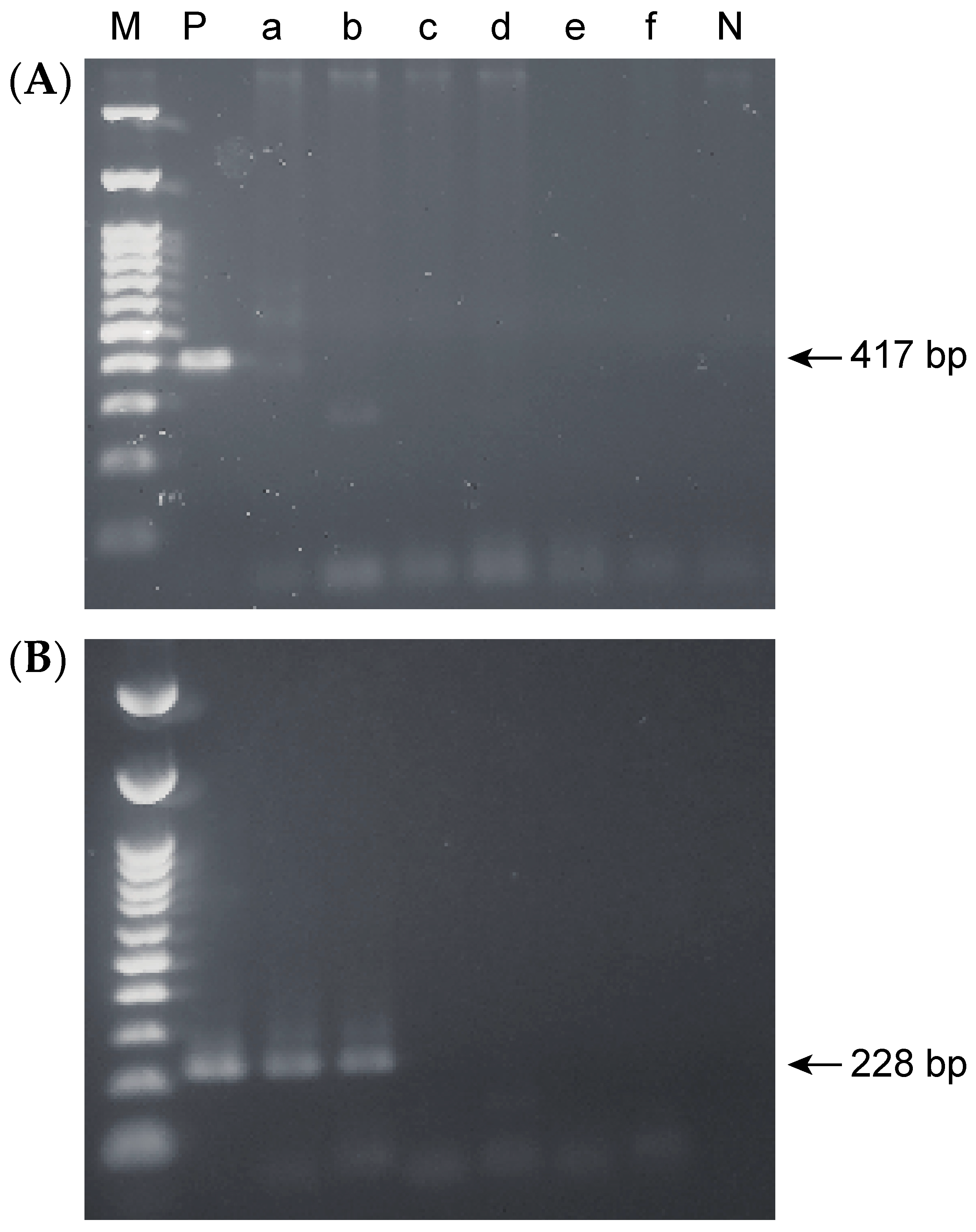
2.3. Accuracy of the Nested PCR Method
2.4. Detection Limits for Single-Step and Nested PCR
3. Discussion
4. Materials and Methods
4.1. Participants
4.2. Sampling
4.3. DNA Extraction and Nested PCR
4.4. Sequencing of Nested PCR Products
4.5. Examination of the Detection Limit of the Single-Step and Nested PCR Methods
4.6. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Marshall, B.J.; Warren, J.R. Unidentified curved bacilli in the stomach of patients with gastritis and peptic ulceration. Lancet 1984, 1, 1311–1315. [Google Scholar] [CrossRef]
- Parsonnet, J.; Friedman, G.D.; Vandersteen, D.P.; Chang, Y.; Vogelman, J.H.; Orentreich, N.; Sibley, R.K. Helicobacter pylori infection and the risk of gastric carcinoma. N. Engl. J. Med. 1991, 325, 1127–1131. [Google Scholar] [CrossRef] [PubMed]
- Nomura, A.; Stemmermann, G.N.; Chyou, P.-H.; Kato, I.; Perez-Perez, G.I.; Blaser, M.J. Helicobacter pylori infection and gastric carcinoma among Japanese Americans in Hawaii. N. Engl. J. Med. 1991, 325, 1132–1136. [Google Scholar] [CrossRef] [PubMed]
- Dooley, C.P.; Cohen, H.; Fitzgibbons, P.L.; Bauer, M.; Appleman, M.D.; Perez-Perez, G.I.; Blaser, M.J. Prevalence of Helicobacter pylori infection and histologic gastritis in asymptomatic persons. N. Engl. J. Med. 1989, 321, 1562–1566. [Google Scholar] [CrossRef] [PubMed]
- Hooi, J.K.Y.; Lai, W.Y.; Ng, W.K.; Suen, M.M.Y.; Underwood, F.E.; Tanyingoh, D.; Malfertheiner, P.; Graham, D.Y.; Wong, V.W.S.; Wu, J.C.Y.; et al. Global prevalence of Helicobactoer pylori infection: Systematic review and meta-analysis. Gastroenteology 2017, 153, 420–429. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vale, F.F.; Vítor, J.M.B. Transmission pathway of Helicobacter pylori: Does food play a role in rural and urban areas? Int. J. Food Microbiol. 2010, 138, 1–12. [Google Scholar] [CrossRef]
- Weyermann, M.; Rothenbacher, D.; Brenner, H. Acquisition of Helicobacter pylori infection in early childhood: Independent contributions of infected mothers, fathers, and siblings. Am. J. Gastroenterol. 2009, 104, 182–189. [Google Scholar] [CrossRef]
- Yee, J.K.C. Are the view of Helicobacter pylori colonized in the oral cavity an illusion? Exp. Mol. Med. 2017, 49, e397. [Google Scholar] [CrossRef] [Green Version]
- Adler, I.; Muiño, A.; Aguas, S.; Harada, L.; Diaz, M.; Lence, A.; Labbrozzi, M.; Muiño, J.M.; Elsner, B.; Avagnina, A.; et al. Helicobacter pylori and oral pathology: Relationship with the gastric infection. World J. Gastroenterol. 2014, 20, 9922–9935. [Google Scholar] [CrossRef]
- Anand, P.S.; Kamath, K.P.; Anil, S. Role of dental plaque, saliva and periodontal disease in Helicobacter pylori infection. World J. Gastroenterol. 2014, 20, 5639–5653. [Google Scholar] [CrossRef]
- Sugimoto, M.; Wu, J.Y.; Abudayyeh, S.; Hoffman, J.; Brahem, H.; Al-khatib, K.; Yamaoka, Y.; Graham, D.Y. Unreliability of results of PCR detection of Helicobacter pylori in clinical or environmental samples. J. Clin. Microbiol. 2009, 47, 738–742. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dowsett, S.A.; Kowolik, M.J. Oral Helicobacter pylori: Can we stomach it? Crit. Rev. Oral Biol. Med. 2003, 14, 226–233. [Google Scholar] [CrossRef] [PubMed]
- Song, Q.; Lange, T.; Spahr, A.; Adler, G.; Bode, G. Characteristic distribution pattern of Helicobacter pylori in dental plaque and saliva detected with nested PCR. J. Med. Microbiol. 2000, 49, 349–353. [Google Scholar] [CrossRef] [PubMed]
- Miyabayashi, H.; Furihata, K.; Shimizu, T.; Ueno, I.; Akamatsu, T. Influence of oral Helicobacter pylori on the success of eradication therapy against gastric Helicobacter pylori. Helicobacter 2000, 5, 30–37. [Google Scholar] [CrossRef]
- Román-Román, A.; Giono-Cerezo, S.; Camorlinga-Ponce, M.; Martínez-Carrillo, D.N.; Loaiza-Loeza, S.; Fernández-Tilapa, G. VacA genotypes of Helicobacter pylori in the oral cavity and stomach of patients with chronic gastritis and gastric ulcer. Enferm. Infecc. Microbiol. Clin. 2013, 31, 130–135. [Google Scholar] [CrossRef]
- Miyamoto, R.; Okuda, M.; Lin, Y.; Murotani, K.; Okumura, A.; Kikuchi, S. Rapidly decreasing prevalence of Helicobacter pylori among Japanese children and adolescents. J. Infect. Chemother. 2019, 25, 526–530. [Google Scholar] [CrossRef]
- Wang, C.; Nishiyama, T.; Kikuchi, S.; Inoue, M.; Sawada, N.; Tsugane, S.; Lin, Y. Changing trends in the prevalence of H. pylori infection in Japan (1908–2003): A systematic review and meta-regression analysis of 170,752 individuals. Sci. Rep. 2017, 7, 15491. [Google Scholar] [CrossRef]
- O’Leary, T.J.; Drake, R.B.; Naylor, J.E. The plaque control record. J. Periodontol. 1972, 43, 38. [Google Scholar] [CrossRef]
- Turesky, S.; Gilmore, N.D.; Glickman, I. Reduced plaque formation by the chloromethyl analogue of victamine C. J. Periodontol. 1970, 41, 41–43. [Google Scholar] [CrossRef] [Green Version]
- Konno, M.; Fujii, N.; Yokota, S.; Sato, K.; Takahashi, M.; Sato, K.; Mino, E.; Sugiyama, T. Five-year follow-up study of mother-to-child transmission of Helicobacter pylori infection detected by a random amplified polymorphic DNA fingerprinting method. J. Clin. Microbiol. 2005, 43, 2246–2250. [Google Scholar] [CrossRef] [Green Version]
- Okuda, M.; Miyashiro, E.; Booka, M.; Tsuji, T.; Nakazawa, T. Helicobacter pylori colonization in the first 3 years of life in Japanese children. Helicobacter 2007, 12, 324–327. [Google Scholar] [CrossRef] [PubMed]
- Ueno, T.; Suzuki, H.; Hirose, M.; Shida, T.; Ikezawa, K.; Matsui, H.; Mizokami, Y.; Yanaka, A. Influence of living environment during childhood on Helicobacter pylori infection in Japanese young adults. Digestion 2019, 24, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Roberts, F.A.; Darveau, R.P. Microbial protection and virulence in periodontal tissue as a function of polymicrobial communities: Symbiosis and dysbiosis. Periodontol. 2000 2015, 69, 18–27. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Marco, M.; Lucrezia, T.; Giuseppe, T.; Vito, C.A.C.; Davide, B.G.; Lucio, M.; Maurizio, P.; Lorenzo, L.M.; Andrea, S. Beyond head and neck cancer: The relationship between oral microbiota and tumour development in distant organs. Front. Cell Infect. Microbiol. 2019, 9, 232. [Google Scholar]
- Kusters, J.G.; van Vliet, A.H.M.; Kuipers, E.J. Pathogenesis of Helicobacter pylori infection. Clin. Microbiol. Rev. 2006, 19, 449–490. [Google Scholar] [CrossRef] [Green Version]
- Scott, D.R.; Marcus, E.A.; Weeks, D.L.; Sachs, G. Mechanisms of acid resistance due to the urease system of Helicobacter pylori. Gastroenterology 2002, 123, 187–195. [Google Scholar] [CrossRef]
- Takahashi, N.; Nyvad, B. The role of bacteria in the caries process: Ecological perspectives. J. Dent. Res. 2011, 90, 294–303. [Google Scholar] [CrossRef]
- Liu, Y.; Lin, H.; Bai, Y.; Qin, X.; Zheng, X.; Sun, Y.; Zhang, Y. Study on the relationship between Helicobacter pylori in the dental plaque and the occurrence of dental caries or oral hygiene index. Helicobacter 2008, 13, 256–260. [Google Scholar] [CrossRef]
- Berg, E.D. The indigenous gastrointestinal microflora. Trends Microbiol. 1996, 4, 430–435. [Google Scholar] [CrossRef]
- Daniluk, T.; Tokajuk, G.; Cylwik-Rokicka, D.; Rożkiewicz, D.; Zaremba, M.L.; Stokowska, W. Aerobic and anaerobic bacteria in subgingival and supragingival plaques of adult patients with periodontal disease. Adv. Med. Sci. 2006, 1, 81–85. [Google Scholar]
- Yamaguchi, H.; Osaki, T.; Takahashi, M.; Taguchi, H.; Kamiya, S. Colony formation by Helicobacter pylori after long-term incubation under anaerobic conditions. FEMS Microbiol. Lett. 1999, 175, 107–111. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kusters, J.G.; Gerrits, M.M.; Van Strijp, J.A.; Vandenbroucke-Grauls, C.M. Coccoid forms of Helicobacter pylori are the morphologic manifestation of cell death. Infect. Immun. 1997, 65, 3672–3679. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hirukawa, S.; Sagara, H.; Kaneto, S.; Kondo, T.; Kiga, K.; Sanada, T.; Kiyono, H.; Mimuro, H. Caracterization of morphological conversion of Helicobacter pylori under anaerobic conditions. Microbiol. Immunol. 2018, 62, 221–228. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, X.M.; Yee, K.C.; Hazeki-Taylor, N.; Li, J.; Fu, H.Y.; Huang, M.L.; Zhang, G.Y. Oral Helicobacter pylori, its relationship to successful eradication of gastric H. pylori and saliva culture confirmation. J. Physiol. Pharmacol. 2014, 65, 559–566. [Google Scholar] [PubMed]
- Song, Q.; Haller, B.; Schmid, R.M.; Adler, G.; Bode, G. Helicobacter pylori in dental plaque: A comparison of different PCR primer sets. Dig. Dis. Sci. 1999, 44, 479–484. [Google Scholar] [CrossRef] [PubMed]
- Song, Q.; Spahr, A.; Schmid, R.M.; Adler, G.; Bode, G. Helicobacter pylori in the oral cavity: High prevalence and great DNA diversity. Dig. Dis. Sci. 2000, 45, 2162–2167. [Google Scholar] [CrossRef]
- Tomb, J.F.; White, O.; Kerlavage, A.R.; Clayton, R.A.; Sutton, G.G.; Fleischmann, R.D.; Ketchum, K.A.; Klenk, H.P.; Gill, S.; Dougherty, B.A.; et al. The complete genome sequence of the gastric pathogen Helicobacter pylori. Nature 1997, 388, 539–547. [Google Scholar] [CrossRef] [Green Version]

| Variable | Male | Female | Total |
|---|---|---|---|
| Gender | |||
| N (%) | 34 (38.6) | 54 (61.4) | 88 (100) |
| Age | |||
| Mean ± SD | 46.7 ± 20.8 | 55.2 ± 20.4 | 52.1 ± 20.8 |
| Medical history N (%) | |||
| Gastric infection carrier | 0 (0) | 3 (0.06) | 3 (0.03) |
| Received eradication therapy | 3 (0.09) | 9 (0.17) | 12 (0.14) |
| Experience not required | 31 (91.2) | 42 (77.8) | 73 (83.0) |
| Individuals (Positive/Total) | Prevalence (%) | p | |
|---|---|---|---|
| Gender | |||
| Male | 10/34 | 29.4 | |
| Female | 22/54 | 40.7 | 0.364 |
| Generation (age) | |||
| Young (24 to 34) | 6/28 | 21.4 | |
| Middle-aged (35 to 64) | 14/29 | 48.3 | |
| Elderly (64 to 91) | 12/31 | 38.7 | 0.103 |
| Individuals (Positive/Total) | Prevalence (%) | p | |
|---|---|---|---|
| Medical history | |||
| Gastric infection carrier | 3/3 | 100 | |
| Received eradication therapy | 9/12 | 75.0 | |
| Experience not required | 20/73 | 27.4 | 0.02 |
| Individuals (Positive/Total) | Prevalence (%) | OR | |
|---|---|---|---|
| Supragingival biofilm | |||
| Lower incisor | 19/88 | 21.6 | 24.0 |
| Upper incisor | 14/88 | 15.9 | 16.5 |
| Lower left molar | 3/88 | 3.4 | 3.1 |
| Upper right molar | 1/88 | 1.1 | 1 |
| Saliva | 4/88 | 4.5 | |
| Tongue | 2/88 | 2.3 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nagata, R.; Ohsumi, T.; Takenaka, S.; Noiri, Y. Current Prevalence of Oral Helicobacter pylori among Japanese Adults Determined Using a Nested Polymerase Chain Reaction Assay. Pathogens 2021, 10, 10. https://doi.org/10.3390/pathogens10010010
Nagata R, Ohsumi T, Takenaka S, Noiri Y. Current Prevalence of Oral Helicobacter pylori among Japanese Adults Determined Using a Nested Polymerase Chain Reaction Assay. Pathogens. 2021; 10(1):10. https://doi.org/10.3390/pathogens10010010
Chicago/Turabian StyleNagata, Ryoko, Tatsuya Ohsumi, Shoji Takenaka, and Yuichiro Noiri. 2021. "Current Prevalence of Oral Helicobacter pylori among Japanese Adults Determined Using a Nested Polymerase Chain Reaction Assay" Pathogens 10, no. 1: 10. https://doi.org/10.3390/pathogens10010010
APA StyleNagata, R., Ohsumi, T., Takenaka, S., & Noiri, Y. (2021). Current Prevalence of Oral Helicobacter pylori among Japanese Adults Determined Using a Nested Polymerase Chain Reaction Assay. Pathogens, 10(1), 10. https://doi.org/10.3390/pathogens10010010





