Molecular Docking and Aberration-Corrected STEM of Palladium Nanoparticles on Viral Templates
Abstract
:1. Introduction
2. Experimental Section
2.1. Recombinant Production of Viral Templates
2.2. Molecular Docking Simulations
2.3. In Situ Synthesis of Pd Nanoparticles
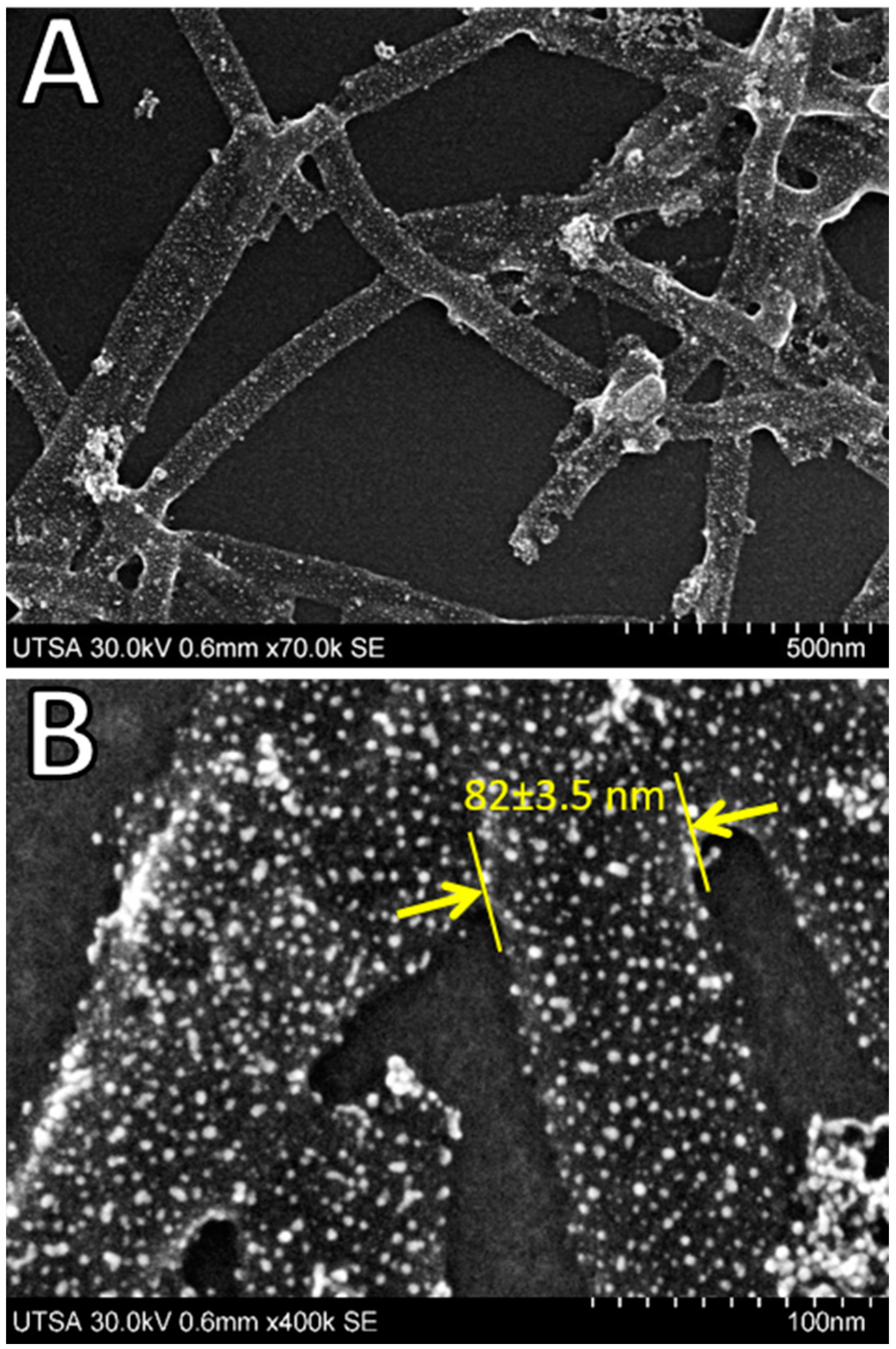
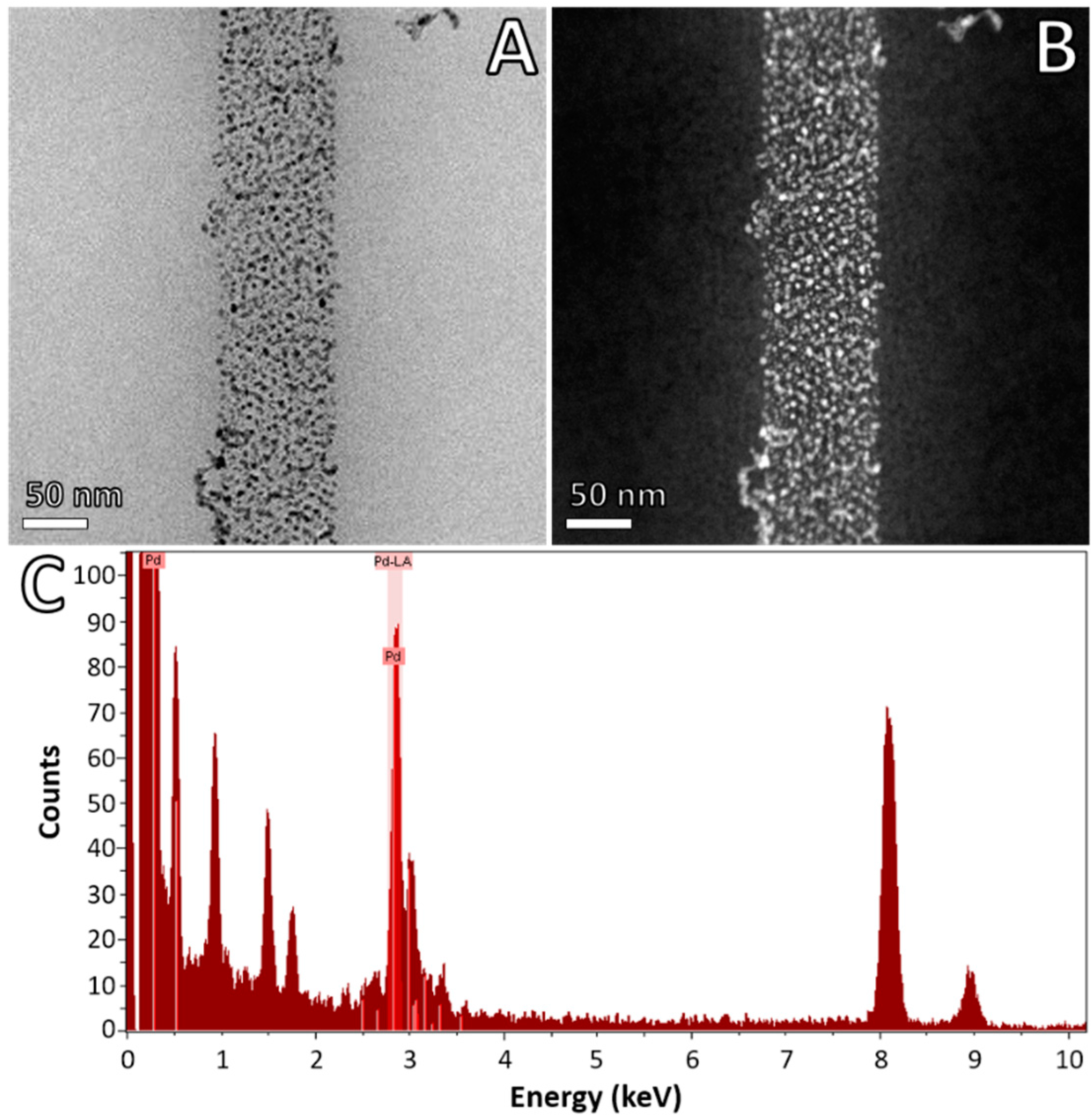
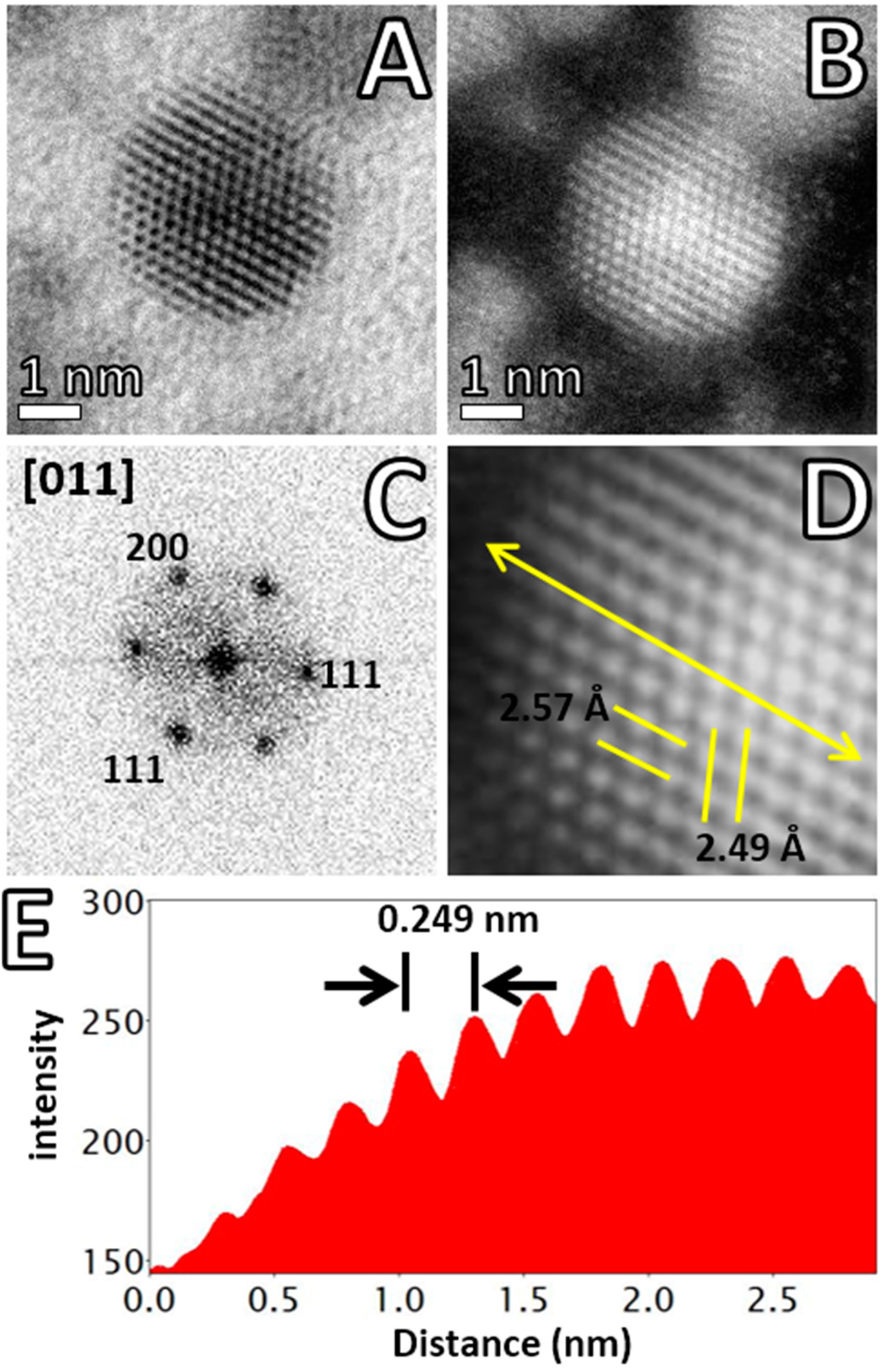
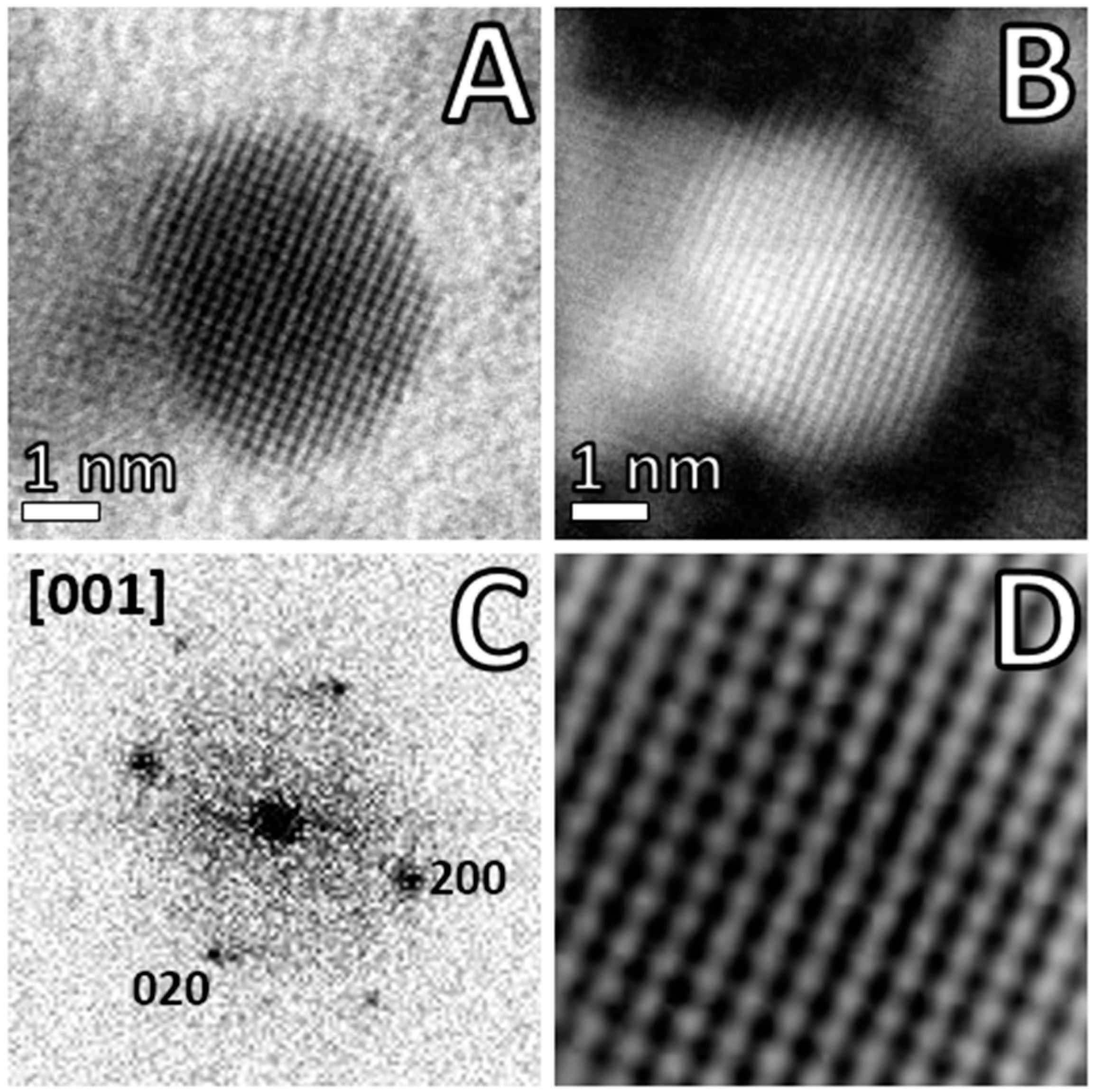
2.4. Advanced Electron Microscopy Imaging
3. Results and Discussion
Molecular Binding Simulation VP6-Pd(II)
4. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Yuan, X.; Dou, X.; Zheng, K.; Xie, J. Recent advances in the synthesis and applications of ultrasmall bimetallic nanoclusters. Part. Part. Syst. Charact. 2015, 32, 613–629. [Google Scholar]
- James, C. The preparation of palladium nanoparticles. Platin. Met. Rev. 2012, 56, 83–98. [Google Scholar]
- Corthey, G.; Rubert, A.A.; Picone, A.L.; Casillas, G.; Giovanetti, L.J.; Ramallo-López, J.M.; Zelaya, E.; Benitez, G.A.; Requejo, F.G.; José-Yacamán, M.; et al. New insights into the chemistry of thiolate-protected palladium nanoparticles. J. Phys. Chem. C 2012, 116, 9830–9837. [Google Scholar]
- Kettemann, F.; Wuithschick, M.; Caputo, G.; Kraehnert, R.; Pinna, N.; Rademann, K.; Polte, J. Reliable palladium nanoparticle syntheses in aqueous solution: The importance of understanding precursor chemistry and growth mechanism. CrystEngComm 2015, 17, 1865–1870. [Google Scholar]
- Scott, R.W.J.; Ye, H.; Henriquez, R.R.; Crooks, R.M. Synthesis, characterization, and stability of dendrimer-encapsulated palladium nanoparticles. Chem. Mater. 2003, 15, 3873–3878. [Google Scholar]
- Dickerson, M.B.; Sandhage, K.H.; Naik, R.R. Protein- and peptide-directed syntheses of inorganic materials. Chem. Rev. 2008, 108, 4935–4978. [Google Scholar]
- Slocik, J.M.; Wright, D.W. Biomimetic mineralization of noble metal nanoclusters. Biomacromolecules 2003, 4, 1135–1141. [Google Scholar]
- Strable, E.; Finn, M.G. Chemical modification of viruses and virus-like particles. Curr. Top. Microbiol. Immunol. 2009, 327, 1–21. [Google Scholar]
- Lim, J.S.; Kim, S.M.; Lee, S.Y.; Stach, E.A.; Culver, J.N.; Harris, M.T. Quantitative study of Au(III) and Pd(II) ion biosorption on genetically engineered Tobacco mosaic virus. J. Colloid Interface Sci. 2010, 342, 455–461. [Google Scholar]
- Lewis, C.L.; Lin, Y.; Yang, C.; Manocchi, A.K.; Yuet, K.P.; Doyle, P.S.; Yi, H. Microfluidic fabrication of hydrogel microparticles containing functionalized viral nanotemplates. Langmuir 2010, 26, 13436–13441. [Google Scholar]
- Manocchi, A.K.; Horelik, N.E.; Lee, B.; Yi, H. Simple, readily controllable palladium nanoparticle formation on surface-assembled viral nanotemplates. Langmuir 2010, 26, 3670–3677. [Google Scholar]
- Manocchi, A.K.; Seifert, S.; Lee, B.; Yi, H. On the thermal stability of surface-assembled viral-metal nanoparticle complexes. Langmuir 2010, 26, 7516–7522. [Google Scholar] [PubMed]
- Manocchi, A.K.; Seifert, S.; Lee, B.; Yi, H. In situ small-angle X-ray scattering analysis of palladium nanoparticle growth on tobacco mosaic virus nanotemplates. Langmuir 2011, 27, 7052–7058. [Google Scholar]
- Yang, C.; Choi, C.H.; Lee, C.S.; Yi, H. A facile synthesis-fabrication strategy for integration of catalytically active viral-palladium nanostructures into polymeric hydrogel microparticles via replica molding. ACS Nano 2013, 7, 5032–5044. [Google Scholar]
- Adigun, O.O.; Freer, A.S.; Miller, J.T.; Loesch-Fries, L.S.; Kim, B.S.; Harris, M.T. Mechanistic study of the hydrothermal reduction of palladium on the Tobacco mosaic virus. J. Colloid Interface Sci. 2015, 450, 1–6. [Google Scholar]
- Oh, D.; Qi, J.; Lu, Y.C.; Zhang, Y.; Shao-Horn, Y.; Belcher, A.M. Biologically enhanced cathode design for improved capacity and cycle life for lithium-oxygen batteries. Nat. Commun. 2013, 4, 2756. [Google Scholar]
- Zhou, J.C.; Soto, C.M.; Chen, M.S.; Bruckman, M.A.; Moore, M.H.; Barry, E.; Ratna, B.R.; Pehrsson, P.E.; Spies, B.R.; Confer, T.S. Biotemplating rod-like viruses for the synthesis of copper nanorods and nanowires. J. Nanobiotechnol. 2012, 10, 18. [Google Scholar]
- Plascencia-Villa, G.; Mena, J.A.; Castro-Acosta, R.M.; Fabian, J.C.; Ramirez, O.T.; Palomares, L.A. Strategies for the purification and characterization of protein scaffolds for the production of hybrid nanobiomaterials. J. Chromatogr. B 2011, 879, 1105–1111. [Google Scholar]
- Plascencia-Villa, G.; Saniger, J.M.; Ascencio, J.A.; Palomares, L.A.; Ramirez, O.T. Use of recombinant rotavirus VP6 nanotubes as a multifunctional template for the synthesis of nanobiomaterials functionalized with metals. Biotechnol. Bioeng. 2009, 104, 871–881. [Google Scholar] [CrossRef] [PubMed]
- Carreno-Fuentes, L.; Ascencio, J.A.; Medina, A.; Aguila, S.; Palomares, L.A.; Ramirez, O.T. Strategies for specifically directing metal functionalization of protein nanotubes: Constructing protein coated silver nanowires. Nanotechnology 2013, 24, 235602. [Google Scholar] [CrossRef] [PubMed]
- Pastor, A.R.; Rodriguez-Limas, W.A.; Contreras, M.A.; Esquivel, E.; Esquivel-Guadarrama, F.; Ramirez, O.T.; Palomares, L.A. The assembly conformation of rotavirus VP6 determines its protective efficacy against rotavirus challenge in mice. Vaccine 2014, 32, 2874–2877. [Google Scholar] [CrossRef] [PubMed]
- Seebeck, B.; Reulecke, I.; Kamper, A.; Rarey, M. Modeling of metal interaction geometries for protein-ligand docking. Proteins 2008, 71, 1237–1254. [Google Scholar] [CrossRef] [PubMed]
- Pennycook, S.J. Scanning transmission electron microscopy: Seeing the atoms more clearly. MRS Bull. 2012, 37, 943–951. [Google Scholar] [CrossRef]
- Chen, J.Z.; Settembre, E.C.; Aoki, S.T.; Zhang, X.; Bellamy, A.R.; Dormitzer, P.R.; Harrison, S.C.; Grigorieff, N. Molecular interactions in rotavirus assembly and uncoating seen by high-resolution cryo-EM. Proc. Natl. Acad. Sci. USA 2009, 106, 10644–10648. [Google Scholar] [CrossRef] [PubMed]
- Mathieu, M.; Petitpas, I.; Navaza, J.; Lepault, J.; Kohli, E.; Pothier, P.; Prasad, B.V.; Cohen, J.; Rey, F.A. Atomic structure of the major capsid protein of rotavirus: Implications for the architecture of the virion. EMBO J. 2001, 20, 1485–1497. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Settembre, E.; Xu, C.; Dormitzer, P.R.; Bellamy, R.; Harrison, S.C.; Grigorieff, N. Near-atomic resolution using electron cryomicroscopy and single-particle reconstruction. Proc. Natl. Acad. Sci. USA 2008, 105, 1867–1872. [Google Scholar] [CrossRef] [PubMed]
- Krieger, E.; Joo, K.; Lee, J.; Raman, S.; Thompson, J.; Tyka, M.; Baker, D.; Karplus, K. Improving physical realism, stereochemistry, and side-chain accuracy in homology modeling: Four approaches that performed well in CASP8. Proteins 2009, 77, 114–122. [Google Scholar] [CrossRef] [PubMed]
- Dolinsky, T.J.; Czodrowski, P.; Li, H.; Nielsen, J.E.; Jensen, J.H.; Klebe, G.; Baker, N.A. PDB2PQR: Expanding and upgrading automated preparation of biomolecular structures for molecular simulations. Nucleic Acids Res. 2007, 35, W522–W525. [Google Scholar] [CrossRef] [PubMed]
- Huey, R.; Morris, G.M.; Olson, A.J.; Goodsell, D.S. A semiempirical free energy force field with charge-based desolvation. J. Comput. Chem. 2007, 28, 1145–1152. [Google Scholar] [CrossRef] [PubMed]
- Morris, G.M.; Goodsell, D.S.; Halliday, R.S.; Huey, R.; Hart, W.E.; Belew, R.K.; Olson, A.J. Automated docking using a Lamarckian genetic algorithm and an empirical binding free energy function. J. Comput. Chem. 1998, 19, 1639–1662. [Google Scholar] [CrossRef]
- Talanova, G.G.; Yatsimirskii, K.B.; Kravchenko, O.V. Peculiarities of K2PdCl4 and K2PtCl4 Complexation with Polymer-Supported Dibenzo-18-crown-6. Ind. Eng. Chem. Res. 2000, 39, 3611–3615. [Google Scholar] [CrossRef]
- Li, J.H.; Byrne, R.H. Amino acid complexation of palladium in seawater. Environ. Sci. Technol. 1990, 24, 1038–1041. [Google Scholar] [CrossRef]
- Odani, A.; Yamauchi, O. Ternary α-amino acid-palladium(II) complexes with ligand-ligand hydrogen bonding. Bull. Chem. Soc. Jpn. 1981, 54, 3773–3779. [Google Scholar] [CrossRef]
- Wilson, E.W.; Martin, R.B. Circular dichroism of palladium(II) complexes of amino acids and peptides. Inorg. Chem. 1970, 9, 528–532. [Google Scholar] [CrossRef]
- Sugahara, M.; Asada, Y.; Shimada, H.; Taka, H.; Kunishima, N. HATODAS II—Heavy-atom database system with potentiality scoring. J. Appl. Crystallogr. 2009, 42, 540–544. [Google Scholar]
- Coppage, R.; Slocik, J.M.; Sethi, M.; Pacardo, D.B.; Naik, R.R.; Knecht, M.R. Elucidation of peptide effects that control the activity of nanoparticles. Angew. Chem. 2010, 49, 3767–3770. [Google Scholar] [CrossRef] [PubMed]
- Lepault, J.; Petitpas, I.; Erk, I.; Navaza, J.; Bigot, D.; Dona, M.; Vachette, P.; Cohen, J.; Rey, F.A. Structural polymorphism of the major capsid protein of rotavirus. EMBO J. 2001, 20, 1498–1507. [Google Scholar] [CrossRef] [PubMed]
- Bedford, N.M.; Ramezani-Dakhel, H.; Slocik, J.M.; Briggs, B.D.; Ren, Y.; Frenkel, A.I.; Petkov, V.; Heinz, H.; Naik, R.R.; Knecht, M.R. Elucidation of peptide-directed palladium surface structure for biologically tunable nanocatalysts. ACS Nano 2015, 9, 5082–5092. [Google Scholar] [CrossRef] [PubMed]
- Bhandari, R.; Knecht, M.R. Isolation of template effects that control the structure and function of nonspherical, biotemplated Pd nanomaterials. Langmuir 2012, 28, 8110–8119. [Google Scholar] [CrossRef] [PubMed]
- José-Yacamán, M.; Marı́n-Almazo, M.; Ascencio, J.A. High resolution TEM studies on palladium nanoparticles. J. Mol. Catal. A Chem. 2001, 173, 61–74. [Google Scholar] [CrossRef]
- Corthey, G.; Olmos-Asar, J.A.; Casillas, G.; Mariscal, M.M.; Mejía-Rosales, S.; Azcárate, J.C.; Larios, E.; José-Yacamán, M.; Salvarezza, R.C.; Fonticelli, M.H. Influence of capping on the atomistic arrangement in palladium nanoparticles at room temperature. J. Phys. Chem. C 2014, 118, 24641–24647. [Google Scholar] [CrossRef]
- Bej, A.; Ghosh, K.; Sarkar, A.; Knight, D.W. Palladium nanoparticles in the catalysis of coupling reactions. RSC Adv. 2016, 6, 11446–11453. [Google Scholar] [CrossRef]
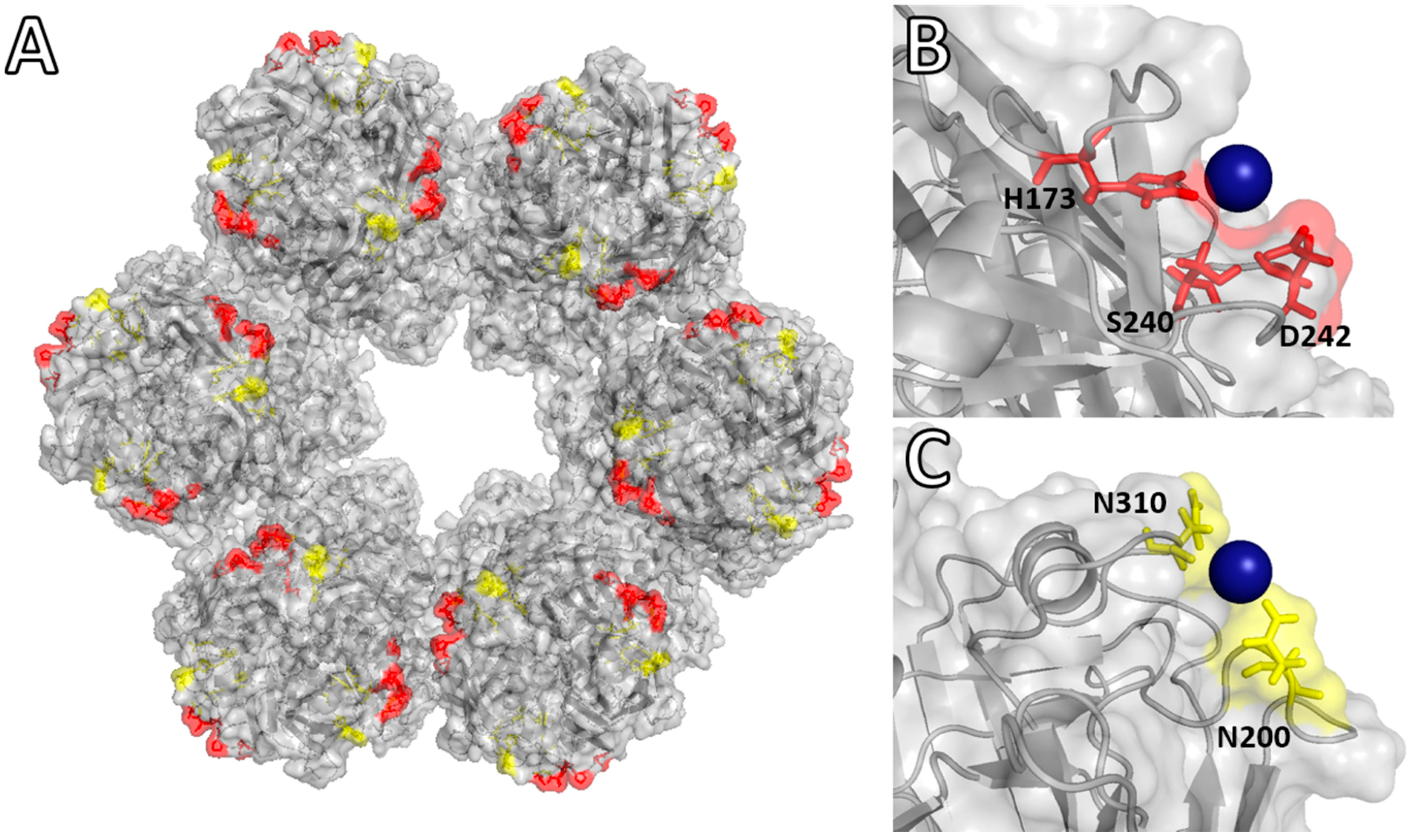
| pH of Simulation | PdCl3(H2O)− | PdCl4− | ||
|---|---|---|---|---|
| Residues | Binding Energy (∆G) | Residues | Binding Energy (∆G) | |
| 5.0 | His173 Ser240 Asp242 | −2.0 kcal·mol−1 | Ser169 Gln170 His173 | −0.80 kcal·mol−1 |
| 8.0 | Asn200 Asn310 | −1.2 kcal·mol−1 | Pro309 Asn310 Ala311 | −0.15 kcal·mol−1 |
© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Carreño-Fuentes, L.; Bahena, D.; Palomares, L.A.; Ramírez, O.T.; José-Yacamán, M.; Plascencia-Villa, G. Molecular Docking and Aberration-Corrected STEM of Palladium Nanoparticles on Viral Templates. Metals 2016, 6, 200. https://doi.org/10.3390/met6090200
Carreño-Fuentes L, Bahena D, Palomares LA, Ramírez OT, José-Yacamán M, Plascencia-Villa G. Molecular Docking and Aberration-Corrected STEM of Palladium Nanoparticles on Viral Templates. Metals. 2016; 6(9):200. https://doi.org/10.3390/met6090200
Chicago/Turabian StyleCarreño-Fuentes, Liliana, Daniel Bahena, Laura A. Palomares, Octavio T. Ramírez, Miguel José-Yacamán, and Germán Plascencia-Villa. 2016. "Molecular Docking and Aberration-Corrected STEM of Palladium Nanoparticles on Viral Templates" Metals 6, no. 9: 200. https://doi.org/10.3390/met6090200
APA StyleCarreño-Fuentes, L., Bahena, D., Palomares, L. A., Ramírez, O. T., José-Yacamán, M., & Plascencia-Villa, G. (2016). Molecular Docking and Aberration-Corrected STEM of Palladium Nanoparticles on Viral Templates. Metals, 6(9), 200. https://doi.org/10.3390/met6090200







