The Skin Microbiome Profile of Contact Sports Athletes—Focus on Sexual Dimorphism and Athlete–Non-Athlete Differences
Abstract
1. Introduction
2. Materials and Methods
2.1. Subjects
2.2. Sampling
2.3. Microbial Analyses
2.4. Statistical Analyses
2.5. Ethical Approval Information
3. Results
4. Discussion
4.1. Commensals—Healthy Skin Microbiome Representatives in Youth
4.2. Pathogens Among Skin Microbiome Representatives in Youth
4.2.1. Sex Differences in Skin Microbiome Profile of Youth
4.2.2. Athlete–Non-Athlete Differences in Skin Microbiome Profile of Youth
- (1)
- The genera Acinetobacter (species are often opportunists in hospitalized patients with impaired immunity, but were also found in contact sports athletes’ skin microbiome indicating the dysbiosis of the wrestlers’ skin) [14] and Brevundimonas (spp. causing infections in immunosuppressed people) [32] had higher relative abundance in wrestlers than members of the other two subgroups—indicating their increased risk for skin infections.
- (2)
- The Micrococcus (species are members of the normal skin microbiome; however, they can lead to skin infections in immunocompromised people) [33] and Enhydrobacter (it was evidenced that correlated with the cheek moisture) [34] genera were found in higher abundances in both wrestlers and kickboxers than in non-athletes.
- (3)
- The Corynebacterium, Cutibacterium, and Streptococcus (infection agents of the skin and the respiratory tract) genera were found in higher relative abundance in kickboxers than in wrestlers or non-athletes (opportunistic pathogen, correlates with skin moisture content) [35].
- (4)
- The Kocuria (pathogen of skin and soft tissue infections), Staphylococcus (Staphylococcus spp. may cause skin infections (e.g., abscesses and boils, cellulitis, or folliculitis), and Escherichia–Shigella (causing infections in the gastrointestinal tract) genera were less represented in athletes than in non-athletes.
- (5)
- Pathogens in wrestlers’ and kickboxers’ skin microbiome and missing from the microbiome of the control group members’ sample were as follows:
- Aliterella sp. (in 29% of athletes)—causing skin infections;
- Arthrobacter (in 29% of athletes)—skin infections;
- Brucella (in 23% of athletes)—humans can acquire the disease through direct contact with infected animals, consuming animal products, symptoms: fever, fatigue, joint and muscle pain;
- Empedobacter sp. (in 39% of athletes) skin infection originating from environmental sources;
- Peptostreptococcus (in 14% of athletes)—can become pathogenic under certain conditions, causing skin and other skin tissue infections.
5. Conclusions
6. Limitations of the Study
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Runacres, A.; Mackintosh, K.A.; McNarry, M.A. Health consequences of an elite sporting career: Long-term detriment or long-term gain? A meta-analysis of 165,000 former athletes. Sports Med. 2021, 51, 289–301. [Google Scholar] [CrossRef]
- Gleeson, M. Immune function in sport and exercise. J. Appl. Physiol. 2007, 103, 693–699. [Google Scholar] [CrossRef]
- Cadegiani, F.A.; Kater, C.E. Hormonal aspects of overtraining syndrome: A systematic review. BMC Sports Sci. Med. Rehabil. 2017, 9, 1–15. [Google Scholar] [CrossRef]
- Kalabiska, I.; Annar, D.; Keki, Z.; Borbas, Z.; Bhattoa, H.P.; Zsakai, A. The oral microbiome profile of water polo players aged 16–20. Sports 2023, 11, 216–226. [Google Scholar] [CrossRef]
- Pecci, M.; Comeau, D.; Chawla, V. Skin conditions in the athlete. Am. J. Sports Med. 2009, 37, 406–418. [Google Scholar] [CrossRef] [PubMed]
- Bassiri-Jahromi, S.; Sadeghi, G.; Asghari Paskiaee, F. Evaluation of the association of superficial dermatophytosis and athletic activities with special reference to its prevention and control. Int. J. Dermatol. 2010, 49, 1159–1164. [Google Scholar] [CrossRef]
- Grice, E.A.; Segre, J.A. The skin microbiome. Nat. Rev. Microbiol. 2011, 9, 244–253. [Google Scholar] [CrossRef]
- Human Microbiome Project Consortium. Structure, function and diversity of the healthy human microbiome. Nature 2012, 486, 207–214. [Google Scholar] [CrossRef] [PubMed]
- Costello, E.K.; Lauber, C.L.; Hamady, M.; Fierer, N.; Gordon, J.I.; Knight, R. Bacterial community variation in human body habitats across space and time. Science 2009, 326, 1694–1697. [Google Scholar] [CrossRef]
- Byrd, A.L.; Belkaid, Y.; Segre, J.A. The human skin microbiome. Nat. Rev. Microbiol. 2018, 16, 143–155. [Google Scholar] [CrossRef] [PubMed]
- Wood, M.; Gibbons, S.M.; Lax, S.; Eshoo-Anton, T.W.; Owens, S.M.; Kennedy, S.; Gilbert, J.A.; Hampton-Marcell, J.T. Athletic equipment microbiota are shaped by interactions with human skin. Microbiome 2015, 3, 25–32. [Google Scholar] [CrossRef]
- Martykanova, D.S.; Davletova, N.C.; Zemlenuhin, I.A.; Mugallimov, S.M.; Ahatov, A.M.; Karamova, N.S.; Malanin, S.Y.; Laikov, A.V.; Markelova, M.I.; Siniagina, M.N.; et al. Characterization of Dysbiotic Changes of Skin Microbiota in Contact Sports Athletes. BioNanoSci 2017, 7, 1–3. [Google Scholar] [CrossRef]
- Nowicka, D.; Bagłaj-Oleszczuk, M.; Maj, J. Infectious diseases of the skin in contact sports. Adv. Clin. Exp. Med. 2020, 29, 1491–1495. [Google Scholar] [CrossRef]
- Martykanova, D.S.; Davletova, N.C.; Zemlenuhin, I.A.; Volchkova, V.I.; Mugallimov, S.M.; Ahatov, A.M.; Laikov, A.V.; Markelova, M.I.; Boulygina, E.A.; Lopukhov, L.V.; et al. Skin microbiota in contact sports athletes and selection of antiseptics for professional hygiene. Biomed. Res. Int. 2019, 2019, 9843781. [Google Scholar] [CrossRef] [PubMed]
- Futatsuya, T.; Ushigami, T.; Nomura, F.; Anzawa, K.; Mochizuki, T.; Cho, O.; Sugita, T. Scalp microbiota in members of a Japanese high school judo team including Trichophyton tonsurans carriers. J. Dermatol. 2020, 47, 1020–1026. [Google Scholar] [CrossRef]
- Dienst, W.L., Jr.; Dightman, L.; Dworkin, M.S.; Thompson, R.K.; Howe, W.B. Pinning down skin infections: Diagnosis treatment and prevention in wrestlers. Physician Sportsmed. 1997, 25, 45–56. [Google Scholar] [CrossRef] [PubMed]
- Adams, B.B. Tinea corporis gladiatorum. J. Am. Acad. Dermatol. 2002, 47, 286–290. [Google Scholar] [CrossRef]
- Na, H.S.; Song, Y.; Yu, Y.; Chung, J. Comparative analysis of primers used for 16S rRNA gene sequencing in oral microbiome studies. Methods Protoc. 2023, 6, 71. [Google Scholar] [CrossRef]
- Kameoka, S.; Motooka, D.; Watanabe, S.; Kubo, R.; Jung, N.; Midorikawa, Y.; Shinozaki, N.O.; Sawai, Y.; Takeda, A.K.; Nakamura, S. Benchmark of 16S rRNA gene amplicon sequencing using Japanese gut microbiome data from the V1–V2 and V3–V4 primer sets. BMC Genom. 2021, 22, 527–536. [Google Scholar] [CrossRef]
- Huang, Z.; Yu, K.; Xiao, Y.; Wang, Y.; Xiao, D.; Wang, D. Comparative Genomic Analysis Reveals Potential Pathogenicity and Slow-Growth Characteristics of Genus Brevundimonas and Description of Brevundimonas pishanensis sp. nov. Microbiol. Spectr. 2022, 10, e0246821. [Google Scholar] [CrossRef]
- Mangutov, E.O.; Kharseeva, G.G.; Alutina, E.L. Corynebacterium spp.—Problematic pathogens of the human respiratory tract (review of literature). Klin. Lab. Diagn. 2021, 66, 502–508. [Google Scholar] [CrossRef]
- Lasek, R.; Szuplewska, M.; Mitura, M.; Decewicz, P.; Chmielowska, C.; Pawłot, A.; Sentkowska, D.; Czarnecki, J.; Bartosik, D. Genome structure of the opportunistic pathogen Paracoccus yeei (Alphaproteobacteria) and identification of putative virulence factors. Front. Microbiol. 2018, 9, 2553. [Google Scholar] [CrossRef]
- Wu, Z.; Lyu, H.; Liang, W.; Jing, X.; Wang, Y.; Ma, X. Microbial community in indoor dusts from university dormitories: Characteristics, potential pathogens and influence factors. Atmos. Pollut. Res. 2021, 12, 321–333. [Google Scholar] [CrossRef]
- Ioannou, P.; Ziogou, A.; Giannakodimos, A.; Giannakodimos, I.; Tsantes, A.G.; Samonis, G. Psychrobacter Infections in Humans–A Narrative Review of Reported Cases. Antibiotics 2025, 14, 140–151. [Google Scholar] [CrossRef]
- Mayslich, C.; Grange, P.A.; Dupin, N. Cutibacterium acnes as an opportunistic pathogen: An update of its virulence-associated factors. Microorganisms 2021, 9, 303–323. [Google Scholar] [CrossRef]
- Ziogou, A.; Giannakodimos, I.; Giannakodimos, A.; Baliou, S.; Ioannou, P. Kocuria Species Infections in Humans–A Narrative Review. Microorganisms 2023, 11, 2362–2374. [Google Scholar] [CrossRef] [PubMed]
- Reynolds, D.; Kollef, M. The epidemiology and pathogenesis and treatment of Pseudomonas aeruginosa infections: An update. Drugs 2021, 81, 2117–2131. [Google Scholar] [CrossRef]
- Miles, S.L.; Holt, K.E.; Mostowy, S. Recent advances in modelling Shigella infection. Trends Microbiol. 2024, 32, 917–924. [Google Scholar] [CrossRef] [PubMed]
- Brouwer, S.; Rivera-Hernandez, T.; Curren, B.F.; Harbison-Price, N.; De Oliveira, D.M.P.; Jespersen, M.G.; Davies, M.R.; Walker, M.J. Pathogenesis, epidemiology and control of Group A Streptococcus infection. Nat. Rev. Microbiol. 2023, 21, 431–447. [Google Scholar] [CrossRef] [PubMed]
- Broadhead, R.; Craeye, L.; Callewaert, C. The future of functional clothing for an improved skin and textile microbiome relationship. Microorganisms 2021, 9, 1192–1199. [Google Scholar] [CrossRef]
- Kortekaas Krohn, I.; Callewaert, C.; Belasri, H.; De Pessemier, B.; Diez Lopez, C.; Mortz, C.G.; O’Mahony, L.; Perez-Gordo, M.; Sokolowska, M.; Unger, Z.; et al. The influence of lifestyle and environmental factors on host resilience through a homeostatic skin microbiota: An EAACI Task Force Report. Allergy 2024, 79, 3269–3284. [Google Scholar] [CrossRef]
- Ryan, M.P.; Pembroke, J.T. Brevundimonas spp.: Emerging global opportunistic pathogens. Virulence 2018, 9, 480–493. [Google Scholar] [CrossRef] [PubMed]
- Timm, C.M.; Loomis, K.; Stone, W.; Mehoke, T.; Brensinger, B.; Pellicore, M.; Staniczenko, P.P.; Charles, C.; Nayak, S.; Karig, D.K. Isolation and characterization of diverse microbial representatives from the human skin microbiome. Microbiome 2020, 8, 58–69. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.H.; Son, S.M.; Park, H.; Kim, B.K.; Choi, I.S.; Kim, H.; Huh, C.S. Taxonomic profiling of skin microbiome and correlation with clinical skin parameters in healthy Koreans. Sci. Rep. 2021, 11, 16269–16283. [Google Scholar] [CrossRef]
- Liang, Z.; Dong, C.B.; Liang, H.; Zhen, Y.X.; Zhou, R.L.; Han, Y.F.; Liang, Z.Q. A microbiome study reveals the potential relationship between the bacterial diversity of a gymnastics hall and human health. Sci. Rep. 2022, 12, 5663–5678. [Google Scholar] [CrossRef] [PubMed]
- Mennitti, C.; Calvanese, M.; Gentile, A.; Vastola, A.; Romano, P.; Ingenito, L.; Gentile, L.; Veneruso, I.; Scarano, C.; La Monica, I.; et al. Skin Microbiome Overview: How Physical Activity Influences Bacteria. Microorganisms 2025, 13, 868–883. [Google Scholar] [CrossRef]
- Ruggiero, M.; Ferrante, L.; Tafuri, D.; Meccariello, R.; Mazzeo, F. Trends in Antidepressant, Anxiolytic, and Cannabinoid Use Among Italian Elite Athletes (2011–2023): A Longitudinal Anti-Doping Analysis. Sports 2025, 13, 233–249. [Google Scholar] [CrossRef]
- Vazquez-Martinez, E.R.; García-Gómez, E.; Camacho-Arroyo, I.; González-Pedrajo, B. Sexual dimorphism in bacterial infections. Biol. Sex. Differ. 2018, 9, 27–46. [Google Scholar] [CrossRef]
- Puttur, F.; Lloyd, C.M. Sex differences in tissue immunity. Science 2024, 384, 159–160. [Google Scholar] [CrossRef]
- Chi, L.; Liu, C.; Gribonika, I.; Gschwend, J.; Corral, D.; Han, S.J.; Lim, A.I.; Rivera, C.A.; Link, V.M.; Wells, A.C.; et al. Sexual dimorphism in skin immunity is mediated by an androgen-ILC2-dendritic cell axis. Science 2024, 384, eadk6200. [Google Scholar] [CrossRef]
- Peterson, A.R.; Nash, E.; Anderson, B.J. Infectious disease in contact sports. Sports Health 2019, 11, 47–58. [Google Scholar] [CrossRef] [PubMed]
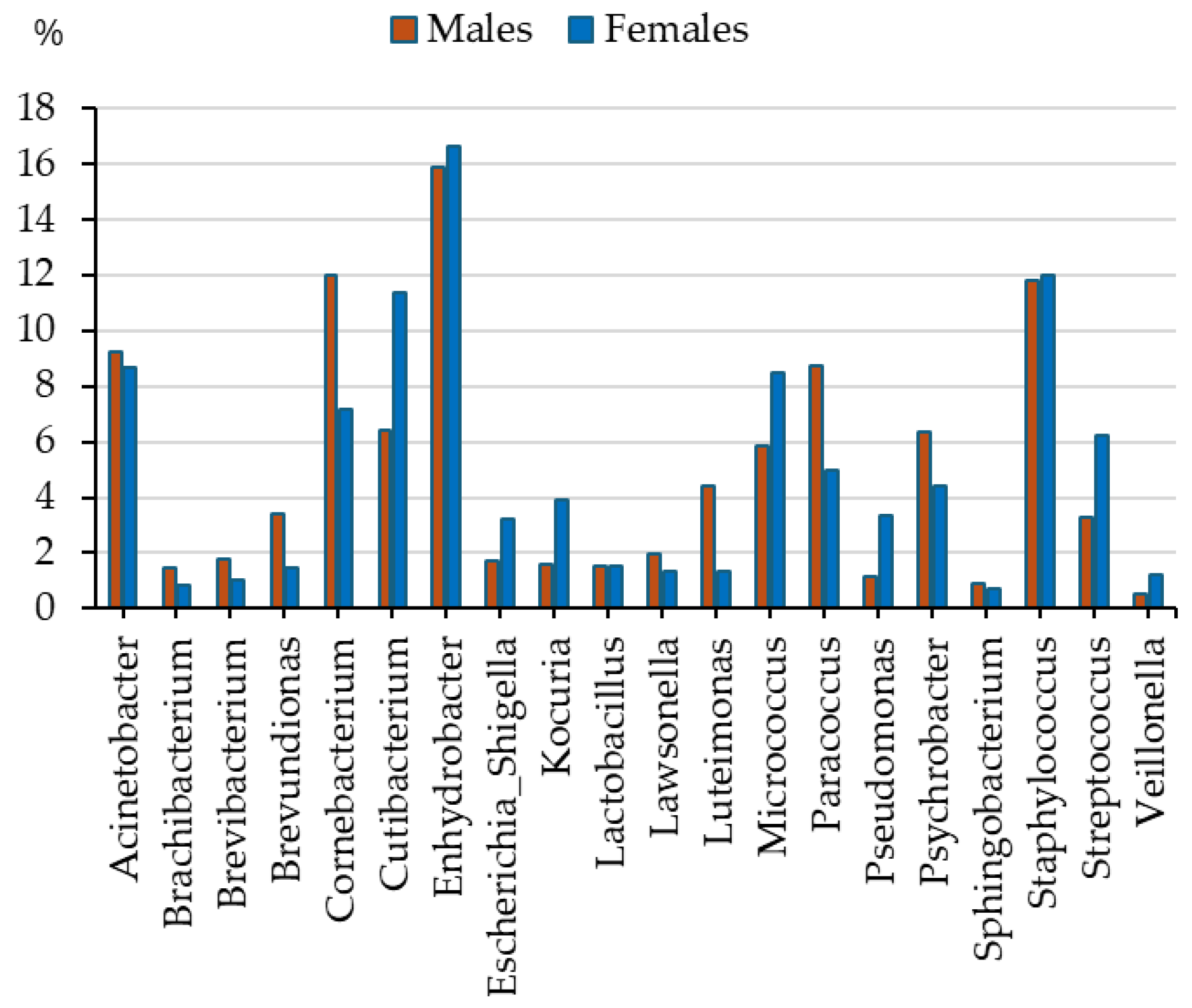
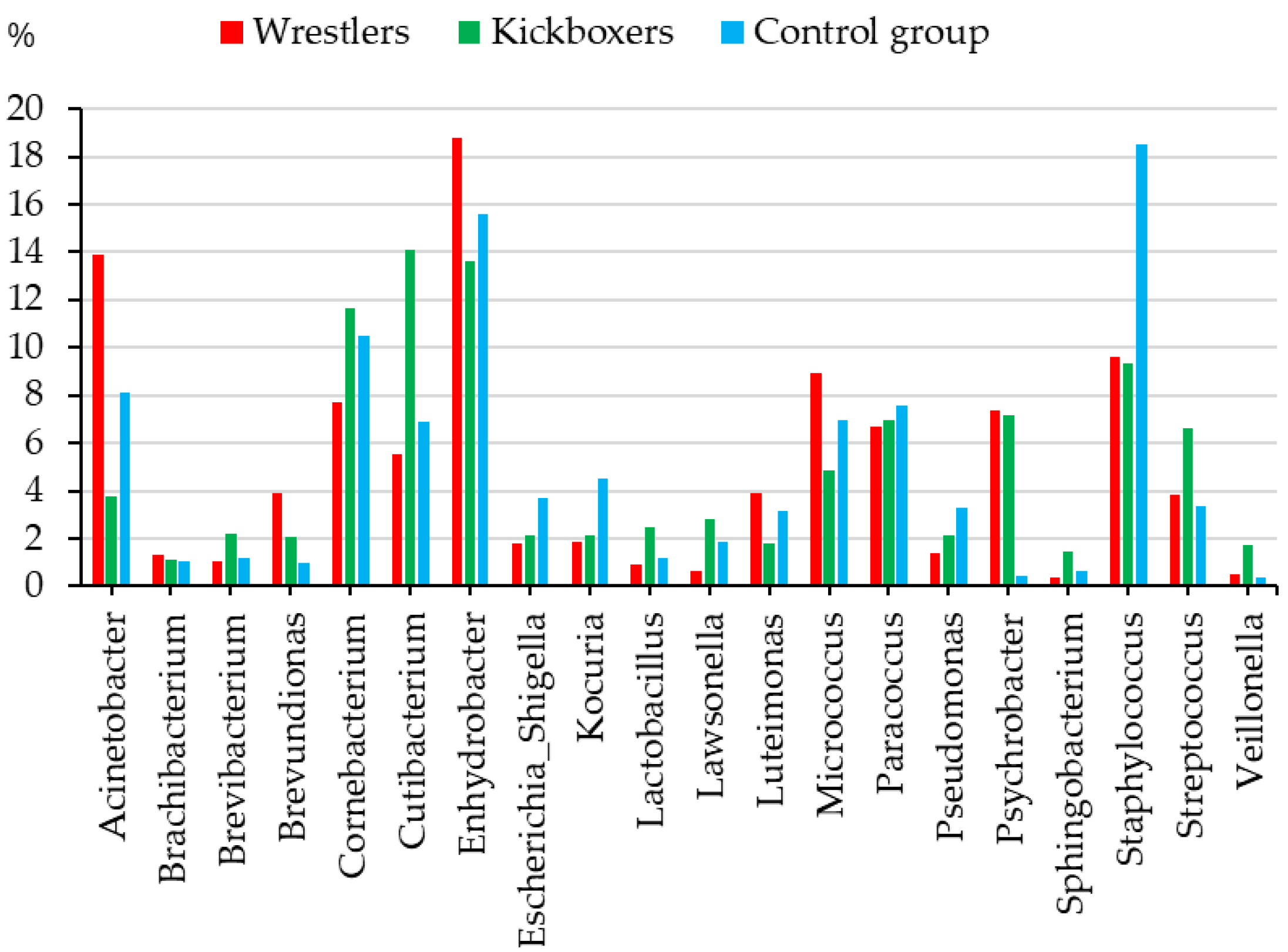
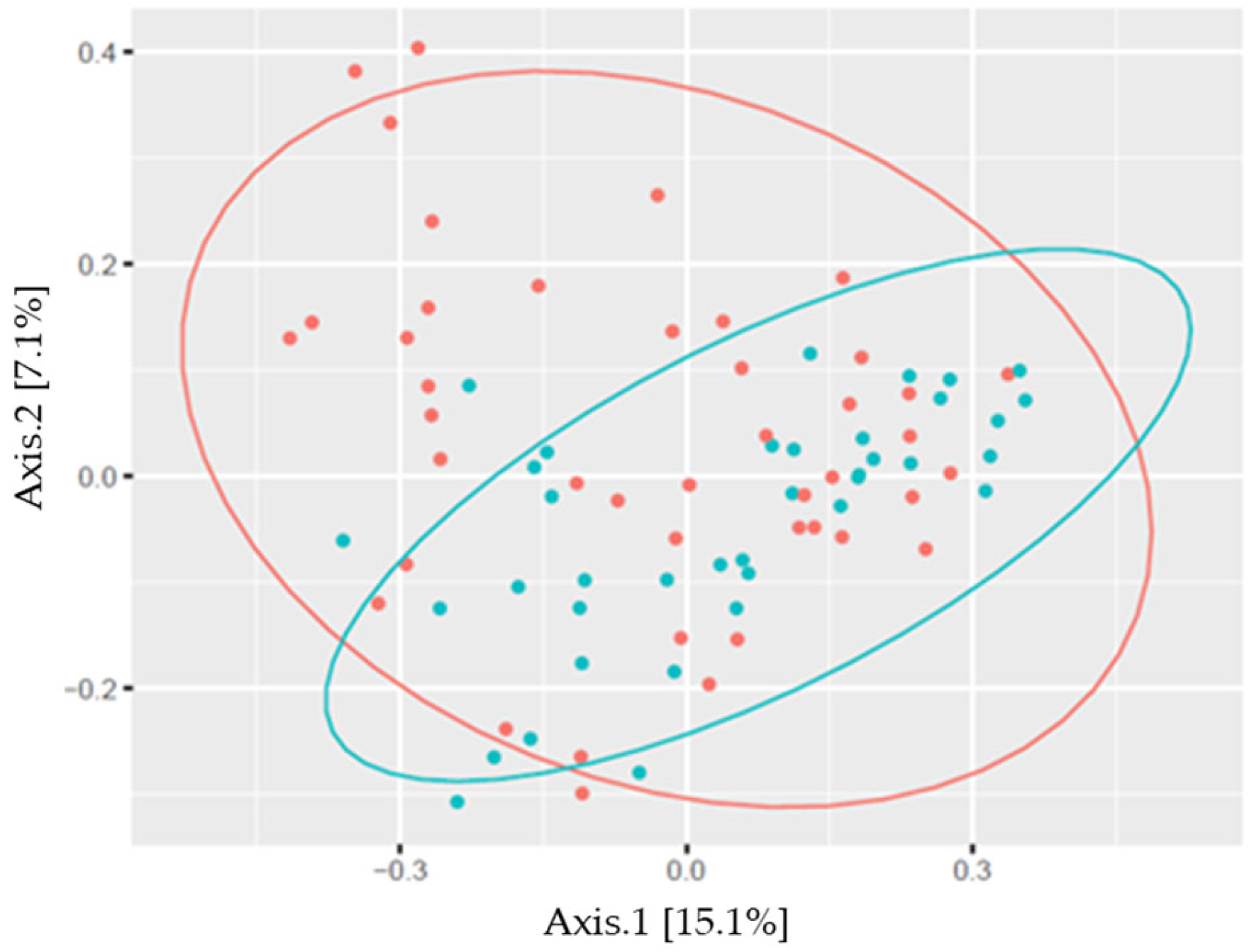
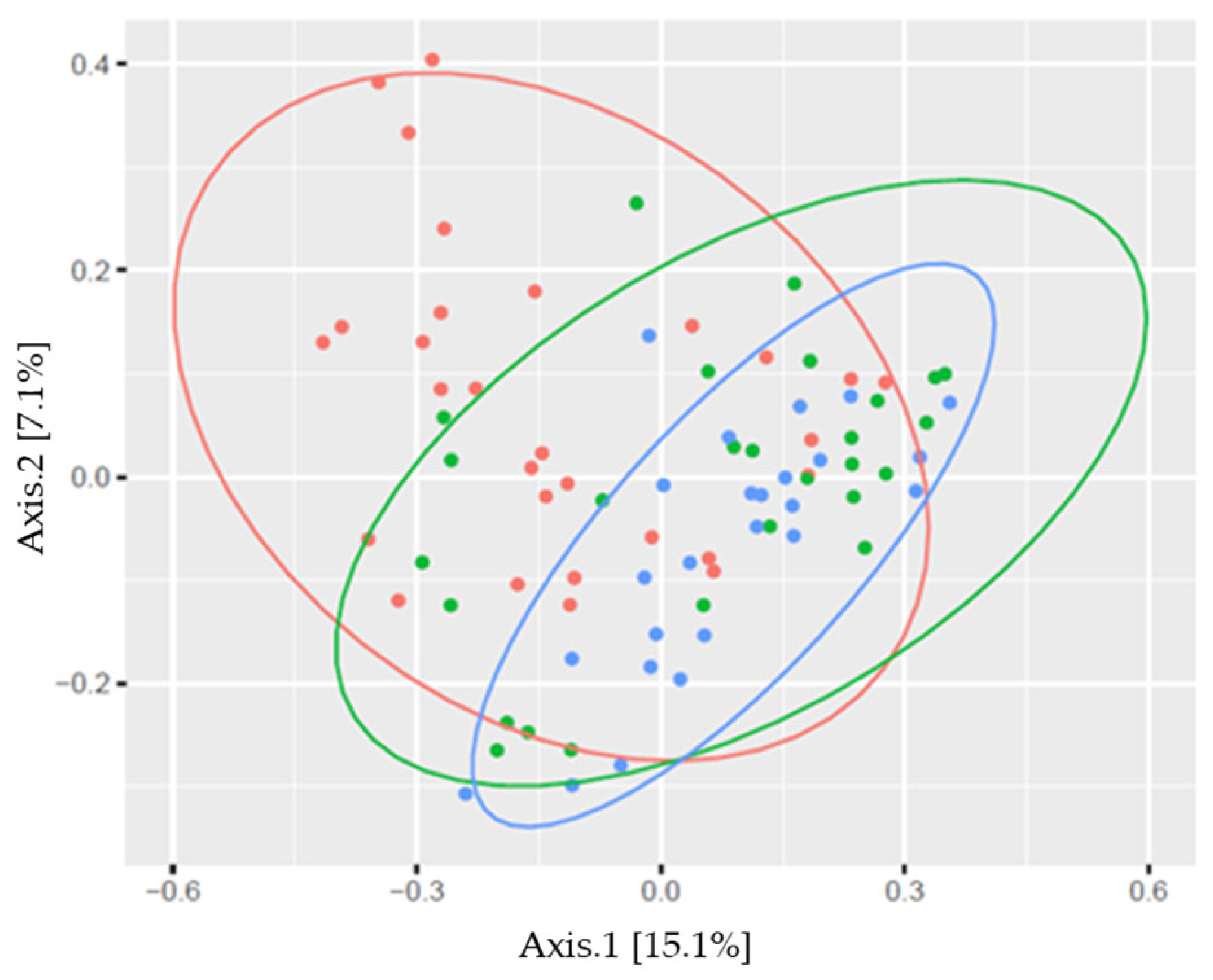
| Bacterial Genus | Males | Females | Athletes | Controls | Together |
|---|---|---|---|---|---|
| Enhydrobacter * | 15.9 (2.3) | 16.6 (3.9) | 16.3 (1.8) | 15.6 (1.2) | 16.2 (2.3) |
| Staphylococcus | 11.8 (3.6) | 12.0 (1.7) | 9.5 (1.2) | 18.5 (3.2) | 11.9 (2.0) |
| Corynebacterium | 12.0 (1.6) | 7.2 (1.0) | 9.6 (1.1) | 10.5 (1.9) | 9.7 (1.0) |
| Acinetobacter | 9.3 (2.5) | 8.7 (1.8) | 9.0 (1.7) | 8.1 (1.2) | 9.0 (1.5) |
| Cutibacterium | 6.4 (1.8) | 11.4 (2.8) | 9.7 (1.7) | 6.9 (1.8) | 8.8 (1.7) |
| Micrococcus | 5.8 (1.0) | 8.5 (0.9) | 7.0 (1.0) | 7.0 (1.6) | 7.1 (0.9) |
| Paracoccus | 8.8 (1.5) | 5.0 (0.9) | 6.8 (0.9) | 7.6 (1.2) | 7.0 (0.9) |
| Psychrobacter | 6.3 (1.3) | 4.4 (1.9) | 7.3 (1.5) | 0.4 (0.2) | 5.4 (1.3) |
| Streptococcus | 3.3 (1.2) | 6.2 (2.8) | 5.2 (1.9) | 3.4 (1.2) | 4.7 (1.6) |
| Luteimonas * | 4.4 (0.9) | 1.4 (0.7) | 2.9 (0.6) | 3.2 (1.9) | 3.0 (0.6) |
| Kocuria | 1.6 (0.5) | 3.9 (1.1) | 2.0 (0.4) | 4.5 (1.6) | 2.7 (0.5) |
| Brevundimonas | 3.4 (0.9) | 1.5 (0.3) | 3.0 (0.6) | 1.0 (0.8) | 2.5 (0.5) |
| Escherichia-Shigella | 1.7 (0.5) | 3.2 (0.7) | 1.9 (0.4) | 3.7 (1.1) | 2.4 (0.4) |
| Pseudomonas | 1.2 (0.3) | 3.4 (0.4) | 1.8 (0.2) | 3.3 (0.8) | 2.2 (0.2) |
| Lawsonella | 1.9 (0.5) | 1.4 (0.4) | 1.7 (0.4) | 1.8 (0.8) | 1.7 (0.3) |
| Lactobacillus | 1.5 (0.8) | 1.5 (0.4) | 1.7 (1.1) | 1.2 (0.3) | 1.5 (0.4) |
| Brevibacterium | 1.8 (0.9) | 1.1 (0.5) | 1.6 (0.6) | 1.2 (0.4) | 1.5 (0.6) |
| Brachybacterium * | 1.5 (0.2) | 0.8 (0.3) | 1.2 (0.2) | 1.1 (0.3) | 1.2 (0.2) |
| M-F | Effect Size | W-KB-C | Effect Size | |
|---|---|---|---|---|
| Acinetobacter | 0.384 | 0.066 (small) | 0.002 w | 0.145 (medium) |
| Brachybacterium | 0.047 m | 0.378 (medium) | 0.559 | 0.015 (small) |
| Brevibacterium | 0.126 | 0.257 (medium) | 0.356 | 0.026 (small) |
| Brevundimonas | 0.015 m | 0.493 (large) | 0.009 w | 0.114 (medium) |
| Corynebacterium | 0.009 m | 0.538 (large) | 0.492 | 0.018 (small) |
| Cutibacterium | 0.063 | 0.346 (medium) | 0.040 c | 0.080 (small) |
| Enhydrobacter | 0.487 | 0.008 (small) | 0.218 | 0.038 (small) |
| Escherichia-Shigella | 0.008 f | 0.551 (large) | 0.172 | 0.044 (small) |
| Kocuria | 0.020 f | 0.466 (large) | 0.318 | 0.029 (small) |
| Lactobacillus | 0.495 | 0.003 (small) | 0.608 | 0.013 (small) |
| Lawsonella | 0.114 | 0.270 (medium) | 0.005 kb | 0.127 (medium) |
| Luteimonas | 0.011 m | 0.522 (large) | 0.290 | 0.031 (small) |
| Micrococcus | 0.050 f | 0.359 (medium) | 0.008 w | 0.115 (medium) |
| Paracoccus | 0.006 m | 0.580 (large) | 0.900 | 0.003 (small) |
| Pseudomonas | 0.004 f | 0.597(large) | 0.461 | 0.020 (small) |
| Psychrobacter | 0.206 | 0.184 (medium) | 0.028 w | 0.088 (small) |
| Sphingobacterium | 0.377 | 0.070 (small) | 0.422 | 0.022 (small) |
| Staphylococcus | 0.468 | 0.018 (small) | 0.178 | 0.043 (small) |
| Streptococcus | 0.032 f | 0.313 (large) | 0.325 | 0.028 (small) |
| Veillonella | 0.194 | 0.193 (medium) | 0.299 | 0.031 (small) |
| Df | Sum sqs | F | R2 | p | |
|---|---|---|---|---|---|
| Sex | 1 | 0.594 | 2.054 | 0.025 | 0.005 |
| Sports | |||||
| W-KB | 1 | 1.047 | 4.000 | 0.069 | <0.001 |
| W-C | 1 | 1.162 | 4.427 | 0.078 | <0.001 |
| KB-C | 1 | 0.400 | 1.287 | 0.025 | 0.099 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kalabiska, I.; Annar, D.; Babszky, G.; Jokai, M.; Borbas, Z.; Hajdu, G.; Ratz-Sulyok, F.Z.; Jang-Kapuy, C.; Palinkas, G.; Bhattoa, H.P.; et al. The Skin Microbiome Profile of Contact Sports Athletes—Focus on Sexual Dimorphism and Athlete–Non-Athlete Differences. Sports 2025, 13, 288. https://doi.org/10.3390/sports13090288
Kalabiska I, Annar D, Babszky G, Jokai M, Borbas Z, Hajdu G, Ratz-Sulyok FZ, Jang-Kapuy C, Palinkas G, Bhattoa HP, et al. The Skin Microbiome Profile of Contact Sports Athletes—Focus on Sexual Dimorphism and Athlete–Non-Athlete Differences. Sports. 2025; 13(9):288. https://doi.org/10.3390/sports13090288
Chicago/Turabian StyleKalabiska, Irina, Dorina Annar, Gergely Babszky, Matyas Jokai, Zoltan Borbas, Gergely Hajdu, Fanny Zselyke Ratz-Sulyok, Csilla Jang-Kapuy, Gergely Palinkas, Harjit Pal Bhattoa, and et al. 2025. "The Skin Microbiome Profile of Contact Sports Athletes—Focus on Sexual Dimorphism and Athlete–Non-Athlete Differences" Sports 13, no. 9: 288. https://doi.org/10.3390/sports13090288
APA StyleKalabiska, I., Annar, D., Babszky, G., Jokai, M., Borbas, Z., Hajdu, G., Ratz-Sulyok, F. Z., Jang-Kapuy, C., Palinkas, G., Bhattoa, H. P., & Zsakai, A. (2025). The Skin Microbiome Profile of Contact Sports Athletes—Focus on Sexual Dimorphism and Athlete–Non-Athlete Differences. Sports, 13(9), 288. https://doi.org/10.3390/sports13090288







