Comparative Mapping of N6-Methyladenine, C5-Methylcytosine, and C5-Hydroxymethylcytosine in a Single Species Reveals Constitutive, Somatic- and Germline-Specific, and Age-Related Genomic Context Distributions and Biological Functions
Abstract
1. Introduction
2. Results
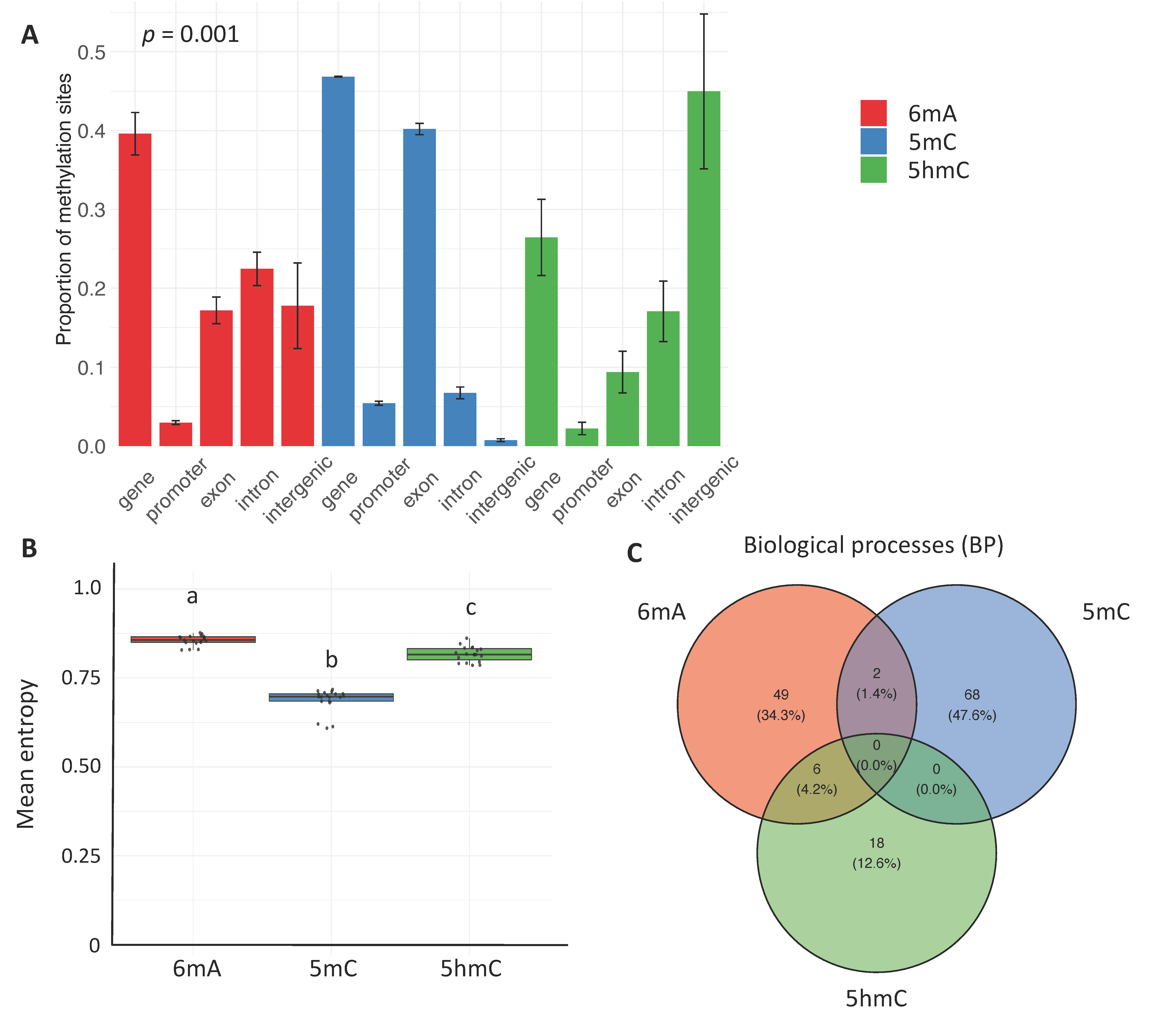
2.1. Constitutive Patterns
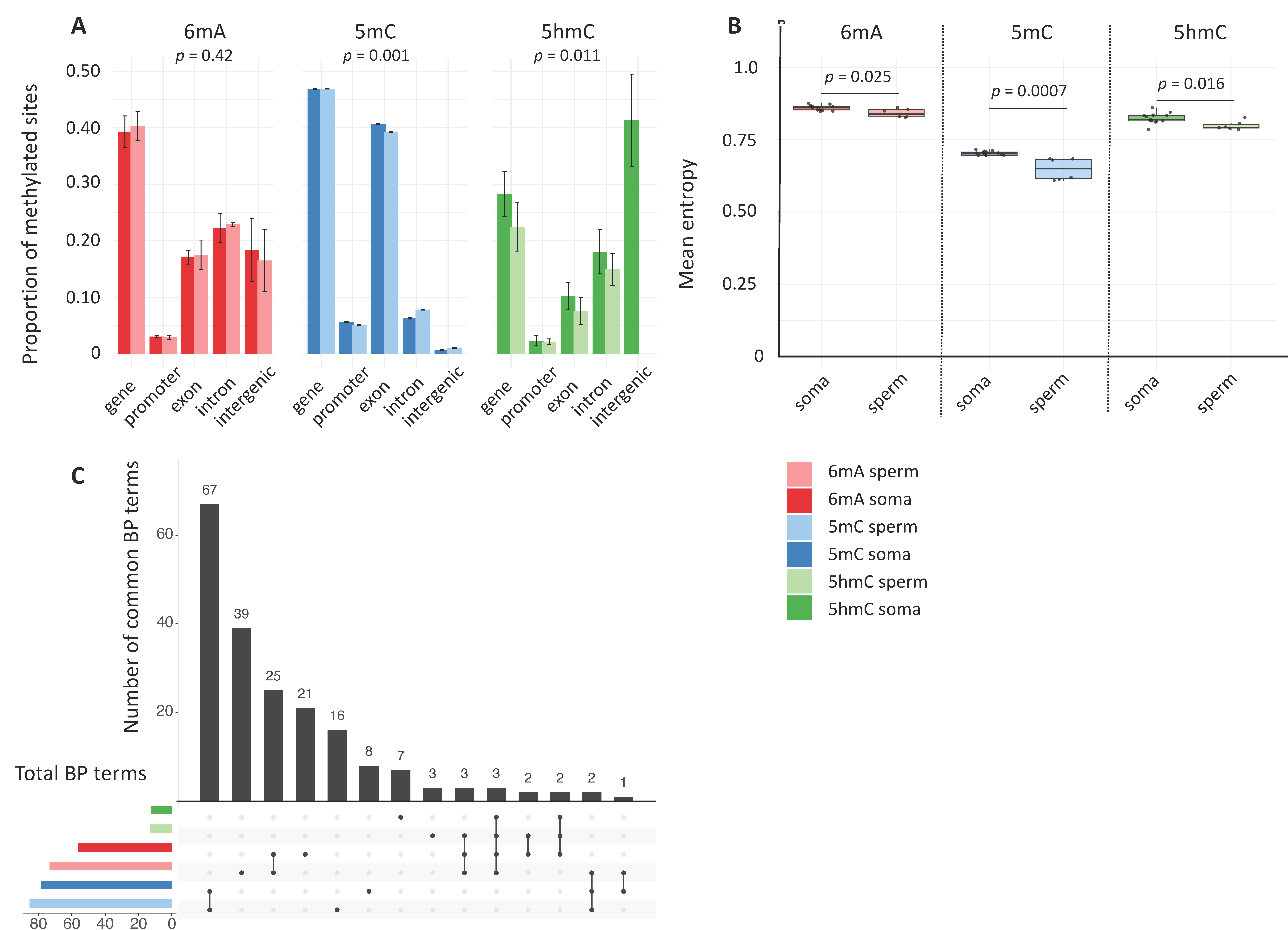
2.2. Somatic- vs. Germline-Specific Signatures
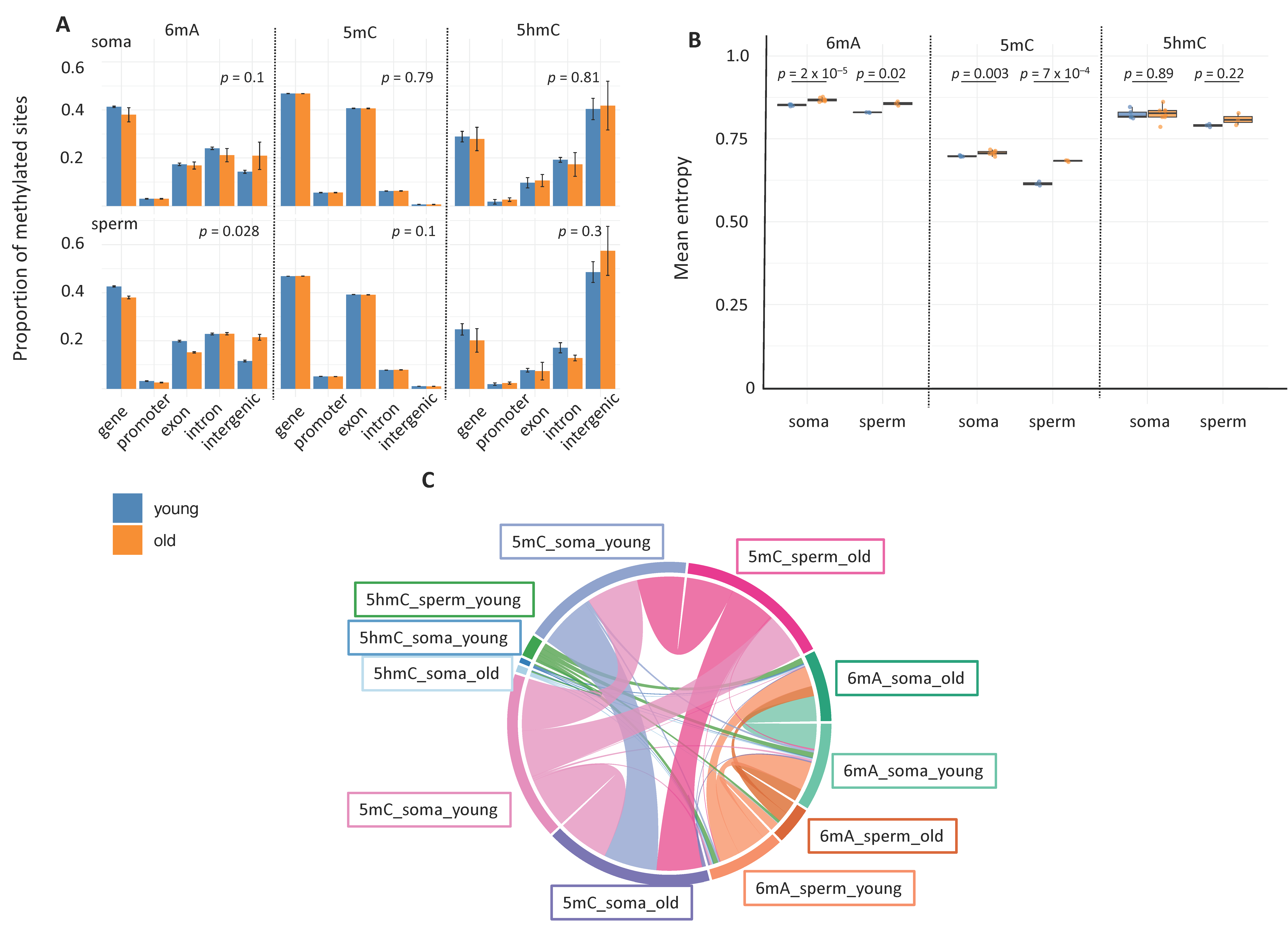
2.3. Age-Related Changes
3. Discussion
4. Materials and Methods
4.1. Model Organism
4.2. Sampling
4.3. DNA Extraction and Library Preparation
4.4. DNA Methylation Analyses
4.5. Statistical Analyses
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| 5mC | C5-methylcytosine |
| 6mA | N6-methyadenine |
| 5hmC | C5-hydroxymethylcytosine |
| GO | Gene ontology |
References
- Zemach, A.; McDaniel, I.E.; Silva, P.; Zilberman, D. Genome-Wide Evolutionary Analysis of Eukaryotic DNA Methylation. Science 2010, 328, 916–919. [Google Scholar] [CrossRef]
- Rodriguez, F.; Yushenova, I.A.; DiCorpo, D.; Arkhipova, I.R. Bacterial N4-Methylcytosine as an Epigenetic Mark in Eukaryotic DNA. Nat. Commun. 2022, 13, 1072. [Google Scholar] [CrossRef]
- Sánchez-Romero, M.A.; Casadesús, J. The Bacterial Epigenome. Nat. Rev. Microbiol. 2020, 18, 7–20. [Google Scholar] [CrossRef] [PubMed]
- Tsagaratou, A.; Äijö, T.; Lio, C.-W.J.; Yue, X.; Huang, Y.; Jacobsen, S.E.; Lähdesmäki, H.; Rao, A. Dissecting the Dynamic Changes of 5-Hydroxymethylcytosine in T-Cell Development and Differentiation. Proc. Natl. Acad. Sci. USA 2014, 111, E3306–E3315. [Google Scholar] [CrossRef] [PubMed]
- Liang, D.; Yan, R.; Long, X.; Ji, D.; Song, B.; Wang, M.; Zhang, F.; Cheng, X.; Sun, F.; Zhu, R.; et al. Distinct Dynamics of Parental 5-Hydroxymethylcytosine during Human Preimplantation Development Regulate Early Lineage gene Expression. Nat. Cell Biol. 2024, 26, 1458–1469. [Google Scholar] [CrossRef] [PubMed]
- López, V.; Fernández, A.F.; Fraga, M.F. The Role of 5-Hydroxymethylcytosine in Development, Aging and Age-Related Diseases. Ageing Res. Rev. 2017, 37, 28–38. [Google Scholar] [CrossRef]
- Boulias, K.; Greer, E.L. Means, Mechanisms and Consequences of Adenine Methylation in DNA. Nat. Rev. Genet. 2022, 23, 411–428. [Google Scholar] [CrossRef]
- Sternglanz, H.; Bugg, C.E. Conformation of N6-Methyladenine, a Base Involved in DNA Modification: Restriction Processes. Science 1973, 182, 833–834. [Google Scholar] [CrossRef]
- Douvlataniotis, K.; Bensberg, M.; Lentini, A.; Gylemo, B.; Nestor, C.E. No Evidence for DNA N6-Methyladenine in Mammals. Sci. Adv. 2020, 6, eaay3335. [Google Scholar] [CrossRef]
- Greer, E.L.; Blanco, M.A.; Gu, L.; Sendinc, E.; Liu, J.; Aristizábal-Corrales, D.; Hsu, C.-H.; Aravind, L.; He, C.; Shi, Y. DNA Methylation on N6-Adenine in C. Elegans. Cell 2015, 161, 868–878. [Google Scholar] [CrossRef]
- Fu, Y.; Luo, G.-Z.; Chen, K.; Deng, X.; Yu, M.; Han, D.; Hao, Z.; Liu, J.; Lu, X.; Doré, L.C.; et al. N6-Methyldeoxyadenosine Marks Active Transcription Start Sites in Chlamydomonas. Cell 2015, 161, 879–892. [Google Scholar] [CrossRef]
- Zhang, G.; Huang, H.; Liu, D.; Cheng, Y.; Liu, X.; Zhang, W.; Yin, R.; Zhang, D.; Zhang, P.; Liu, J.; et al. N6-Methyladenine DNA Modification in Drosophila. Cell 2015, 161, 893–906. [Google Scholar] [CrossRef]
- Koziol, M.J.; Bradshaw, C.R.; Allen, G.E.; Costa, A.S.H.; Frezza, C.; Gurdon, J.B. Identification of Methylated Deoxyadenosines in Vertebrates Reveals Diversity in DNA Modifications. Nat. Struct. Mol. Biol. 2016, 23, 24–30. [Google Scholar] [CrossRef]
- Wu, T.P.; Wang, T.; Seetin, M.G.; Lai, Y.; Zhu, S.; Lin, K.; Liu, Y.; Byrum, S.D.; Mackintosh, S.G.; Zhong, M.; et al. DNA Methylation on N6-Adenine in Mammalian Embryonic Stem Cells. Nature 2016, 532, 329–333. [Google Scholar] [CrossRef]
- Liu, J.; Zhu, Y.; Luo, G.-Z.; Wang, X.; Yue, Y.; Wang, X.; Zong, X.; Chen, K.; Yin, H.; Fu, Y.; et al. Abundant DNA 6mA Methylation during Early Embryogenesis of Zebrafish and Pig. Nat. Commun. 2016, 7, 13052. [Google Scholar] [CrossRef] [PubMed]
- Liang, Z.; Shen, L.; Cui, X.; Bao, S.; Geng, Y.; Yu, G.; Liang, F.; Xie, S.; Lu, T.; Gu, X.; et al. DNA N6-Adenine Methylation in Arabidopsis Thaliana. Dev. Cell 2018, 45, 406–416.e3. [Google Scholar] [CrossRef]
- Mondo, S.J.; Dannebaum, R.O.; Kuo, R.C.; Louie, K.B.; Bewick, A.J.; LaButti, K.; Haridas, S.; Kuo, A.; Salamov, A.; Ahrendt, S.R.; et al. Widespread adenine N6-Methylation of Active Genes in Fungi. Nat. Genet. 2017, 49, 964–968. [Google Scholar] [CrossRef] [PubMed]
- Fernandes, S.B.; Grova, N.; Roth, S.; Duca, R.C.; Godderis, L.; Guebels, P.; Mériaux, S.B.; Lumley, A.I.; Bouillaud-Kremarik, P.; Ernens, I.; et al. N6-Methyladenine in Eukaryotic DNA: Tissue Distribution, Early Embryo Development, and Neuronal Toxicity. Front. Genet. 2021, 12, 657171. [Google Scholar] [CrossRef]
- Ma, C.; Niu, R.; Huang, T.; Shao, L.-W.; Peng, Y.; Ding, W.; Wang, Y.; Jia, G.; He, C.; Li, C.-Y.; et al. N6-Methyldeoxyadenine Is a Transgenerational Epigenetic Signal for Mitochondrial Stress Adaptation. Nat. Cell Biol. 2019, 21, 319–327. [Google Scholar] [CrossRef] [PubMed]
- Kigar, S.L.; Chang, L.; Guerrero, C.R.; Sehring, J.R.; Cuarenta, A.; Parker, L.L.; Bakshi, V.P.; Auger, A.P. N6-Methyladenine Is an Epigenetic Marker of Mammalian Early Life Stress. Sci. Rep. 2017, 7, 18078. [Google Scholar] [CrossRef]
- Yao, B.; Cheng, Y.; Wang, Z.; Li, Y.; Chen, L.; Huang, L.; Zhang, W.; Chen, D.; Wu, H.; Tang, B.; et al. DNA N6-Methyladenine is Dynamically Regulated in the Mouse Brain Following Environmental Stress. Nat. Commun. 2017, 8, 1122. [Google Scholar] [CrossRef]
- Renard, T.; Boseret, M.; Aron, S. Epigenetic Age Prediction Using N6-Methyladenine in the Bumblebee Bombus Terrestris. bioRxiv 2025, 2025.06.20.660749. [Google Scholar] [CrossRef]
- Zhang, L.; Rong, W.; Ma, J.; Li, H.; Tang, X.; Xu, S.; Wang, L.; Wan, L.; Zhu, Q.; Jiang, B.; et al. Comprehensive Analysis of DNA 5-Methylcytosine and N6-Adenine Methylation by Nanopore Sequencing in Hepatocellular Carcinoma. Front. Cell Dev. Biol. 2022, 10, 827391. [Google Scholar] [CrossRef]
- Wu, C.; Yao, S.; Li, X.; Chen, C.; Hu, X. Genome-Wide Prediction of DNA Methylation Using DNA Composition and Sequence Complexity in Human. Int. J. Mol. Sci. 2017, 18, 420. [Google Scholar] [CrossRef]
- Scala, G.; Federico, A.; Palumbo, D.; Cocozza, S.; Greco, D. DNA Sequence Context as a Marker of CpG Methylation Instability in Normal and Cancer Tissues. Sci. Rep. 2020, 10, 1721. [Google Scholar] [CrossRef]
- Zhang, X.; Gan, Y.; Zou, G.; Guan, J.; Zhou, S. Genome-Wide Analysis of Epigenetic Dynamics across Human Developmental Stages and Tissues. BMC Genom. 2019, 20, 221. [Google Scholar] [CrossRef] [PubMed]
- Kerepesi, C.; Zhang, B.; Lee, S.-G.; Trapp, A.; Gladyshev, V.N. Epigenetic Clocks Reveal a Rejuvenation Event during Embryogenesis Followed by Aging. Sci. Adv. 2021, 7, eabg6082. [Google Scholar] [CrossRef] [PubMed]
- Atlasi, Y.; Stunnenberg, H.G. The Interplay of Epigenetic Marks during Stem Cell Differentiation and Development. Nat. Rev. Genet. 2017, 18, 643–658. [Google Scholar] [CrossRef]
- Seale, K.; Horvath, S.; Teschendorff, A.; Eynon, N.; Voisin, S. Making Sense of the Ageing Methylome. Nat. Rev. Genet. 2022, 23, 585–605. [Google Scholar] [CrossRef]
- Horvath, S. DNA Methylation Age of Human Tissues and Cell Types. Genome Biol. 2013, 14, 3156. [Google Scholar] [CrossRef]
- O’Brown, Z.K.; Greer, E.L. N6-Methyladenine: A Conserved and Dynamic DNA Mark. In DNA Methyltransferases—Role and Function; Jeltsch, A., Jurkowska, R.Z., Eds.; Springer International Publishing: Cham, Switzerland, 2016; pp. 213–246. ISBN 978-3-319-43624-1. [Google Scholar]
- O’Brown, Z.K.; Boulias, K.; Wang, J.; Wang, S.Y.; O’Brown, N.M.; Hao, Z.; Shibuya, H.; Fady, P.-E.; Shi, Y.; He, C.; et al. Sources of Artifact in Measurements of 6mA and 4mC abundance in Eukaryotic Genomic DNA. BMC Genom. 2019, 20, 445. [Google Scholar] [CrossRef] [PubMed]
- Hannum, G.; Guinney, J.; Zhao, L.; Zhang, L.; Hughes, G.; Sadda, S.; Klotzle, B.; Bibikova, M.; Fan, J.-B.; Gao, Y.; et al. Genome-wide Methylation Profiles Reveal Quantitative Views of Human Aging Rates. Mol. Cell 2013, 49, 359–367. [Google Scholar] [CrossRef]
- Chan, J.; Rubbi, L.; Pellegrini, M. DNA Methylation Entropy Is a Biomarker for Aging. Aging 2025, 17, 685–698. [Google Scholar] [CrossRef]
- Wang, X.; Li, Z.; Zhang, Q.; Li, B.; Lu, C.; Li, W.; Cheng, T.; Xia, Q.; Zhao, P. DNA Methylation on N6-Adenine in Lepidopteran Bombyx mori. Biochim. Biophys. Acta BBA-Gene Regul. Mech. 2018, 1861, 815–825. [Google Scholar] [CrossRef]
- Zhang, Y.; Zhang, Q.; Yang, X.; Gu, X.; Chen, J.; Shi, T. 6mA DNA Methylation on Genes in Plants Is Associated with Gene Complexity, Expression and Duplication. Plants 2023, 12, 1949. [Google Scholar] [CrossRef] [PubMed]
- Lax, C.; Mondo, S.J.; Martínez, J.F.; Muszewska, A.; Baumgart, L.A.; Pérez-Ruiz, J.A.; Carrillo-Marín, P.; LaButti, K.; Lipzen, A.; Zhang, Y.; et al. Symmetric Adenine Methylation Is an Essential DNA Modification in the Early-Diverging Fungus Rhizopus Microsporus. Nat. Commun. 2025, 16, 3843. [Google Scholar] [CrossRef]
- Renard, T.; Martinet, B.; De Souza Araujo, N.; Aron, S. DNA Methylation Extends Lifespan in the Bumblebee Bombus Terrestris. Proc. R. Soc. B Biol. Sci. 2023, 290, 20232093. [Google Scholar] [CrossRef] [PubMed]
- He, B.; Zhang, C.; Zhang, X.; Fan, Y.; Zeng, H.; Liu, J.; Meng, H.; Bai, D.; Peng, J.; Zhang, Q.; et al. Tissue-Specific 5-Hydroxymethylcytosine Landscape of the Human Genome. Nat. Commun. 2021, 12, 4249. [Google Scholar] [CrossRef]
- Cui, X.-L.; Nie, J.; Ku, J.; Dougherty, U.; West-Szymanski, D.C.; Collin, F.; Ellison, C.K.; Sieh, L.; Ning, Y.; Deng, Z.; et al. A Human Tissue Map of 5-Hydroxymethylcytosines Exhibits Tissue Specificity through Gene and Enhancer Modulation. Nat. Commun. 2020, 11, 6161. [Google Scholar] [CrossRef]
- Uribe-Lewis, S.; Carroll, T.; Menon, S.; Nicholson, A.; Manasterski, P.J.; Winton, D.J.; Buczacki, S.J.A.; Murrell, A. 5-Hydroxymethylcytosine and Gene Activity in Mouse Intestinal Differentiation. Sci. Rep. 2020, 10, 546. [Google Scholar] [CrossRef]
- Shah, K.; Cao, W.; Ellison, C.E. Adenine Methylation in Drosophila Is Associated with the Tissue-Specific Expression of Developmental and Regulatory Genes. G3 GenesGenomesGenetics 2019, 9, 1893–1900. [Google Scholar] [CrossRef]
- Barau, J.; Teissandier, A.; Zamudio, N.; Roy, S.; Nalesso, V.; Hérault, Y.; Guillou, F.; Bourc’his, D. The DNA Methyltransferase DNMT3C Protects Male Germ Cells from Transposon Activity. Science 2016, 354, 909–912. [Google Scholar] [CrossRef] [PubMed]
- Leismann, J.; Kanta, S.-E.; Amar, I.; Szczepińska, A.; Mielnicka, M.; Petrosino, G.; Busch, A.; Scheibe, M.; Wang, P.; Wang, Y.; et al. DNA Methylation at Retrotransposons Protects the Germline by Preventing NRF1-Mediated Activation. EMBO Rep. 2025, 26, 4312–4339. [Google Scholar] [CrossRef]
- Deniz, Ö.; Frost, J.M.; Branco, M.R. Regulation of Transposable Elements by DNA Modifications. Nat. Rev. Genet. 2019, 20, 417–431. [Google Scholar] [CrossRef]
- Bourc’his, D.; Bestor, T.H. Meiotic Catastrophe and Retrotransposon Reactivation in Male Germ Cells Lacking Dnmt3L. Nature 2004, 431, 96–99. [Google Scholar] [CrossRef] [PubMed]
- Lismer, A.; Kimmins, S. Emerging Evidence That the Mammalian Sperm Epigenome Serves as a Template for Embryo Development. Nat. Commun. 2023, 14, 2142. [Google Scholar] [CrossRef] [PubMed]
- Greeson, K.W.; Crow, K.M.S.; Edenfield, R.C.; Easley, C.A. Inheritance of Paternal Lifestyles and Exposures through Sperm DNA Methylation. Nat. Rev. Urol. 2023, 20, 356–370. [Google Scholar] [CrossRef]
- Wang, X.-X.; Sun, B.-F.; Jiao, J.; Chong, Z.-C.; Chen, Y.-S.; Wang, X.-L.; Zhao, Y.; Zhou, Y.-M.; Li, D. Genome-Wide 5-Hydroxymethylcytosine Modification Pattern is a Novel Epigenetic Feature of Globozoospermia. Oncotarget 2015, 6, 6535–6543. [Google Scholar] [CrossRef]
- Guo, W.; Li, X.; Qin, K.; Zhang, P.; He, J.; Liu, Y.; Yang, X.; Wu, S. Nanopore Sequencing Demonstrates the Roles of Spermatozoal DNA N6-Methyladenine in Mediating Transgenerational Lipid Metabolism Disorder Induced by Excessive Folate Consumpton. Poult. Sci. 2024, 103, 103953. [Google Scholar] [CrossRef]
- Bewick, A.J.; Vogel, K.J.; Moore, A.J.; Schmitz, R.J. Evolution of DNA Methylation across Insects. Mol. Biol. Evol. 2017, 34, 654–665. [Google Scholar] [CrossRef]
- Boulet, M.; Gilbert, G.; Renaud, Y.; Schmidt-Dengler, M.; Plantié, E.; Bertrand, R.; Nan, X.; Jurkowski, T.; Helm, M.; Vandel, L.; et al. Adenine Methylation Is Very Scarce in the Drosophila Genome and Not Erased by the Ten-Eleven Translocation Dioxygenase. eLife 2023, 12, RP91655. [Google Scholar] [CrossRef] [PubMed]
- Paynter, E.; Baer-Imhoof, B.; Linden, M.; Lee-Pullen, T.; Heel, K.; Rigby, P.; Baer, B. Flow Cytometry as a Rapid and Reliable Method to Quantify Sperm Viability in the Honeybee Apis mellifera. Cytom. Part A 2014, 85, 463–472. [Google Scholar] [CrossRef] [PubMed]
- Heidelbach, S.; Dall, S.M.; Bøjer, J.S.; Nissen, J.; van der Maas, L.N.L.; Sereika, M.; Kirkegaard, R.H.; Jensen, S.I.; Kousgaard, S.J.; Thorlacius-Ussing, O.; et al. Nanomotif: Leveraging DNA Methylation Motifs for Genome Recovery and Host Association of Plasmids in Metagenomes from Complex Microbial Communities. bioRxiv 2025, 2024.04.29.591623. [Google Scholar] [CrossRef]
- Quinlan, A.R.; Hall, I.M. BEDTools: A Flexible Suite of Utilities for Comparing Genomic Features. Bioinformatics 2010, 26, 841–842. [Google Scholar] [CrossRef]
- Walsh, A.T.; Triant, D.A.; Le Tourneau, J.J.; Shamimuzzaman, M.; Elsik, C.G. Hymenoptera Genome Database: New Genomes and Annotation Datasets for Improved Go Enrichment and Orthologue Analyses. Nucleic Acids Res. 2022, 50, D1032–D1039. [Google Scholar] [CrossRef]
- Alexa, A.; Rahnenfuhrer, J. Gene Set Enrichment Analysis with topGO. 2016. Available online: https://bioconductor.statistik.tu-dortmund.de/packages/3.3/bioc/vignettes/topGO/inst/doc/topGO.pdf (accessed on 15 September 2025).
- Oksanen, J.; Simpson, G.L.; Blanchet, F.G.; Kindt, R.; Legendre, P.; Minchin, P.R.; O’Hara, R.B.; Solymos, P.; Stevens, M.H.H.; Szoecs, E.; et al. vegan: Community Ecology Package 2001, 2.7-1. Available online: https://mirror.ibcp.fr/pub/CRAN/web/packages/vegan/vegan.pdf (accessed on 15 September 2025).
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Renard, T.; Aron, S. Comparative Mapping of N6-Methyladenine, C5-Methylcytosine, and C5-Hydroxymethylcytosine in a Single Species Reveals Constitutive, Somatic- and Germline-Specific, and Age-Related Genomic Context Distributions and Biological Functions. Epigenomes 2025, 9, 35. https://doi.org/10.3390/epigenomes9030035
Renard T, Aron S. Comparative Mapping of N6-Methyladenine, C5-Methylcytosine, and C5-Hydroxymethylcytosine in a Single Species Reveals Constitutive, Somatic- and Germline-Specific, and Age-Related Genomic Context Distributions and Biological Functions. Epigenomes. 2025; 9(3):35. https://doi.org/10.3390/epigenomes9030035
Chicago/Turabian StyleRenard, Thibaut, and Serge Aron. 2025. "Comparative Mapping of N6-Methyladenine, C5-Methylcytosine, and C5-Hydroxymethylcytosine in a Single Species Reveals Constitutive, Somatic- and Germline-Specific, and Age-Related Genomic Context Distributions and Biological Functions" Epigenomes 9, no. 3: 35. https://doi.org/10.3390/epigenomes9030035
APA StyleRenard, T., & Aron, S. (2025). Comparative Mapping of N6-Methyladenine, C5-Methylcytosine, and C5-Hydroxymethylcytosine in a Single Species Reveals Constitutive, Somatic- and Germline-Specific, and Age-Related Genomic Context Distributions and Biological Functions. Epigenomes, 9(3), 35. https://doi.org/10.3390/epigenomes9030035




