Epigenetics of Mitochondria-Associated Genes in Striated Muscle
Abstract
:1. Introduction
2. Results
2.1. Gene Selection Strategy
2.2. COX6A2 and COX7A1, Highly and Preferentially Expressed Genes in SkM and Heart, Are Embedded in Promoter Chromatin Surrounded by Enhancer Chromatin in These Tissues
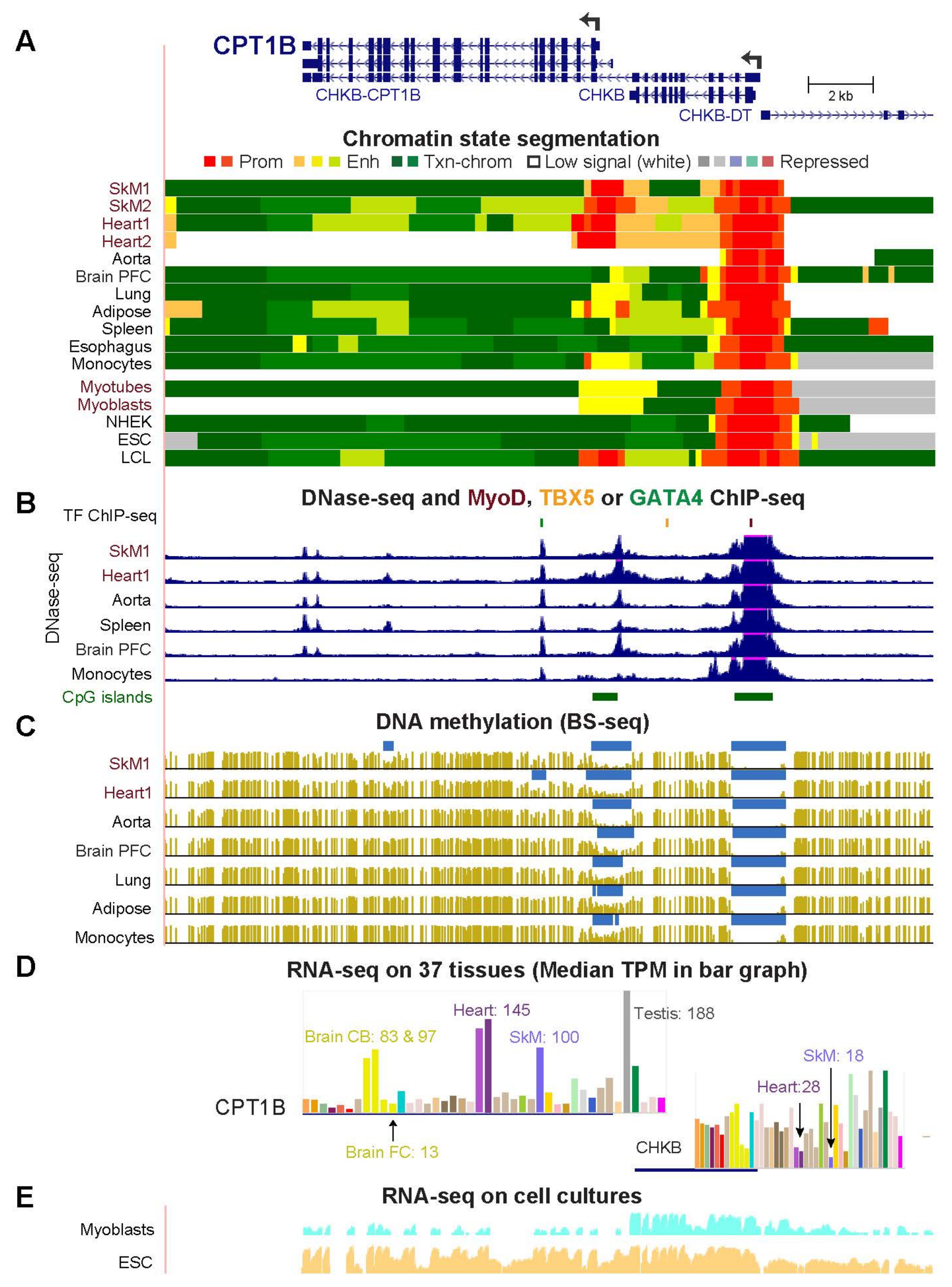
2.3. COQ10A, HADHB, and CPT1B Have Adjacent Mitochondria-Associated Genes That Share Enhancer or Promoter Chromatin with Them in Skeletal Muscle and Heart
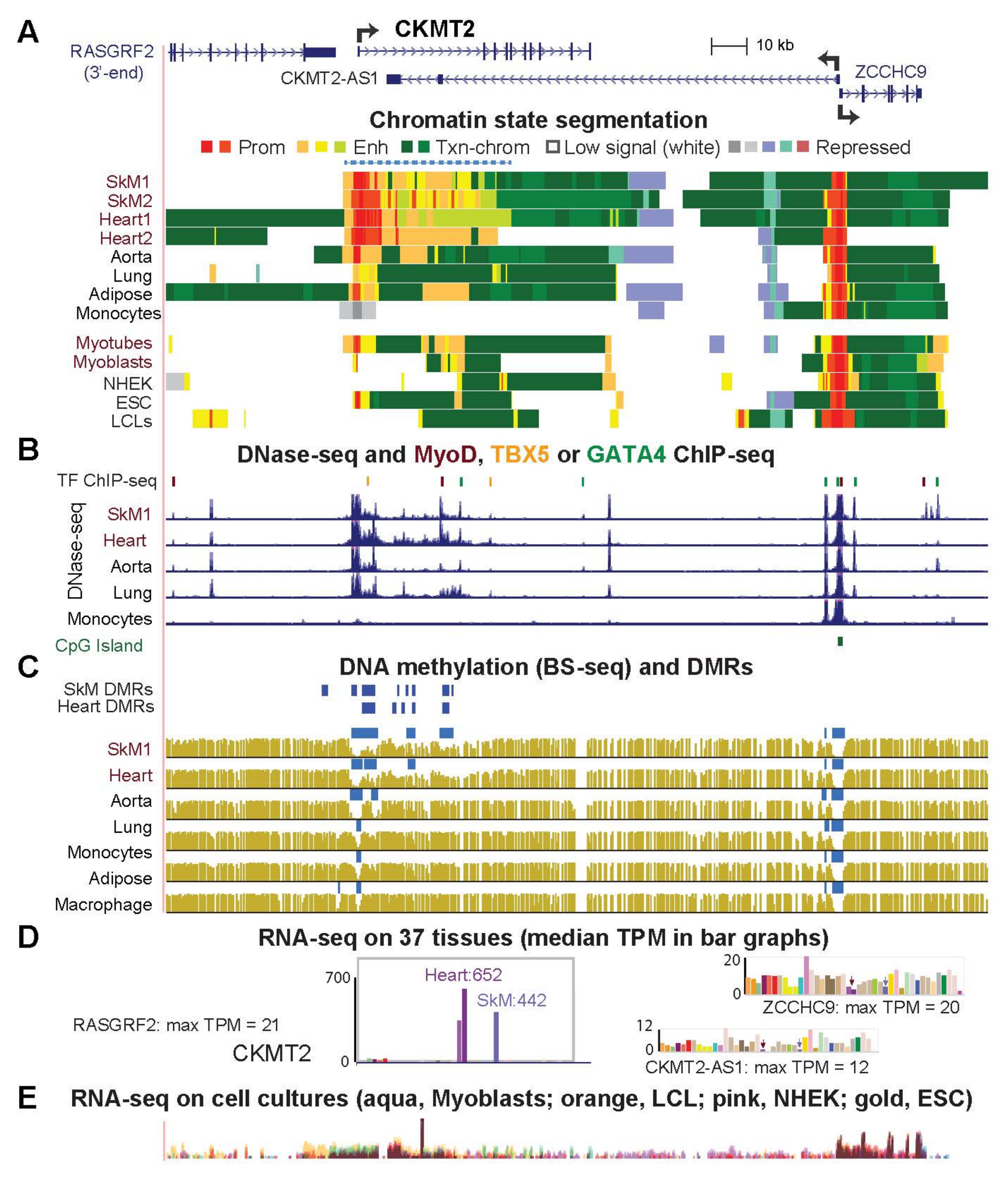
2.4. Highly Specific Expression in Striated Muscle of CKMT2, SLC25A4 and ACO2 Is Associated with a Super-Enhancer over the Gene Body
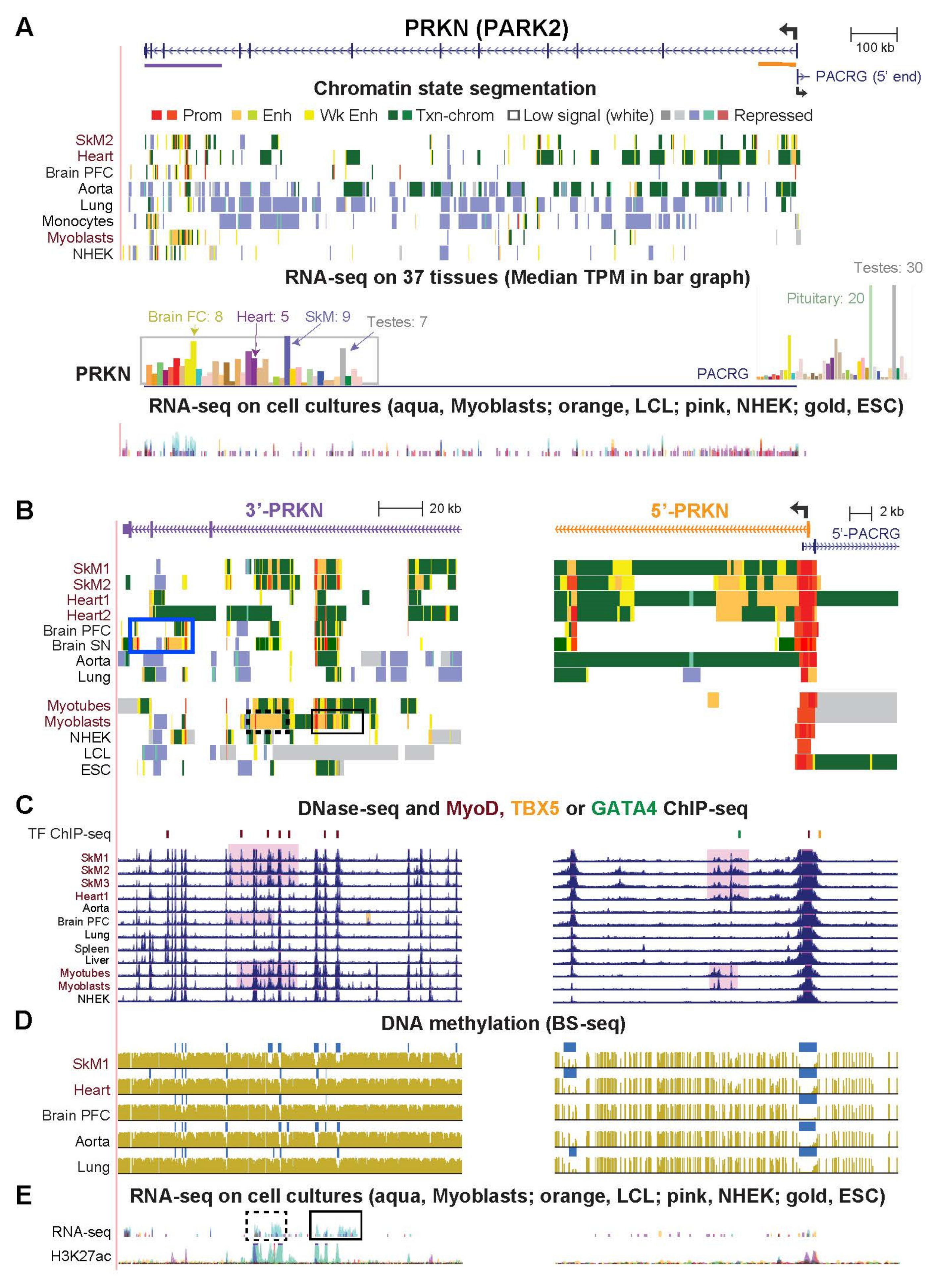
2.5. PRKN/PARK2, a Gene Important for Mitophagy, Has Striated Muscle-Associated and Brain-Associated Enhancer Chromatin and Encodes Myoblast/Skeletal-Related Antisense RNA at Its 3′ End
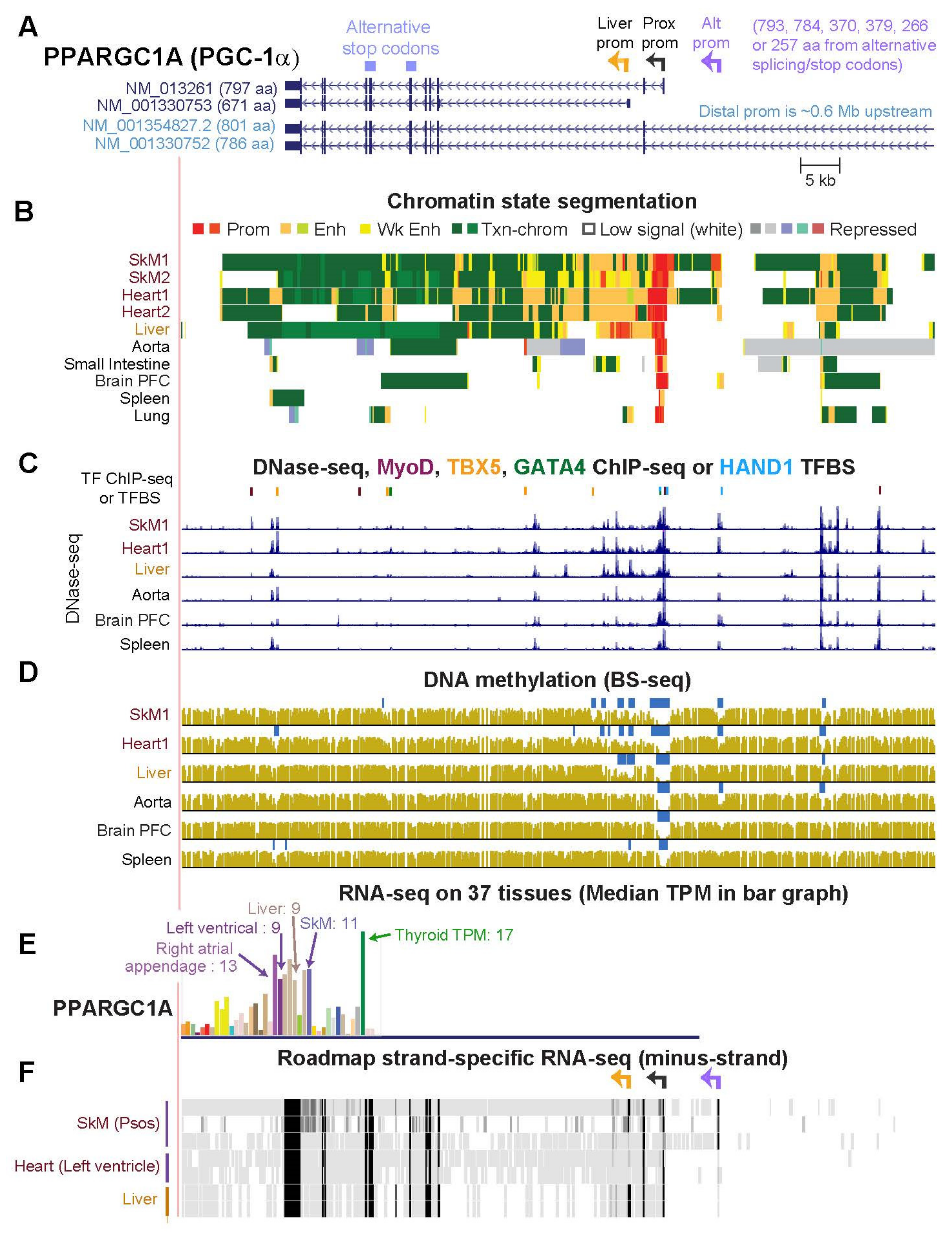
2.6. PPARGC1A, a Key Gene for Mitochondrial Biogenesis, Generates Multiple RNA Isoforms Whose Tissue Specificity Is Elucidated by Tissue-Specific Epigenetics and Novel Transcripts
3. Discussion
4. Materials and Methods
4.1. RNA-Seq for Tissues and Cells
4.2. Epigenomics
4.3. Transcription Factor Binding
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Benhar, H.; Idri, A.; Fernández-Alemán, J.L. Data preprocessing for heart disease classification: A systematic literature review. Comput. Methods Programs Biomed. 2020, 195, 105635. [Google Scholar] [CrossRef] [PubMed]
- Sweeney, H.L.; Hammers, D.W. Muscle Contraction. Cold Spring Harb. Perspect. Biol. 2018, 10, a023200. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Terry, E.E.; Zhang, X.; Hoffmann, C.; Hughes, L.D.; Lewis, S.A.; Li, J.; Wallace, M.J.; Riley, L.A.; Douglas, C.M.; Gutierrez-Monreal, M.A.; et al. Transcriptional profiling reveals extraordinary diversity among skeletal muscle tissues. Elife 2018, 7, e34613. [Google Scholar] [CrossRef] [PubMed]
- Bergmann, O.; Zdunek, S.; Felker, A.; Salehpour, M.; Alkass, K.; Bernard, S.; Sjostrom, S.L.; Szewczykowska, M.; Jackowska, T.; Dos Remedios, C.; et al. Dynamics of Cell Generation and Turnover in the Human Heart. Cell 2015, 161, 1566–1575. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Taylor-Weiner, H.; Grigsby, C.L.; Ferreira, D.M.S.; Dias, J.M.; Stevens, M.M.; Ruas, J.L.; Teixeira, A.I. Modeling the transport of nuclear proteins along single skeletal muscle cells. Proc. Natl. Acad. Sci. USA 2020, 117, 2978–2986. [Google Scholar] [CrossRef] [Green Version]
- Wardle, F.C. Master control: Transcriptional regulation of mammalian Myod. J. Muscle Res. Cell Motil. 2019, 40, 211–226. [Google Scholar] [CrossRef] [Green Version]
- Miquerol, L.; Kelly, R.G. Organogenesis of the vertebrate heart. Wiley Interdiscip. Rev. Dev. Biol. 2013, 2, 17–29. [Google Scholar] [CrossRef]
- Shouman, S.; Zaher, A.; Abdelhameed, A.; Elshaboury, S.; Sakr, S.; Fouda, B.E.; Mohamed, H.; El-Badri, N. Cardiac Progenitor Cells. Adv. Exp. Med. Biol. 2021, 1312, 51–73. [Google Scholar]
- Swedlund, B.; Lescroart, F. Cardiopharyngeal Progenitor Specification: Multiple Roads to the Heart and Head Muscles. Cold Spring Harb. Perspect. Biol. 2020, 12, a036731. [Google Scholar] [CrossRef]
- Mackey, A.L.; Magnan, M.; Chazaud, B.; Kjaer, M. Human skeletal muscle fibroblasts stimulate in vitro myogenesis and in vivo muscle regeneration. J. Physiol. 2017, 595, 5115–5127. [Google Scholar] [CrossRef] [Green Version]
- Bhattacharya, D.; Scimè, A. Mitochondrial function in muscle stem cell fates. Front. Cell Dev. Biol. 2020, 8, 480. [Google Scholar] [CrossRef]
- Glancy, B.; Balaban, R.S. Energy metabolism design of the striated muscle cell. Physiol. Rev. 2021, 101, 1561–1607. [Google Scholar] [CrossRef]
- Boengler, K.; Kosiol, M.; Mayr, M.; Schulz, R.; Rohrbach, S. Mitochondria and ageing: Role in heart, skeletal muscle and adipose tissue. J. Cachexia-Sarcopenia Muscle 2017, 8, 349–369. [Google Scholar] [CrossRef] [Green Version]
- Schiaffino, S.; Reggiani, C.; Murgia, M. Fiber type diversity in skeletal muscle explored by mass spectrometry-based single fiber proteomics. Histol. Histopathol. 2020, 35, 239–246. [Google Scholar]
- Nunnari, J.; Suomalainen, A. Mitochondria: In sickness and in health. Cell 2012, 148, 1145–1159. [Google Scholar] [CrossRef] [Green Version]
- Bose, A.; Beal, M.F. Mitochondrial dysfunction in Parkinson’s disease. J. Neurochem. 2016, 139 (Suppl. 1), 216–231. [Google Scholar] [CrossRef]
- Zhang, R.; Krigman, J.; Luo, H.; Ozgen, S.; Yang, M.; Sun, N. Mitophagy in cardiovascular homeostasis. Mech. Ageing Dev. 2020, 188, 111245. [Google Scholar] [CrossRef]
- Thompson, K.; Majd, H.; Dallabona, C.; Reinson, K.; King, M.S.; Alston, C.L.; He, L.; Lodi, T.; Jones, S.A.; Fattal-Valevski, A.; et al. Recurrent de novo dominant mutations in SLC25A4 cause severe early-onset mitochondrial disease and loss of mitochondrial DNA copy number. Am. J. Hum. Genet. 2016, 99, 860–876. [Google Scholar] [CrossRef] [Green Version]
- Dorn, G.W., 2nd; Vega, R.B.; Kelly, D.P. Mitochondrial biogenesis and dynamics in the developing and diseased heart. Genes Dev. 2015, 29, 1981–1991. [Google Scholar] [CrossRef] [Green Version]
- Doblado, L.; Lueck, C.; Rey, C.; Samhan-Arias, A.K.; Prieto, I.; Stacchiotti, A.; Monsalve, M. Mitophagy in Human Diseases. Int. J. Mol. Sci. 2021, 22, 3903. [Google Scholar] [CrossRef]
- Moore-Morris, T.; van Vliet, P.P.; Andelfinger, G.; Puceat, M. Role of epigenetics in cardiac development and congenital diseases. Physiol. Rev. 2018, 98, 2453–2475. [Google Scholar] [CrossRef] [Green Version]
- Lacey, M.; Baribault, C.; Ehrlich, K.C.; Ehrlich, M. Data showing atherosclerosis-associated differentially methylated regions are often at enhancers. Data Brief 2019, 23, 103812. [Google Scholar] [CrossRef]
- Ehrlich, K.C.; Lacey, M.; Ehrlich, M. Epigenetics of Skeletal Muscle-Associated Genes in the ASB, LRRC, TMEM, and OSBPL Gene Families. Epigenomes 2020, 4, 1. [Google Scholar] [CrossRef] [Green Version]
- Napoli, C.; Benincasa, G.; Donatelli, F.; Ambrosio, G. Precision medicine in distinct heart failure phenotypes: Focus on clinical epigenetics. Am. Heart J. 2020, 224, 113–128. [Google Scholar] [CrossRef]
- Liu, L.; Ding, C.; Fu, T.; Feng, Z.; Lee, J.E.; Xiao, L.; Xu, Z.; Yin, Y.; Guo, Q.; Sun, Z.; et al. Histone methyltransferase MLL4 controls myofiber identity and muscle performance through MEF2 interaction. J. Clin. Investig. 2020, 130, 4710–4725. [Google Scholar] [CrossRef]
- Myers, R.M.; Stamatoyannopoulos, J.; Snyder, M.; Dunham, I.; Hardison, R.C.; Bernstein, B.E.; Gingeras, T.R.; Kent, W.J.; Birney, E.; Wold, B.; et al. A user’s guide to the encyclopedia of DNA elements (ENCODE). PLoS Biol. 2011, 9, e1001046. [Google Scholar]
- Roadmap_Epigenomics_Consortium; Kundaje, A.; Meuleman, W.; Ernst, J.; Bilenky, M.; Yen, A.; Heravi-Moussavi, A.; Kheradpour, P.; Zhang, Z.; Wang, J.; et al. Integrative analysis of 111 reference human epigenomes. Nature 2015, 518, 317–330. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- GTEx_Consortium. Human genomics. The Genotype-Tissue Expression (GTEx) pilot analysis: Multitissue gene regulation in humans. Science 2015, 348, 648–660. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lindskog, C.; Linné, J.; Fagerberg, L.; Hallström, B.M.; Sundberg, C.J.; Lindholm, M.; Huss, M.; Kampf, C.; Choi, H.; Liem, D.A.; et al. The human cardiac and skeletal muscle proteomes defined by transcriptomics and antibody-based profiling. BMC Genom. 2015, 16, 475. [Google Scholar] [CrossRef] [PubMed]
- Stelzer, G.; Rosen, N.; Plaschkes, I.; Zimmerman, S.; Twik, M.; Fishilevich, S.; Stein, T.I.; Nudel, R.; Lieder, I.; Mazor, Y.; et al. The GeneCards Suite: From Gene Data Mining to Disease Genome Sequence Analyses. Curr. Protoc. Bioinform. 2016, 54, 1–30. [Google Scholar] [CrossRef] [PubMed]
- Timón-Gómez, A.; Nývltová, E.; Abriata, L.A.; Vila, A.J.; Hosler, J.; Barrientos, A. Mitochondrial cytochrome c oxidase biogenesis: Recent developments. Semin. Cell Dev. Biol. 2018, 76, 163–178. [Google Scholar] [CrossRef]
- Haeussler, M.; Zweig, A.S.; Tyner, C.; Speir, M.L.; Rosenbloom, K.R.; Raney, B.J.; Lee, C.M.; Lee, B.T.; Hinrichs, A.S.; Gonzalez, J.N.; et al. The UCSC Genome Browser database: 2019 update. Nucleic Acids Res. 2019, 47, D853–D858. [Google Scholar] [CrossRef] [Green Version]
- Song, Q.; Decato, B.; Hong, E.E.; Zhou, M.; Fang, F.; Qu, J.; Garvin, T.; Kessler, M.; Zhou, J.; Smith, A.D. A reference methylome database and analysis pipeline to facilitate integrative and comparative epigenomics. PLoS ONE 2013, 8, e81148. [Google Scholar]
- Puig, R.R.; Boddie, P.; Khan, A.; Castro-Mondragon, J.A.; Mathelier, A. UniBind: Maps of high-confidence direct TF-DNA interactions across nine species. BMC Genom. 2021, 22, 482. [Google Scholar] [CrossRef]
- Shintaku, J.; Peterson, J.M.; Talbert, E.E.; Gu, J.M.; Ladner, K.J.; Williams, D.R.; Mousavi, K.; Wang, R.; Sartorelli, V.; Guttridge, D.C. MyoD Regulates Skeletal Muscle Oxidative Metabolism Cooperatively with Alternative NF-κB. Cell Rep. 2016, 17, 514–526. [Google Scholar] [CrossRef] [Green Version]
- Garry, G.A.; Bassel-Duby, R.; Olson, E.N. Direct reprogramming as a route to cardiac repair. Semin. Cell Dev. Biol. 2021. [Google Scholar] [CrossRef]
- Breckenridge, R.A.; Zuberi, Z.; Gomes, J.; Orford, R.; Dupays, L.; Felkin, L.E.; Clark, J.E.; Magee, A.I.; Ehler, E.; Birks, E.J.; et al. Overexpression of the transcription factor Hand1 causes predisposition towards arrhythmia in mice. J. Mol. Cell Cardiol. 2009, 47, 133–141. [Google Scholar] [CrossRef]
- Padmanabhan, A.; Alexanian, M.; Linares-Saldana, R.; González-Terán, B.; Andreoletti, G.; Huang, Y.; Connolly, A.J.; Kim, W.; Hsu, A.; Duan, Q.; et al. BRD4 (Bromodomain-Containing Protein 4) Interacts with GATA4 (GATA Binding Protein 4) to Govern Mitochondrial Homeostasis in Adult Cardiomyocytes. Circulation 2020, 142, 2338–2355. [Google Scholar] [CrossRef]
- Steimle, J.D.; Moskowitz, I.P. TBX5: A Key Regulator of Heart Development. Curr. Top. Dev. Biol. 2017, 122, 195–221. [Google Scholar]
- Khan, A.; Zhang, X. dbSUPER: A database of super-enhancers in mouse and human genome. Nucleic Acids Res. 2016, 44, D164–D171. [Google Scholar] [CrossRef] [Green Version]
- Gouspillou, G.; Godin, R.; Piquereau, J.; Picard, M.; Mofarrahi, M.; Mathew, J.; Purves-Smith, F.M.; Sgarioto, N.; Hepple, R.T.; Burelle, Y.; et al. Protective role of Parkin in skeletal muscle contractile and mitochondrial function. J. Physiol. 2018, 596, 2565–2579. [Google Scholar] [CrossRef]
- Geisler, S.; Holmström, K.M.; Skujat, D.; Fiesel, F.C.; Rothfuss, O.C.; Kahle, P.J.; Springer, W. PINK1/Parkin-mediated mitophagy is dependent on VDAC1 and p62/SQSTM1. Nat. Cell Biol. 2010, 12, 119–131. [Google Scholar] [CrossRef]
- Soyal, S.M.; Zara, G.; Ferger, B.; Felder, T.K.; Kwik, M.; Nofziger, C.; Dossena, S.; Schwienbacher, C.; Hicks, A.A.; Pramstaller, P.P.; et al. The PPARGC1A locus and CNS-specific PGC-1α isoforms are associated with Parkinson’s Disease. Neurobiol. Dis. 2019, 121, 34–46. [Google Scholar] [CrossRef]
- Soyal, S.; Krempler, F.; Oberkofler, H.; Patsch, W. PGC-1alpha: A potent transcriptional cofactor involved in the pathogenesis of type 2 diabetes. Diabetologia 2006, 49, 1477–1488. [Google Scholar] [CrossRef]
- Chambers, J.M.; Wingert, R.A. PGC-1α in Disease: Recent Renal Insights into a Versatile Metabolic Regulator. Cells 2020, 9, 2234. [Google Scholar] [CrossRef]
- Martínez-Redondo, V.; Pettersson, A.T.; Ruas, J.L. The hitchhiker’s guide to PGC-1α isoform structure and biological functions. Diabetologia 2015, 58, 1969–1977. [Google Scholar] [CrossRef] [Green Version]
- Léveillé, M.; Besse-Patin, A.; Jouvet, N.; Gunes, A.; Sczelecki, S.; Jeromson, S.; Khan, N.P.; Baldwin, C.; Dumouchel, A.; Correia, J.C.; et al. PGC-1α isoforms coordinate to balance hepatic metabolism and apoptosis in inflammatory environments. Mol. Metab. 2020, 34, 72–84. [Google Scholar] [CrossRef] [PubMed]
- Kwik, M.; Hainzl, S.; Oppelt, J.; Tichy, B.; Koller, U.; Bernardinelli, E.; Steiner, M.; Zara, G.; Nofziger, C.; Weis, S.; et al. Selective activation of CNS and reference PPARGC1A promoters Is associated with distinct gene programs relevant for neurodegenerative diseases. Int. J. Mol. Sci. 2021, 22, 3296. [Google Scholar] [CrossRef] [PubMed]
- Popov, D.V.; Lysenko, E.A.; Kuzmin, I.V.; Vinogradova, V.; Grigoriev, A.I. Regulation of PGC-1α isoform expression in skeletal muscles. Acta Nat. 2015, 7, 48–59. [Google Scholar] [CrossRef]
- Ruas, J.L.; White, J.P.; Rao, R.R.; Kleiner, S.; Brannan, K.T.; Harrison, B.C.; Greene, N.P.; Wu, J.; Estall, J.L.; Irving, B.A.; et al. A PGC-1α isoform induced by resistance training regulates skeletal muscle hypertrophy. Cell 2012, 151, 1319–1331. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schiaffino, S.; Reggiani, C.; Akimoto, T.; Blaauw, B. Molecular Mechanisms of Skeletal Muscle Hypertrophy. J. Neuromuscul. Dis. 2021, 8, 169–183. [Google Scholar] [CrossRef]
- Martínez-Redondo, V.; Jannig, P.R.; Correia, J.C.; Ferreira, D.M.; Cervenka, I.; Lindvall, J.M.; Sinha, I.; Izadi, M.; Pettersson-Klein, A.T.; Agudelo, L.Z.; et al. Peroxisome Proliferator-activated Receptor γ Coactivator-1 α Isoforms Selectively Regulate Multiple Splicing Events on Target Genes. J. Biol. Chem. 2016, 291, 15169–15184. [Google Scholar] [CrossRef] [Green Version]
- Soyal, S.M.; Felder, T.K.; Auer, S.; Hahne, P.; Oberkofler, H.; Witting, A.; Paulmichl, M.; Landwehrmeyer, G.B.; Weydt, P.; Patsch, W. A greatly extended PPARGC1A genomic locus encodes several new brain-specific isoforms and influences Huntington disease age of onset. Hum. Mol. Genet. 2012, 21, 3461–3473. [Google Scholar] [CrossRef] [Green Version]
- Cordeiro, A.V.; Bricola, R.S.; Braga, R.R.; Lenhare, L.; Silva, V.R.R.; Anaruma, C.P.; Katashima, C.K.; Crisol, B.M.; Simabuco, F.M.; Silva, A.S.R.; et al. Aerobic exercise training Induces the mitonuclear iImbalance and UPRmt in the skeletal muscle of aged mice. J. Gerontol. A Biol. Sci. Med. Sci. 2020, 75, 2258–2261. [Google Scholar] [CrossRef]
- Sondheimer, N.; Hewson, S.; Cameron, J.M.; Somers, G.R.; Broadbent, J.D.; Ziosi, M.; Quinzii, C.M.; Naini, A.B. Novel recessive mutations in COQ4 cause severe infantile cardiomyopathy and encephalopathy associated with CoQ(10) deficiency. Mol. Genet. Metab. Rep. 2017, 12, 23–27. [Google Scholar] [CrossRef]
- Park, H.D.; Kim, S.R.; Ki, C.S.; Lee, S.Y.; Chang, Y.S.; Jin, D.K.; Park, W.S. Two novel HADHB gene mutations in a Korean patient with mitochondrial trifunctional protein deficiency. Ann. Clin. Lab. Sci. 2009, 39, 399–404. [Google Scholar]
- Dagher, R.; Massie, R.; Gentil, B.J. MTP deficiency caused by HADHB mutations: Pathophysiology and clinical manifestations. Mol. Genet. Metab. 2021, 133, 1–7. [Google Scholar] [CrossRef]
- Ji, S.; You, Y.; Kerner, J.; Hoppel, C.L.; Schoeb, T.R.; Chick, W.S.; Hamm, D.A.; Sharer, J.D.; Wood, P.A. Homozygous carnitine palmitoyltransferase 1b (muscle isoform) deficiency is lethal in the mouse. Mol. Genet. Metab. 2008, 93, 314–322. [Google Scholar] [CrossRef] [Green Version]
- Gladyck, S.; Aras, S.; Hüttemann, M.; Grossman, L.I. Regulation of COX assembly and function by twin CX(9)C proteins-Implications for human disease. Cells 2021, 10, 197. [Google Scholar] [CrossRef]
- Whittington, H.J.; Ostrowski, P.J.; McAndrew, D.J.; Cao, F.; Shaw, A.; Eykyn, T.R.; Lake, H.A.; Tyler, J.; Schneider, J.E.; Neubauer, S.; et al. Over-expression of mitochondrial creatine kinase in the murine heart improves functional recovery and protects against injury following ischaemia-reperfusion. Cardiovasc. Res. 2018, 114, 858–869. [Google Scholar] [CrossRef]
- Eryilmaz, I.E.; Cecener, G.; Erer, S.; Egeli, U.; Tunca, B.; Zarifoglu, M.; Elibol, B.; Bora Tokcaer, A.; Saka, E.; Demirkiran, M.; et al. Epigenetic approach to early-onset Parkinson’s disease: Low methylation status of SNCA and PARK2 promoter regions. Neurol. Res. 2017, 39, 965–972. [Google Scholar] [CrossRef]
- Krämer, A.I.; Handschin, C. How epigenetic modifications drive the expression and mediate the action of PGC-1α in the regulation of metabolism. Int. J. Mol. Sci. 2019, 20, 5449. [Google Scholar] [CrossRef] [Green Version]
- Lochmann, T.L.; Thomas, R.R.; Bennett, J.P., Jr.; Taylor, S.M. Epigenetic modifications of the PGC-1α promoter during exercise induced expression in mice. PLoS ONE 2015, 10, e0129647. [Google Scholar] [CrossRef]
- Ronn, T.; Poulsen, P.; Hansson, O.; Holmkvist, J.; Almgren, P.; Nilsson, P.; Tuomi, T.; Isomaa, B.; Groop, L.; Vaag, A.; et al. Age influences DNA methylation and gene expression of COX7A1 in human skeletal muscle. Diabetologia 2008, 51, 1159–1168. [Google Scholar] [CrossRef] [Green Version]
- Marshall, L.L.; Killinger, B.A.; Ensink, E.; Li, P.; Li, K.X.; Cui, W.; Lubben, N.; Weiland, M.; Wang, X.; Gordevicius, J.; et al. Epigenomic analysis of Parkinson’s disease neurons identifies Tet2 loss as neuroprotective. Nat. Neurosci. 2020, 23, 1203–1214. [Google Scholar] [CrossRef]
- Patel, B.V.; Yao, F.; Howenstine, A.; Takenaka, R.; Hyatt, J.A.; Sears, K.E.; Shewchuk, B.M. Emergent Coordination of the CHKB and CPT1B Genes in Eutherian Mammals: Implications for the Origin of Brown Adipose Tissue. J. Mol. Biol. 2020, 432, 6127–6145. [Google Scholar] [CrossRef]
- Kadenbach, B. Complex IV-The regulatory center of mitochondrial oxidative phosphorylation. Mitochondrion 2021, 58, 296–302. [Google Scholar] [CrossRef]
- Boczonadi, V.; Giunta, M.; Lane, M.; Tulinius, M.; Schara, U.; Horvath, R. Investigating the role of the physiological isoform switch of cytochrome c oxidase subunits in reversible mitochondrial disease. Int. J. Biochem. Cell Biol. 2015, 63, 32–40. [Google Scholar] [CrossRef]
- Roudbar, M.A.; Mousavi, S.F.; Ardestani, S.S.; Lopes, F.B.; Momen, M.; Gianola, D.; Khatib, H. Prediction of biological age and evaluation of genome-wide dynamic methylomic changes throughout human aging. G3 Genes Genomes Genet. 2021, 11, jkab112. [Google Scholar] [CrossRef]
- Gumeni, S.; Papanagnou, E.D.; Manola, M.S.; Trougakos, I.P. Nrf2 activation induces mitophagy and reverses Parkin/Pink1 knock down-mediated neuronal and muscle degeneration phenotypes. Cell Death Dis. 2021, 12, 671. [Google Scholar] [CrossRef] [PubMed]
- Leduc-Gaudet, J.P.; Reynaud, O.; Hussain, S.N.; Gouspillou, G. Parkin overexpression protects from ageing-related loss of muscle mass and strength. J. Physiol. 2019, 597, 1975–1991. [Google Scholar] [CrossRef] [PubMed]
- Shin, J.H.; Ko, H.S.; Kang, H.; Lee, Y.; Lee, Y.I.; Pletinkova, O.; Troconso, J.C.; Dawson, V.L.; Dawson, T.M. PARIS (ZNF746) repression of PGC-1α contributes to neurodegeneration in Parkinson’s disease. Cell 2011, 144, 689–702. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mizuno, Y. More than 20 years of the discovery of Park2. Neurosci. Res. 2020, 159, 3–8. [Google Scholar] [CrossRef] [PubMed]
- Leach, J.P.; Heallen, T.; Zhang, M.; Rahmani, M.; Morikawa, Y.; Hill, M.C.; Segura, A.; Willerson, J.T.; Martin, J.F. Hippo pathway deficiency reverses systolic heart failure after infarction. Nature 2017, 550, 260–264. [Google Scholar] [CrossRef] [Green Version]
- Sin, J.; Andres, A.M.; Taylor, D.J.; Weston, T.; Hiraumi, Y.; Stotland, A.; Kim, B.J.; Huang, C.; Doran, K.S.; Gottlieb, R.A. Mitophagy is required for mitochondrial biogenesis and myogenic differentiation of C2C12 myoblasts. Autophagy 2016, 12, 369–380. [Google Scholar] [CrossRef] [Green Version]
- Lampert, M.A.; Orogo, A.M.; Najor, R.H.; Hammerling, B.C.; Leon, L.J.; Wang, B.J.; Kim, T.; Sussman, M.A.; Gustafsson, Å.B. BNIP3L/NIX and FUNDC1-mediated mitophagy is required for mitochondrial network remodeling during cardiac progenitor cell differentiation. Autophagy 2019, 15, 1182–1198. [Google Scholar] [CrossRef]
- Song, H.; Tian, X.; Liu, D.; Liu, M.; Liu, Y.; Liu, J.; Mei, Z.; Yan, C.; Han, Y. CREG1 improves the capacity of the skeletal muscle response to exercise endurance via modulation of mitophagy. Autophagy 2021, 1–17. [Google Scholar] [CrossRef]
- Sartorelli, V.; Lauberth, S.M. Enhancer RNAs are an important regulatory layer of the epigenome. Nat. Struct. Mol. Biol. 2020, 27, 521–528. [Google Scholar] [CrossRef]
- Murillo-González, F.E.; García-Aguilar, R.; Vega, L.; Elizondo, G. Regulation of Parkin expression as the key balance between neural survival and cancer cell death. Biochem. Pharmacol. 2021, 190, 114650. [Google Scholar] [CrossRef]
- Vega, R.B.; Huss, J.M.; Kelly, D.P. The coactivator PGC-1 cooperates with peroxisome proliferator-activated receptor alpha in transcriptional control of nuclear genes encoding mitochondrial fatty acid oxidation enzymes. Mol. Cell. Biol. 2000, 20, 1868–1876. [Google Scholar] [CrossRef] [Green Version]
- Wagner, A.E.; Ernst, I.M.; Birringer, M.; Sancak, O.; Barella, L.; Rimbach, G. A combination of lipoic acid plus coenzyme Q10 induces PGC1α, a master switch of energy metabolism, improves stress response, and increases cellular glutathione levels in cultured C2C12 skeletal muscle cells. Oxidative Med. Cell. Longev. 2012, 2012, 835970. [Google Scholar] [CrossRef] [Green Version]
- Lin, J.; Wu, H.; Tarr, P.T.; Zhang, C.Y.; Wu, Z.; Boss, O.; Michael, L.F.; Puigserver, P.; Isotani, E.; Olson, E.N.; et al. Transcriptional co-activator PGC-1 alpha drives the formation of slow-twitch muscle fibres. Nature 2002, 418, 797–801. [Google Scholar] [CrossRef]
- Allan, R.; Morton, J.P.; Close, G.L.; Drust, B.; Gregson, W.; Sharples, A.P. PGC-1α alternative promoter (Exon 1b) controls augmentation of total PGC-1α gene expression in response to cold water immersion and low glycogen availability. Eur. J. Appl. Physiol. 2020, 120, 2487–2493. [Google Scholar] [CrossRef]
- Larson, C.; Opichka, M.; McGlynn, M.L.; Collins, C.W.; Slivka, D. Exercise- and Cold-Induced Human PGC-1α mRNA Isoform Specific Responses. Int. J. Environ. Res. Public Health 2020, 17, 5740. [Google Scholar] [CrossRef]
- Banerji, C.R.S.; Panamarova, M.; Pruller, J.; Figeac, N.; Hebaishi, H.; Fidanis, E.; Saxena, A.; Contet, J.; Sacconi, S.; Severini, S.; et al. Dynamic transcriptomic analysis reveals suppression of PGC1α/ERRα drives perturbed myogenesis in facioscapulohumeral muscular dystrophy. Hum. Mol. Genet. 2019, 28, 1244–1259. [Google Scholar] [CrossRef]
- Soyal, S.M.; Bonova, P.; Kwik, M.; Zara, G.; Auer, S.; Scharler, C.; Strunk, D.; Nofziger, C.; Paulmichl, M.; Patsch, W. The Expression of CNS-Specific PPARGC1A Transcripts Is Regulated by Hypoxia and a Variable GT Repeat Polymorphism. Mol. Neurobiol. 2020, 57, 752–764. [Google Scholar] [CrossRef] [Green Version]
- Terragni, J.; Zhang, G.; Sun, Z.; Pradhan, S.; Song, L.; Crawford, G.E.; Lacey, M.; Ehrlich, M. Notch signaling genes: Myogenic DNA hypomethylation and 5-hydroxymethylcytosine. Epigenetics 2014, 9, 842–850. [Google Scholar] [CrossRef] [Green Version]
- Ehrlich, K.C.; Baribault, C.; Ehrlich, M. Epigenetics of muscle- and brain-specific expression of KLHL family genes. Int. J. Mol. Sci. 2020, 21, 8394. [Google Scholar] [CrossRef]
- Ang, Y.S.; Rivas, R.N.; Ribeiro, A.J.S.; Srivas, R.; Rivera, J.; Stone, N.R.; Pratt, K.; Mohamed, T.M.A.; Fu, J.D.; Spencer, C.I.; et al. Disease Model of GATA4 Mutation Reveals Transcription Factor Cooperativity in Human Cardiogenesis. Cell 2016, 167, 1734–1749.e1722. [Google Scholar] [CrossRef] [Green Version]




| Gene | TPM b SkM | TPM Heart | TPM Ratios: SkM/Heart | TPM Ratios: SkM/Aorta | Median TPM of Other Tissues | TPM Ratios: SkM/Median Other Tissues | TPM Ratios: Heart/Median Other Tissues |
|---|---|---|---|---|---|---|---|
| ACADS c | 112.8 | 60.1 | 1.9 | 5.2 | 30.7 | 3.7 | 2.0 |
| ACO2 (Fig. S4) | 272.4 | 252.4 | 1.1 | 5.4 | 53.8 | 5.1 | 4.7 |
| CHCHD10 c | 229.7 | 248.5 | 0.9 | 7.5 | 45.7 | 5.0 | 5.4 |
| CKMT2 (Fig. 5) | 441.9 | 652.4 | 0.7 | 17.7 | 5.6 | 79.5 | 117.4 |
| COQ10A (Fig. 3) | 65.1 | 79.1 | 0.8 | 5.0 | 14.0 | 4.6 | 5.6 |
| COX5A c | 389.6 | 421.1 | 0.9 | 5.4 | 88.3 | 4.4 | 4.8 |
| COX6A2 (Fig. 2) | 2283.9 | 1593.7 | 1.4 | 556.2 | 1.0 | 2263.4 | 1579.4 |
| COX7A1 (Fig. S1) | 704.8 | 584.3 | 1.2 | 4.3 | 56.5 | 12.5 | 10.3 |
| COX10 c | 28.6 | 19.1 | 1.5 | 3.8 | 9.5 | 3.0 | 2.0 |
| CPT1B (Fig. 4) | 100.5 | 144.6 | 0.7 | 13.8 | 20.9 | 4.8 | 6.9 |
| G0S2 c | 65.0 | 84.9 | 0.8 | 3.6 | 22.8 | 2.8 | 3.7 |
| GOT1 c | 212.3 | 304.6 | 0.7 | 10.5 | 32.7 | 6.5 | 9.3 |
| GOT2 c | 280.0 | 251.9 | 1.1 | 4.7 | 74.9 | 3.7 | 3.4 |
| HADHB (Fig. S2) | 375.3 | 307.5 | 1.2 | 5.6 | 77.1 | 4.9 | 4.0 |
| IDH2 c | 442.8 | 311.5 | 1.4 | 13.5 | 54.1 | 8.2 | 5.8 |
| LDHD c | 47.1 | 58.5 | 0.8 | 4.5 | 12.0 | 3.9 | 4.9 |
| MDH2 c | 361.2 | 210.3 | 1.7 | 3.4 | 104.6 | 3.5 | 2.0 |
| NDUFB9 c | 299.5 | 188.2 | 1.6 | 3.2 | 82.8 | 3.6 | 2.3 |
| NDUFS1 c | 50.6 | 42.4 | 1.2 | 3.1 | 16.3 | 3.1 | 2.6 |
| NNT c | 69.7 | 52.5 | 1.3 | 4.2 | 16.4 | 4.3 | 3.2 |
| OGDH c | 222.2 | 201.4 | 1.1 | 3.1 | 65.6 | 3.4 | 3.1 |
| PPARGC1A (Figs. 7, 8, S7 and S8) | 11.0 | 9.4 | 1.2 | 24.5 | 2.7 | 4.2 | 3.5 |
| PRKN/PARK2 (Figs. 6 and S5) | 8.7 | 4.8 | 1.8 | 3.4 | 2.4 | 3.6 | 2.0 |
| SLC25A4 (Fig. S3) | 370.5 | 563.8 | 0.7 | 3.4 | 24.6 | 15.0 | 22.9 |
| TMEM143 c | 34.2 | 21.1 | 1.6 | 4.9 | 8.5 | 4.0 | 2.5 |
| UQCRFS1 c | 133.8 | 102.0 | 1.3 | 4.0 | 39.5 | 3.4 | 2.6 |
| VDAC1 (Fig. S6) | 390.9 | 199.3 | 2.0 | 3.7 | 91.0 | 4.3 | 2.2 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ehrlich, K.C.; Deng, H.-W.; Ehrlich, M. Epigenetics of Mitochondria-Associated Genes in Striated Muscle. Epigenomes 2022, 6, 1. https://doi.org/10.3390/epigenomes6010001
Ehrlich KC, Deng H-W, Ehrlich M. Epigenetics of Mitochondria-Associated Genes in Striated Muscle. Epigenomes. 2022; 6(1):1. https://doi.org/10.3390/epigenomes6010001
Chicago/Turabian StyleEhrlich, Kenneth C., Hong-Wen Deng, and Melanie Ehrlich. 2022. "Epigenetics of Mitochondria-Associated Genes in Striated Muscle" Epigenomes 6, no. 1: 1. https://doi.org/10.3390/epigenomes6010001
APA StyleEhrlich, K. C., Deng, H.-W., & Ehrlich, M. (2022). Epigenetics of Mitochondria-Associated Genes in Striated Muscle. Epigenomes, 6(1), 1. https://doi.org/10.3390/epigenomes6010001






