Validation of Stable Reference Genes for RT-qPCR Normalization in Oxycetonia jucunda (Coleoptera: Scarabaeidae)
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Insect Source
2.2. Tissue Sample Collection
2.3. RNA Extraction and cDNA Synthesis
2.4. Quantitative Real-Time PCR Analysis
2.5. Analysis of the Stability of Candidate Reference Genes
2.6. Validation of Reference Gene Stability
2.7. Statistical Analysis
3. Results
3.1. Specificity and Amplification Efficiency of RT-qPCR Primers
3.2. ΔCt Method
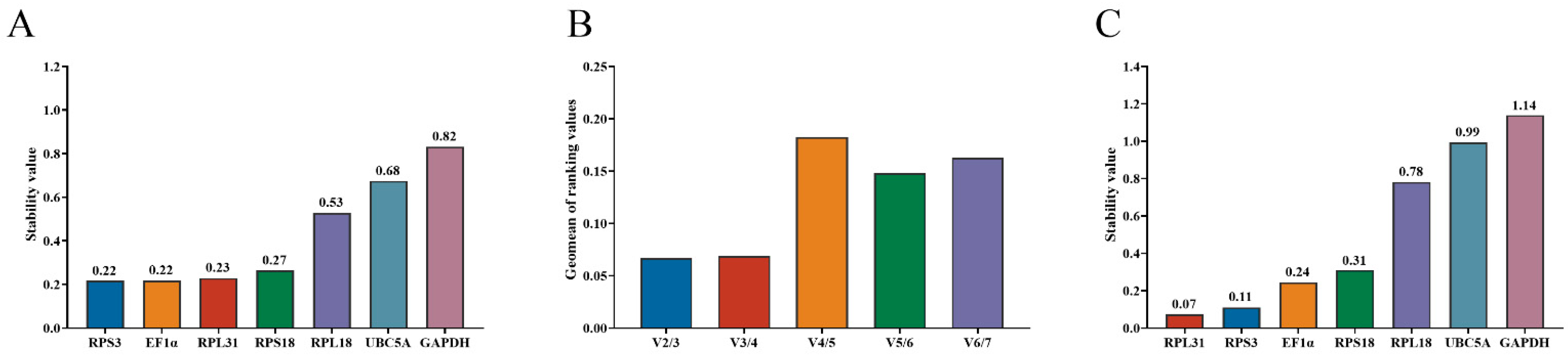
3.3. GeNorm and NormFinder Analyses
3.4. BestKeeper Analysis
3.5. RefFinder Analysis
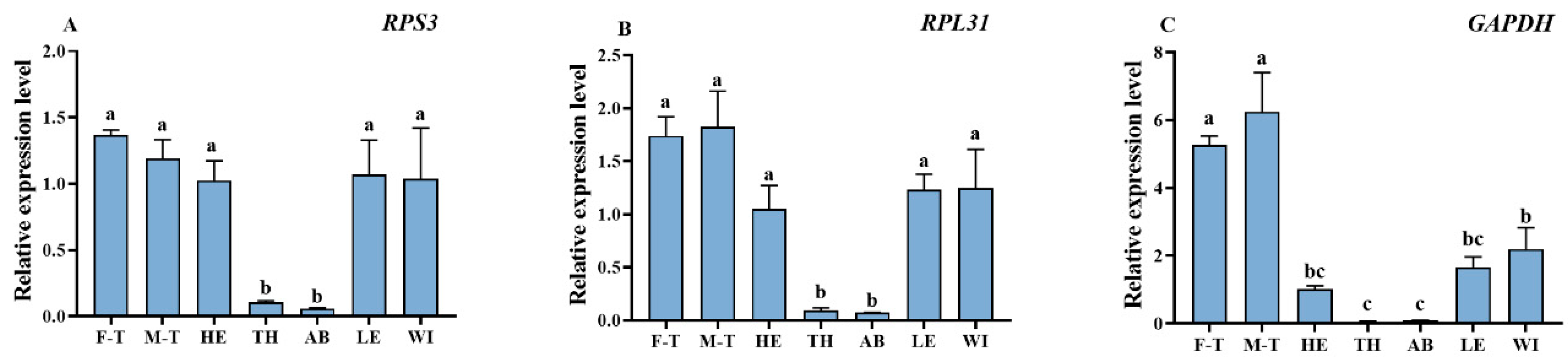
3.6. Validation of the Stability of Reference Genes
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Miao, M.; Li, J.Q. Harm of Oxycetonia jucunda Faldermann to apple trees. For. Pest Dis. 2016, 35, 20–22. [Google Scholar]
- Ni, T.L.; Li, Z.Y.; Wang, M.; Guo, G.Y. Occurrence and control of Oxycetonia jucunda Faldermann. J. Hebei For. Sci. Technol. 2003, 43. [Google Scholar] [CrossRef]
- Zhen, H.W.; Wang, R.Z.; Wang, R.X.; Gu, W.W. Experiment on trapping Oxycetonia jucunda with Sophora japonica. J. Hebei For. Sci. Technol. 2005, 5–6. [Google Scholar] [CrossRef]
- Mulvey, J.; Cresswell, J.E. Time-dependent effects on bumble bees of dietary exposures to farmland insecticides (imidacloprid, thiamethoxam and fipronil). Pest Manag. Sci. 2020, 76, 2846–2853. [Google Scholar] [CrossRef] [PubMed]
- Sun, X.H.; Sun, S.M.; Qiao, J.Z.; Zhang, J.J. Study on the Outbreak Patterns and Biological Characteristics of Apple Orchard Beetles in Henan Province. Huazhong Insect Res. 2013, 9, 289–293. [Google Scholar]
- Farder-Gomes, C.F.; Grella, T.C.; Malaspina, O.; Nocelli, R.F.C. Exposure to sublethal concentrations of imidacloprid, pyraclostrobin, and glyphosate harm the behavior and fat body cells of the stingless bee Scaptotrigona postica. Sci. Total Environ. 2024, 907, 168072. [Google Scholar] [CrossRef] [PubMed]
- Cedden, D.; Güney, G.; Rostás, M.; Bucher, G. Optimizing dsRNA sequences for RNAi in pest control and research with the dsRIP web platform. BMC Biol. 2025, 23, 114. [Google Scholar] [CrossRef] [PubMed]
- List, F.; Tarone, A.M.; Zhu-Salzman, K.; Vargo, E.L. RNA meets toxicology: Efficacy indicators from the experimental design of RNAi studies for insect pest management. Pest Manag. Sci. 2022, 78, 3215–3225. [Google Scholar] [CrossRef]
- Lu, Q.; Cui, H.; Li, W.; Liu, T.; Chen, Q.; Yang, Q. Synthetic nanoscale RNAi constructs as pesticides for the control of Locusta migratoria. J. Agric. Food Chem. 2022, 70, 10762–10770. [Google Scholar] [CrossRef]
- Aly, S.M.; Sabri, D.M. Next generation sequencing (NGS): A golden tool in forensic toolkit. Arch. Med. Sadowej Kryminol. 2015, 65, 260–271. [Google Scholar] [CrossRef]
- Castellanos, N.L.; Smagghe, G.; Taning, C.N.T.; Oliveira, E.E.; Christiaens, O. Risk assessment of RNAi-based pesticides to non-target organisms: Evaluating the effects of sequence similarity in the parasitoid wasp Telenomus podisi. Sci. Total Environ. 2022, 832, 154746. [Google Scholar] [CrossRef]
- Parsons, K.H.; Mondal, M.H.; McCormick, C.L.; Flynt, A.S. Guanidinium-functionalized interpolyelectrolyte complexes enabling RNAi in resistant insect pests. Biomacromolecules 2018, 19, 1111–1117. [Google Scholar] [CrossRef]
- Lee, D.E.; Lee, S.H.; Kim, J.H. Validation of reliable reference genes for comparison of gene expression across species in the Anopheles hyrcanus group. Sci. Rep. 2025, 15, 9037. [Google Scholar] [CrossRef]
- Vandesompele, J.; De Preter, K.; Pattyn, F.; Poppe, B.; Van Roy, N.; De Paepe, A.; Speleman, F. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002, 3, research0034.1. [Google Scholar] [CrossRef] [PubMed]
- Bustin, S.A. Absolute quantification of mRNA using real-time reverse transcription polymerase chain reaction assays. J. Mol. Endocrinol. 2000, 25, 169–193. [Google Scholar] [CrossRef]
- Bustin, S.A.; Benes, V.; Nolan, T.; Pfaffl, M.W. Quantitative real-time RT-PCR—A perspective. J. Mol. Endocrinol. 2005, 34, 597–601. [Google Scholar] [CrossRef]
- VanGuilder, H.D.; Vrana, K.E.; Freeman, W.M. Twenty-five years of quantitative PCR for gene expression analysis. Biotechniques 2008, 44, 619–626. [Google Scholar] [CrossRef] [PubMed]
- Lee, P.D.; Sladek, R.; Greenwood, C.M.T.; Hudson, T.J. Control genes and variability: Absence of ubiquitous reference transcripts in diverse mammalian expression studies. Genome Res. 2002, 12, 292–297. [Google Scholar] [CrossRef] [PubMed]
- Nolan, T.; Hands, R.E.; Bustin, S.A. Quantification of mRNA using real-time RT-PCR. Nat. Protoc. 2006, 1, 1559–1582. [Google Scholar] [CrossRef]
- Yuan, F.; Xie, Z.; Li, Z.; Lian, P.; Wei, C. Screening of reference genes for gene expression study in different tissues from the transcriptome data of the vector leafhopper Psammotettix striatus. Gene 2024, 927, 148696. [Google Scholar] [CrossRef]
- Xin, H.-J.; Liu, C.-Y.; Yan, F.; Wang, L.-D.; Zhang, H.-H.; Shen, C.-H.; Zhai, Q. Reference gene selection for quantitative gene expression analysis in Argynnis hyperbius. Insects 2025, 16, 1008. [Google Scholar] [CrossRef]
- Yang, C.; Pan, H.; Noland, J.E.; Zhang, D.; Zhang, Z.; Liu, Y.; Zhou, X. Selection of reference genes for RT-qPCR analysis in a predatory biological control agent, Coleomegilla maculata (Coleoptera: Coccinellidae). Sci. Rep. 2015, 5, 18201. [Google Scholar] [CrossRef]
- Yang, C.; Preisser, E.L.; Zhang, H.; Liu, Y.; Dai, L.; Pan, H.; Zhou, X. Selection of reference genes for RT-qPCR analysis in Coccinella septempunctata to assess un-intended effects of RNAi transgenic plants. Front. Plant Sci. 2016, 7, 1672. [Google Scholar] [CrossRef]
- Zhang, B.; Liu, J.; Yuan, G.; Chen, X.; Gao, X. Selection and evaluation of potential reference genes for gene expression analysis in greenbug (Schizaphis graminum Rondani). J. Integr. Agric. 2018, 17, 2054–2065. [Google Scholar] [CrossRef]
- Huggett, J.; Dheda, K.; Bustin, S.; Zumla, A. Real-time RT-PCR normalisation; strategies and considerations. Genes Immun. 2005, 6, 279–284. [Google Scholar] [CrossRef] [PubMed]
- Bansal, R.; Mittapelly, P.; Chen, Y.; Mamidala, P.; Zhao, C.; Michel, A. Quantitative RT-PCR gene evaluation and RNA interference in the brown marmorated stink bug. PLoS ONE 2016, 11, e0152730. [Google Scholar] [CrossRef]
- Radonić, A.; Thulke, S.; Mackay, I.M.; Landt, O.; Siegert, W.; Nitsche, A. Guideline to reference gene selection for quantitative real-time PCR. Biochem. Biophys. Res. Commun. 2004, 313, 856–862. [Google Scholar] [CrossRef] [PubMed]
- Shakeel, M.; Rodriguez, A.; Tahir, U.B.; Jin, F. Gene expression studies of reference genes for quantitative real-time PCR: An overview in insects. Biotechnol. Lett. 2018, 40, 227–236. [Google Scholar] [CrossRef]
- Silver, N.; Best, S.; Jiang, J.; Thein, S.L. Selection of housekeeping genes for gene expression studies in human reticulocytes using real-time PCR. BMC Mol. Biol. 2006, 7, 33. [Google Scholar] [CrossRef]
- Andersen, C.L.; Jensen, J.L.; Ørntoft, T.F. Normalization of real-time quantitative reverse transcription-PCR data: A model-based variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer data sets. Cancer Res. 2004, 64, 5245–5250. [Google Scholar] [CrossRef]
- Pfaffl, M.W.; Tichopad, A.; Prgomet, C.; Neuvians, T.P. Determination of stable housekeeping genes, differentially regulated target genes and sample integrity: BestKeeper—Excel-based tool using pair-wise correlations. Biotechnol. Lett. 2004, 26, 509–515. [Google Scholar] [CrossRef]
- Xie, F.; Wang, J.; Zhang, B. RefFinder: A web-based tool for comprehensively analyzing and identifying reference genes. Funct. Integr. Genom. 2023, 23, 125. [Google Scholar] [CrossRef]
- Valasek, M.A.; Repa, J.J. The power of real-time PCR. Adv. Physiol. Educ. 2005, 29, 151–159. [Google Scholar] [CrossRef]
- Gutierrez, L.; Mauriat, M.; Guénin, S.; Pelloux, J.; Lefebvre, J.; Louvet, R.; Rusterucci, C.; Moritz, T.; Guerineau, F.; Bellini, C.; et al. The lack of a systematic validation of reference genes: A serious pitfall undervalued in reverse transcription-polymerase chain reaction (RT-PCR) analysis in plants. Plant Biotechnol. J. 2008, 6, 609–618. [Google Scholar] [CrossRef]
- Kozera, B.; Rapacz, M. Reference genes in real-time PCR. J. Appl. Genet. 2013, 54, 391–406. [Google Scholar] [CrossRef]
- Hildyard, J.C.W.; Finch, A.M.; Wells, D.J. Identification of qPCR reference genes suitable for normalizing gene expression in the mdx mouse model of Duchenne muscular dystrophy. PLoS ONE 2019, 14, e0211384. [Google Scholar] [CrossRef]
- Li, H.; Chang, F.; Cui, X.; Xi, B.; Li, G.; Liu, D.; Niu, K. Stability evaluation of reference genes in Gynaephora qinghaiensis (Lepidoptera: Lymantriidae) for qRT-PCR normalization. Insects 2025, 16, 1019. [Google Scholar] [CrossRef] [PubMed]
- Lü, J.; Yang, C.; Zhang, Y.; Pan, H. Selection of reference genes for the normalization of RT-qPCR data in gene expression studies in insects: A systematic review. Front. Physiol. 2018, 9, 1560. [Google Scholar] [CrossRef] [PubMed]
- Shi, C.; Yang, F.; Zhu, X.; Du, E.; Yang, Y.; Wang, S.; Wu, Q.; Zhang, Y. Evaluation of housekeeping genes for quantitative real-time PCR analysis of Bradysia odoriphaga (Diptera: Sciaridae). Int. J. Mol. Sci. 2016, 17, 1034. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; An, S.; Li, Z.; Wu, F.; Yang, Q.; Liu, Y.; Cao, J.; Zhang, H.; Zhang, Q.; Liu, X. Identification and validation of reference genes for normalization of gene expression analysis using qRT-PCR in Helicoverpa armigera (Lepidoptera: Noctuidae). Gene 2015, 555, 393–402. [Google Scholar] [CrossRef]
- Fu, W.; Xie, W.; Zhang, Z.; Wang, S.; Wu, Q.; Liu, Y.; Zhou, X.; Zhou, X.; Zhang, Y. Exploring valid reference genes for quantitative real-time PCR analysis in Plutella xylostella (Lepidoptera: Plutellidae). Int. J. Biol. Sci. 2013, 9, 792–802. [Google Scholar] [CrossRef]
- Yang, X.; Pan, H.; Yuan, L.; Zhou, X. Reference gene selection for RT-qPCR analysis in Harmonia axyridis, a global invasive lady beetle. Sci. Rep. 2018, 8, 2689. [Google Scholar] [CrossRef]
- Lord, J.C.; Hartzer, K.; Toutges, M.; Oppert, B. Evaluation of quantitative PCR reference genes for gene expression studies in Tribolium castaneum after fungal challenge. J. Microbiol. Methods 2010, 80, 219–221. [Google Scholar] [CrossRef]
- Luo, J.; Ma, C.; Li, Z.; Zhu, B.; Zhang, J.; Lei, C.; Jin, S.; Hull, J.J.; Chen, L. Assessment of suitable reference genes for qRT-PCR analysis in Adelphocoris suturalis. J. Integr. Agric. 2018, 17, 2745–2757. [Google Scholar] [CrossRef]
- Chen, C.; Li, S.; Zhu, H.; Fan, B.; Wang, Y.; Hao, D. Identification and evaluation of reference genes for gene expression analysis in the weevil pest Pagiophloeus tsushimanus using RT-qPCR. J. Asia-Pac. Entomol. 2020, 23, 336–344. [Google Scholar] [CrossRef]
- Sirover, M.A. New insights into an old protein: The functional diversity of mammalian glyceraldehyde-3-phosphate dehydrogenase. Biochim. Biophys. Acta 1999, 1432, 159–184. [Google Scholar] [CrossRef] [PubMed]
- Nazari, F.; Parham, A.; Maleki, A.F. GAPDH, β-actin and Β2-microglobulin, as three common reference genes, are not reliable for gene expression studies in equine adipose- and marrow-derived mesenchymal stem cells. J. Anim. Sci. Technol. 2015, 57, 18. [Google Scholar] [CrossRef] [PubMed]
- Muronetz, V.I.; Medvedeva, M.V.; Sevostyanova, I.A.; Schmalhausen, E.V. Modification of glyceraldehyde-3-phosphate dehydrogenase with nitric oxide: Role in signal transduction and development of apoptosis. Biomolecules 2021, 11, 1656. [Google Scholar] [CrossRef]
- Fu, X.; Meyer-Rochow, V.B. Selection and validation of suitable reference genes for RT-qPCR analysis in the rare aquatic firefly Aquatica leii (Coleoptera: Lampyridae). Insects 2021, 12, 359. [Google Scholar] [CrossRef]
- Zhang, S.; Chen, C.; Xie, T.; Ye, S. Identification and validation of reference genes for qRT-PCR studies of the obligate aphid pathogenic fungus Pandora neoaphidis during different developmental stages. PLoS ONE 2017, 12, e0179930. [Google Scholar] [CrossRef]
- Cao, Y.; Li, B.; Chen, N.; Yang, D.; Li, L.; Liu, T. Evaluation of reference genes for quantitative reverse transcription polymerase chain reaction in Bactrocera dorsalis (Diptera: Tephritidae) subjected to various phytosanitary treatments. Insects 2021, 12, 945. [Google Scholar] [CrossRef] [PubMed]
- Hu, H.; Ye, X.; Wang, H.; Ji, R. Selection of reference genes for normalization of real-time PCR data in Calliptamus italicus (Orthoptera: Acrididae) under different temperature conditions. J. Insect Sci. 2019, 19, 7. [Google Scholar] [CrossRef] [PubMed]
- Sharath Chandra, G.; Asokan, R.; Manamohan, M.; Krishna Kumar, N.K.; Sita, T. Evaluation of reference genes for quantitative real-time PCR normalization in cotton bollworm Helicoverpa armigera. Mol. Biol. 2014, 48, 813–822. [Google Scholar] [CrossRef]
- Yuan, M.; Lu, Y.; Zhu, X.; Wan, H.; Shakeel, M.; Zhan, S.; Jin, B.-R.; Li, J. Selection and evaluation of potential reference genes for gene expression analysis in the brown planthopper, Nilaparvata lugens (Hemiptera: Delphacidae) using reverse-transcription quantitative PCR. PLoS ONE 2014, 9, e86503. [Google Scholar] [CrossRef]

| Gene | Efficiency (%) | R2 |
|---|---|---|
| RPL18 | 108.6 | 0.991 |
| RPS31 | 107.3 | 0.997 |
| UBC5A | 106.1 | 0.997 |
| GAPDH | 95.6 | 0.995 |
| EF-1α | 95.5 | 0.998 |
| RPS3 | 98.6 | 0.998 |
| RPS18 | 96.5 | 0.998 |
| Parameter | GAPDH | EF1α | RPS3 | RPS18 | UBC5A | RPL18 | RPS31 |
|---|---|---|---|---|---|---|---|
| std dev [+/− CP] | 1.1 | 0.38 | 0.3 | 0.19 | 0.76 | 0.88 | 0.38 |
| CV [% CP] | 5.64 | 2.12 | 1.53 | 0.92 | 3.1 | 3.64 | 1.95 |
| p-value | 0.056 | 0.02 | 0.001 | 0.045 | 0.464 | 0.041 | 0.006 |
| Method | 1 | 2 | 3 | 4 | 5 | 6 | 7 |
|---|---|---|---|---|---|---|---|
| Delta CT | RPS3 | RPS31 | EF1α | RPS18 | RPL18 | UBC5A | GAPDH |
| BestKeeper | RPS18 | RPS3 | RPS31 | EF1α | UBC5A | RPL18 | GAPDH |
| Normfinder | RPS31 | RPS3 | EF1α | RPS18 | RPL18 | UBC5A | GAPDH |
| Genorm | EF1α|RPS3 | RPS31 | RPS18 | RPL18 | UBC5A | GAPDH | |
| OVERALL | RPS3 | RPS31 | EF1α | RPS18 | RPL18 | UBC5A | GAPDH |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2026 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license.
Share and Cite
Zhao, S.-H.; Yue, Y.; Yu, R.-T.; Gao, Q.; Zhao, J.-Q.; Zhang, S.-P.; Zhou, N.; Xu, G.-L. Validation of Stable Reference Genes for RT-qPCR Normalization in Oxycetonia jucunda (Coleoptera: Scarabaeidae). Insects 2026, 17, 57. https://doi.org/10.3390/insects17010057
Zhao S-H, Yue Y, Yu R-T, Gao Q, Zhao J-Q, Zhang S-P, Zhou N, Xu G-L. Validation of Stable Reference Genes for RT-qPCR Normalization in Oxycetonia jucunda (Coleoptera: Scarabaeidae). Insects. 2026; 17(1):57. https://doi.org/10.3390/insects17010057
Chicago/Turabian StyleZhao, Shi-Hang, Yang Yue, Rui-Tao Yu, Qi Gao, Jia-Qiang Zhao, Sheng-Ping Zhang, Nan Zhou, and Guo-Liang Xu. 2026. "Validation of Stable Reference Genes for RT-qPCR Normalization in Oxycetonia jucunda (Coleoptera: Scarabaeidae)" Insects 17, no. 1: 57. https://doi.org/10.3390/insects17010057
APA StyleZhao, S.-H., Yue, Y., Yu, R.-T., Gao, Q., Zhao, J.-Q., Zhang, S.-P., Zhou, N., & Xu, G.-L. (2026). Validation of Stable Reference Genes for RT-qPCR Normalization in Oxycetonia jucunda (Coleoptera: Scarabaeidae). Insects, 17(1), 57. https://doi.org/10.3390/insects17010057





