Simple Summary
The Asian spongy moth is one of the most severe forest pests, having experienced dramatic outbreaks that have led to severe defoliation in Korea. We assessed the genetic diversity of this species. We collected 123 sequences from Korea and adjacent countries, and the collecting locations cover the whole distribution of the Asian spongy moth in Korea. Our results revealed low nucleotide diversity and high haplotype diversity within Korea. Two haplogroups, the Middle and Southern haplogroups, were identified in Korea. Their genetic relationships with other haplotypes and their geographic distribution suggest that Korean populations have been influenced by human activities such as international trade.
Abstract
In this study, we assessed the genetic diversity of the Asian spongy moth, Lymantria dispar asiatica Vnukovskii (Lepidoptera: Erebidae), in Korea. We obtained 123 sequences of the species, including those from 26 regions in Korea. We analyzed the genetic diversity within the Korean samples (n = 87) and haplotype networks between the Korean and global samples (123 sequences from 26 regions in Korea and five other countries) using median-joining (MJ) network analysis. The results showed low nucleotide diversity and high haplotype diversity (π = 0.00159; Hd = 0.660). The neutrality tests were also significantly negative. The MJ network recovered a star-shaped network with diverse populations in Korea, with 12 haplotypes and a dominating haplotype, H07, in all regions. Two haplogroups, Middle and Southern, were identified. The Middle haplogroup included haplotypes from Shandong, China, indicating shared populations between the two regions. In contrast, the Southern haplogroup, primarily found in ship and harbor samples, likely originated from invasive populations. This pattern reflects the influence of human activities such as international trade, highlighting the importance of strict monitoring at ports to prevent the introduction of invasive pests and to support effective forest pest management.
1. Introduction
Lymantria dispar (Lepidoptera: Erebidae) is a polyphagous insect that feeds on more than 100 families of deciduous trees and conifers [1,2,3]. Infested trees suffer severe defoliation and, in some cases, may die [2]. Due to its broad host range and voracious feeding habits, an outbreak of the Asian spongy moth, Lymantria dispar asiatica Vnukovskij, in 2020 caused severe damage to forests in the Korean Peninsula [4]. In addition, the European subspecies, Lymantria dispar dispar (Linnaeus), has accidentally invaded non-native regions, such as North America [5,6,7,8]. The introduced population has become a serious pest, and this species has been listed among the world’s most severe alien species [9].
Because of the ecological and economic impacts, numerous studies have focused on understanding its genetic characteristics. George [10] revealed that the Canadian populations originated from France. Bogdanowicz et al. [11] identified four groups using mitochondrial DNA among global populations: Okinawa (Japan), Hokkaido (Japan), the rest of Japan + mainland Asia, and Europe + Tunisia + North America. Keena et al. [12] separated the spongy moth into three groups: North American, European/Siberian, and Asian. deWaard et al. [13] constructed a COI barcode reference library and identified three clades within Lymantria dispar: North America + France (L. d. dispar), Europe + West Asia (L. d. dispar), and East Asia (L. d. asiatica/japonica). Qian et al. [14] examined COI gene differences among subspecies, supporting the division into three subspecies. Wu et al. [5] analyzed the genetic structure, identified four distinct clusters (three subspecies and the North American population), and reported increased generic variation in East Asia. Kang et al. [15] focused on populations in Far East Asia, revealing three genetic groups within Eastern Asia (central Korea and adjacent regions, southern Korea, and Hokkaido) and suggesting a mixed genetic pattern linked to expansions during the Würm glacial period. Zhao et al. [16] analyzed Chinese populations and found that none of the three subspecies were monophyletic. They also revealed four clusters within China and an independent Black Sea–Caspian genetic lineage. Zahiri et al. [17] studied the global phylogeography of Lymantria dispar using mitochondrial and nuclear genes, identifying three mtDNA lineages, Transcaucasia, East Asia + Japan, and Europe + Central Asia, suggesting the Transcaucasia region as the ancestral area for the entire dispar group.
While numerous studies have examined the genetic characteristics of this species, only a few have included Korean populations. Currently, there is no research based on samples collected across South Korea. Given that the Korean Peninsula may serve as a geographic bridge connecting the Japanese archipelago and the Asian continent, understanding the genetic diversity of the Korean population is important. In addition to this geographic role, the Korean population has expanded and dispersed beyond its original habitats due to recent outbreaks. Moreover, due to their habit of ovipositing on various substrates, such as tree trunks and cargo, spongy moth egg masses are easily transported internationally via cargo ships and other vessels. Invasive populations from other countries have been detected on vessels during quarantine inspections, suggesting the possibility of interactions between native and alien populations. Therefore, we expect that both the outbreak and introductions of alien populations have affected the genetic diversity of this species in Korea. In this study, we aim to characterize the genetic diversity of the Korean population and to examine the relationships between the Korean and global populations. This assessment is essential for understanding population relationships, monitoring the effects of recent outbreaks, and informing management strategies to prevent further spread.
2. Materials and Methods
We obtained COI sequences of L. dispar asiatica from 17 different regions in Korea and also downloaded them from GenBank (NCBI) (Table 1 and Figure 1). To minimize sampling bias, no more than five sequences from the same locality were included. The origins of ship-collected samples were verified by the Animal and Plant Quarantine Agency, Korea (APQA). In total, 123 sequences from 26 regions in Korea and five other countries were analyzed.

Table 1.
Geographic origin and number of samples analyzed.
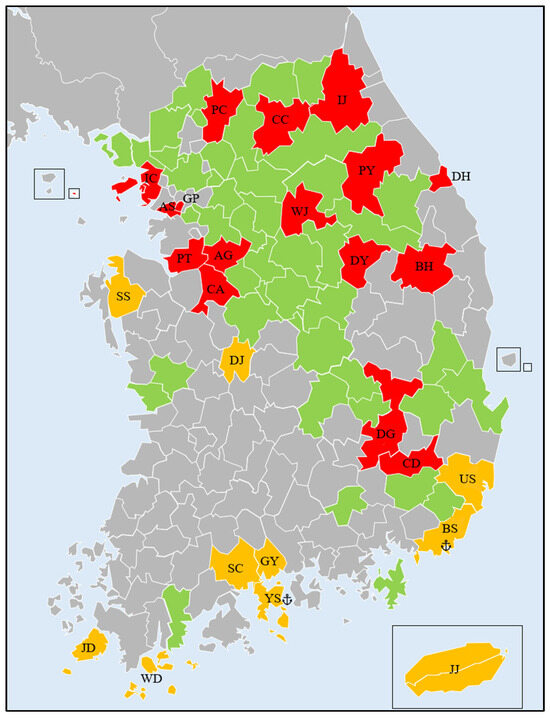
Figure 1.
Sampling locations in Korea. The green areas represent the known distribution of Lymantria dispar asiatica, as reported by Jung et al. [20]. The red colors indicate sampling locations within this known distribution range. The orange colors show sampling locations outside the previously reported distribution. Anchor marks indicate harbors.
DNA extraction was performed using the QIAamp DNA Mini Kit (Qiagen, Hilden, Germany), and PCR was conducted using Solg 2X Taq PCR Pre-mix (SolGent Co., Ltd., Daejeon, Republic of Korea) with the primer set LepF1/LepR1 [21] to amplify the cytochrome oxidase I (COI) gene. PCR products were sequenced by Macrogen Inc. (Seoul, Republic of Korea), and all sequences were submitted to NCBI GenBank (Supplementary Table S1).
Indices of genetic diversity and neutrality statistics (Tajima’s D, Fu and Li’s D*, and Fu and Li’s F*) were calculated using DnaSP v6.12.03 software [22]. Median-joining (MJ) network analysis [23] was performed using NETWORK ver. 10.2.0.0 (http://www.fluxus-engineering.com), with transversion being weighted twice as transition in the analysis, and highly variable characters were down-weighted to resolve high-dimensional cubes and large network cycles. The analysis was conducted using an epsilon value of 70 to ensure a full median network based on the range of genetic distances in the dataset. Maximum parsimony (MP) calculation [24] was applied to generate the shortest network tree.
Phylogenetic analysis was performed using RAxML v.8.2.12 [25] through raxmlGUI 2.0.13 [26]. Codon position partitioning and the substitution model (GTR + G) were selected based on results from PartitionFinder 2 v.2.1.1 [27].
3. Results
The Korean samples of Lymantria dispar asiatica (n = 84; L = 657 bp) showed low nucleotide diversity (π = 0.00159 ± 0.00022 SE) but high haplotype diversity (Hd = 0.660 ± 0.053 SD). Neutrality tests were significantly negative (Tajima’s D = −1.84565, p < 0.05; Fu and Li’s D* = −3.11568, p < 0.05; Fu and Li’s F* = −3.16555, p < 0.02).
The MJ analysis identified 20 haplotypes of Lymantria dispar asiatica (Figure 2 and Table 2; see also Supplementary Table S2 for detailed information). Among these, 12 haplotypes were found within Korean sequences. The most common haplotype, H07, appeared in 85 sequences from all countries, except Kyrgyzstan. This haplotype spanned 21 regions across South Korea, representing the entire country. The haplotype from Kyrgyzstan was the most genetically distant from H07, differing by four characters. Within the Korean sequences, the most distant haplotype was H20, which differed by three characters from H07.
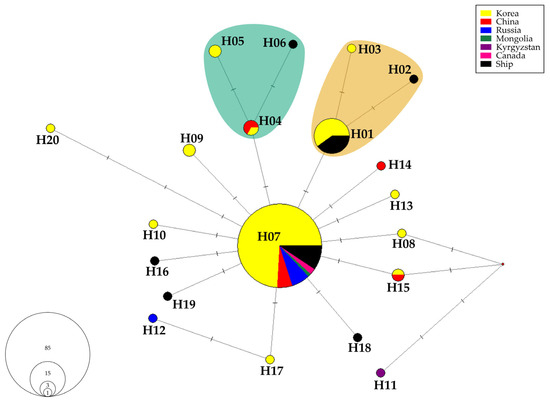
Figure 2.
MJ network of Lymantria dispar asiatica haplotypes in Korea and adjacent countries. Haplotypes are color-coded by the country of collection. Mutational differences between haplotypes are indicated along the branches. The Southern haplogroup is shown as the orange cluster, and the Middle haplogroup is shown as the teal cluster.

Table 2.
Haplogroups and haplotypes of Lymantria dispar asiatica, with their distribution.
The second most frequent haplotype, H01, was identified in 15 sequences from Korea and ships. H01 was present in the southern regions of Korea, including Busan, Cheongdo, Daegu, Suncheon, and Yeosu (Figure 3). H01 differed by one character from haplotypes H02 and H03, which were collected from the southern regions. These three closely related haplotypes were designated as the Southern haplogroup.
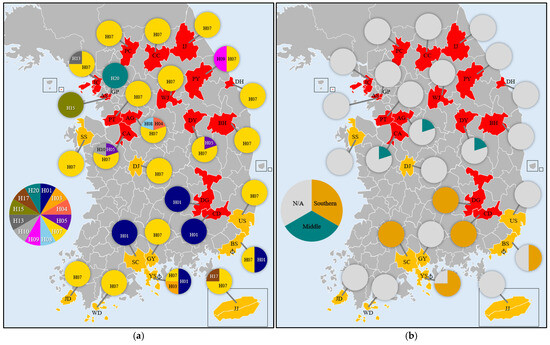
Figure 3.
Distribution of haplotypes and haplogroups of Lymantria dispar asiatica in Korea. (a) Haplotype distribution: each color represents a distinct haplotype. (b) Haplogroup distribution: orange indicates the Southern haplogroup and teal indicates the Middle haplogroup. Anchor marks indicate harbors.
Haplotypes H04 and H05, collected from similar latitudes in Korea, differed by one character. Haplotype H06 also differed from H04 by one character (Figure 3). These three haplotypes (H04, H05, and H06) were classified as the Middle haplogroup.
To further assess the clustering of haplotypes, ML analysis was conducted. The resulting ML tree showed distinct clustering of the two haplogroups, although the backbone of the tree was not supported (Supplementary Figure S1).
4. Discussion
The results revealed 12 haplotypes from Korea, highlighting the diverse populations of Lymantria dispar asiatica in the region. Despite this diversity, the genetic variation within Korean samples appeared low. Most sequences were grouped under a single haplotype, H07, while the remaining sequences were distinct, exhibiting several differences from H07. To minimize bias from the sample size, we included a maximum of five sequences per region. Nevertheless, haplotype H07 was present across all regions of Korea and in neighboring countries. This suggests that multiple invasive populations may have been introduced into Korea through human activities such as international trade and travel.
The low genetic diversity observed in Korean L. dispar asiatica populations may be attributed to the outbreak of this species in 2020. Due to their flight capability and broad range of host plants, these moths rapidly spread across the Korean Peninsula. This extensive dispersal likely contributed to the reduced genetic diversity within the population. This interpretation is supported by the pattern of low nucleotide diversity, high haplotype diversity, significantly negative neutrality tests (Tajima’s D, Fu and Li’s D*, and F*), and a star-shaped haplotype network.
The Southern haplogroup primarily consisted of samples collected from ships and harbors, suggesting a potential origin from invasive haplotypes. In contrast, the Middle haplogroup comprised sequences from Korea’s central region and the Shandong region of China. The proximity of Shandong to Korea, with its peninsula extending into the Yellow Sea, suggests a shared population across these regions. This geographical connection suggests that L. dispar asiatica populations in central Korea may be genetically similar to those in Shandong, regardless of their origin.
A previous study by Kang et al. [15] investigated the genetic structure of this species in Korea and neighboring countries, identifying three haplogroups: Group 1 (Korean in-land and adjacent areas), Group 2 (southern Korea), and Group 3 (Hokkaido). Although the specific genetic differences between groups were not detailed, the distinction between southern and inland haplotypes was evident. The presence of the southern haplotype group aligns with the Southern haplogroup identified in our study.
Haplotype H11, originating from Kyrgyzstan, exhibited the greatest genetic distance from the dominant H07 haplotype. Zahiri et al. [17] described three major lineages of Lymantria dispar, irrespective of subspecies: the “Europe + Central Asia lineage,” the “East Asia + Japan lineage,” and the “Transcaucasia lineage.” Although our study focused exclusively on L. d. asiatica, the sampling range spanned from Central to East Asia. The genetic divergence observed between H11 and the other haplotypes supports the lineage distinctions proposed by Zahiri et al. [17].
5. Conclusions
This study characterized the genetic diversity of Lymantria dispar asiatica populations in Korea. The results revealed low nucleotide diversity but high haplotype diversity, suggesting that these populations have expanded recently and have been affected by alien populations. These findings emphasize the need for strict monitoring at ports to prevent the introduction of invasive pests and the importance of forest pest management. This study was limited to mitochondrial COI fragments. Future studies should include nuclear markers like microsatellites or SNPs to achieve higher resolution. In addition, broader sampling across East Asia and temporal monitoring of haplotypes should be conducted to better understand and track ongoing changes in haplotype composition.
Supplementary Materials
The following supporting information can be downloaded at https://www.mdpi.com/article/10.3390/insects16090958/s1: Figure S1: ML tree of Lymantria dispar asiatica; Table S1: Sample information and accession numbers of Lymantria dispar asiatica; Table S2: Designation of haplogroups and haplotypes in Lymantria dispar asiatica.
Author Contributions
Conceptualization, S.J. and H.-S.L.; investigation, J.B. and H.-M.B.; resources, J.B., H.-M.B., S.C., M.K., G.J., E.K., and J.P.; data curation, H.-M.B., H.-S.L., and S.J.; writing, J.B.; visualization, J.B.; supervision, S.J.; project administration, H.-S.L.; funding acquisition, S.J. All authors have read and agreed to the published version of the manuscript.
Funding
This research was supported by the Research and Development (R&D) project (Z-1543086-2024-25-02) on the animal and plant quarantine inspection technology of the Animal and Plant Quarantine Agency in the Republic of Korea.
Data Availability Statement
All sequences used in this study are available in NCBI GenBank.
Conflicts of Interest
The authors declare no conflicts of interest.
Abbreviations
The following abbreviations are used in this manuscript:
| AG | Anseong-si |
| AS | Ansan-si |
| BH | Bonghwa-gun |
| BS | Busan |
| CA | Cheonan-si |
| CB | Chungcheongbuk-do |
| CC | Chuncheon-si |
| CD | Cheongdo-gun |
| CN | Chungcheongnam-do |
| DG | Daegu |
| DH | Donghae-si |
| DJ | Daejeon |
| DY | Danyang-gun |
| GB | Gyeongsangbuk-do |
| GG | Gyeonggi-do |
| GP | Gunpo-si |
| GW | Gangwon-do |
| GY | Gwangyang-si |
| IC | Incheon |
| IJ | Inje-gun |
| JD | Jindo-gun |
| JJ | Jeju island |
| JN | Jeollanam-do |
| PC | Pocheon-si |
| PT | Pyeongtaek-si |
| PY | Pyeongchang-gun |
| SC | Suncheon-si |
| SS | Seosan-si |
| US | Ulsan |
| WD | Wando-gun |
| WJ | Wonju-si |
| YS | Yeosu-si |
References
- Savotikov, I.F.; Smetnik, A.I.; Orlinskii, A.D. Situation of the Asian form of gypsy moth (Lymantria dispar) in Russia and in the world. EPPO Bull. 1995, 25, 617–622. [Google Scholar] [CrossRef]
- Marr, J. Gypsy Moth Monitoring Program: Town of Pelham: 2020 Population Assessments and 2021 Forecasts; Bioforest: Sault Ste. Marie, ON, Canada, 2021; pp. 1–42. [Google Scholar]
- Ahn, S.; Lee, S.; Kim, J.; Nam, Y.; Choi, S.; Jung, J.-K. Occurrence of Lepidopteran Insects in Urban Forests. Korean J. Appl. Entomol. 2022, 61, 481–496. [Google Scholar] [CrossRef]
- Jung, J.-K.; Nam, Y.; Kim, D.; Lee, S.-H.; Lim, J.-H.; Choi, W.I.; Kim, E.-S. Tree-crown Defoliation caused by Outbreak of Forest Insect Pests in Korea during 2020. Korean J. Appl. Entomol. 2020, 59, 409–410. [Google Scholar] [CrossRef]
- Wu, Y.; Molongoski, J.J.; Winograd, D.F.; Bogdanowicz, S.M.; Louyakis, A.S.; Lance, D.R.; Mastro, V.C.; Harrison, R.G. Genetic structure, admixture and invasion success in a Holarctic defoliator, the gypsy moth (Lymantria dispar, Lepidoptera: Erebidae). Mol. Ecol. 2015, 24, 1275–1291. [Google Scholar] [CrossRef] [PubMed]
- Pogue, M.G.; Schaefer, P.W. A Review of Selected Species of Lymantria Hübner (1819) (Lepidoptera: Noctuidae: Lymantriinae) from Subtropical and Temperate Regions of Asia, Including the Descriptions of Three New Species, Some Potentially Invasive to North America; Forest Health Technology Enterprise Team, U.S. Department of Agriculture: Washington, DC, USA, 2007; p. 221.
- Roy, A.S.; McNamara, D.G.; Smith, I.M. Situation of Lymantria dispar in Europe. Bull. OEPP/EPPO Bull. 1995, 25, 611–616. [Google Scholar] [CrossRef]
- Leonard, D.E. Recent Developments in Ecology and Control of the Gypsy Moth. Annu. Rev. Entomol. 1974, 19, 197–229. [Google Scholar] [CrossRef]
- Lowe, S.; Browne, M.; Boudjelas, S.; De Poorter, M. 100 of the World’s Worst Invasive Alien Species A selection from the Global Invasive Species Database; ISSG: Auckland, New Zealand, 2000; p. 12. [Google Scholar]
- George, C. Allozyme variation in natural populations of Lymantria dispar (Lepidoptera). Genet. Sel. Evol. 1984, 16, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Bogdanowicz, S.M.; Schaefer, P.W.; Harrison, R.G. Mitochondrial DNA variation among worldwide populations of gypsy moths, Lymantria dispar. Mol. Phylogenet. Evol. 2000, 15, 487–495. [Google Scholar] [CrossRef]
- Keena, M.A.; Côté, M.-J.; Grinberg, P.S.; Wallner, W.E. World Distribution of Female Flight and Genetic Variation in Lymantria dispar (Lepidoptera: Lymantriidae). Environ. Entomol. 2008, 37, 636–649. [Google Scholar] [CrossRef]
- deWaard, J.R.; Mitchell, A.; Keena, M.A.; Gopurenko, D.; Boykin, L.M.; Armstrong, K.F.; Pogue, M.G.; Lima, J.; Floyd, R.; Hanner, R.H.; et al. Towards a global barcode library for Lymantria (Lepidoptera: Lymantriinae) tussock moths of biosecurity concern. PLoS ONE 2010, 5, e14280. [Google Scholar] [CrossRef]
- Qian, L.; An, Y.; Song, J.; Xu, M.; Ye, J.; Wu, C.; Li, B.; Hao, D. COI gene geographic variation of Gypsy moth (Lepidoptera: Lymantriidae) and a TaqMan PCR diagnostic assay. DNA Barcodes 2014, 2, 10–16. [Google Scholar] [CrossRef]
- Kang, T.H.; Han, S.H.; Lee, H.S. Genetic structure and demographic history of Lymantria dispar (Linnaeus, 1758) (Lepidoptera: Erebidae) in its area of origin and adjacent areas. Ecol. Evol. 2017, 7, 9162–9178. [Google Scholar] [CrossRef]
- Zhao, J.; Wu, Y.; Kurenshchikov, D.K.; Ilyinykh, A.V.; Shi, J. Underestimated mitochondrial diversity in gypsy moth Lymantria dispar from Asia. Agric. For. Entomol. 2019, 21, 235–242. [Google Scholar] [CrossRef]
- Zahiri, R.; Christian Schmidt, B.; Schintlmeister, A.; Yakovlev, R.V.; Rindos, M. Global phylogeography reveals the origin and the evolutionary history of the gypsy moth (Lepidoptera, Erebidae). Mol. Phylogenet. Evol. 2019, 137, 1–13. [Google Scholar] [CrossRef]
- Kang, T.H.; Lee, K.-s.; Lee, H.-s. DNA Barcoding of the Korean Lymantria Hubner, 1819 (Lepidoptera: Erebidae: Lymantriinae) for Quarantine Inspection. J. Econ. Entomol. 2015, 108, 1596–1611. [Google Scholar] [CrossRef]
- Stewart, D.; Zahiri, R.; Djoumad, A.; Freschi, L.; Lamarche, J.; Holden, D.; Cervantes, S.; Ojeda, D.I.; Potvin, A.; Nisole, A.; et al. A Multi-Species TaqMan PCR Assay for the Identification of Asian Gypsy Moths (Lymantria spp.) and Other Invasive Lymantriines of Biosecurity Concern to North America. PLoS One 2016, 11, e0160878. [Google Scholar] [CrossRef]
- Jung, J.-K.; Lee, C.Y.; Nam, Y.; Jung, B.N.; Park, J.H.; Han, H.R.; Lee, S.H. 매미나방 예찰 및 방제 요령; National Institute of Forest Science: Seoul, Republic of Korea, 2021; p. 15. [Google Scholar]
- Hebert, P.D.N.; Penton, E.H.; Burns, J.M.; Janzen, D.H.; Hallwachs, W. Ten species in one: DNA barcoding reveals cryptic species in neotropical skipper butterfly Astraptes fulgerator. Proc. Natl. Acad. Sci. USA 2004, 101, 14812–14817. [Google Scholar] [CrossRef]
- Rozas, J.; Ferrer-Mata, A.; Sanchez-DelBarrio, J.C.; Guirao-Rico, S.; Librado, P.; Ramos-Onsins, S.E.; Sanchez-Gracia, A. DnaSP 6: DNA Sequence Polymorphism Analysis of Large Data Sets. Mol. Biol. Evol. 2017, 34, 3299–3302. [Google Scholar] [CrossRef] [PubMed]
- Bandelt, H.-J.; Forster, P.; Röhl, A. Median-Joining Networks for Inferring Intraspecific Phylogenies. Mol. Biol. Evol. 1999, 16, 37–48. [Google Scholar] [CrossRef] [PubMed]
- Polzin, T.; Daneshmand, S.V. On Steiner trees and minimum spanning trees in hypergraphs. Oper. Res. Lett. 2003, 31, 12–20. [Google Scholar] [CrossRef]
- Stamatakis, A. RAxML version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 2014, 30, 1312–1313. [Google Scholar] [CrossRef] [PubMed]
- Edler, D.; Klein, J.; Antonelli, A.; Silvestro, D.; Matschiner, M. raxmlGUI 2.0: A graphical interface and toolkit for phylogenetic analyses using RAxML. Methods Ecol. Evol. 2020, 12, 373–377. [Google Scholar] [CrossRef]
- Lanfear, R.; Frandsen, P.B.; Wright, A.M.; Senfeld, T.; Calcott, B. PartitionFinder 2: New Methods for Selecting Partitioned Models of Evolution for Molecular and Morphological Phylogenetic Analyses. Mol. Biol. Evol. 2017, 34, 772–773. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).