A Review of the Newly Recorded Genus Proceroplatus Edwards, 1925 (Diptera: Keroplatidae) in China with Two New Species, and Its Characterization and Phylogenetic Implication of Mitogenomes
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Morphological and Molecular Identification
2.2. Mitochondrial Genome Assembly, Annotation, and Analysis
2.3. Phylogenetic Analyses
3. Results
3.1. Generic Characters
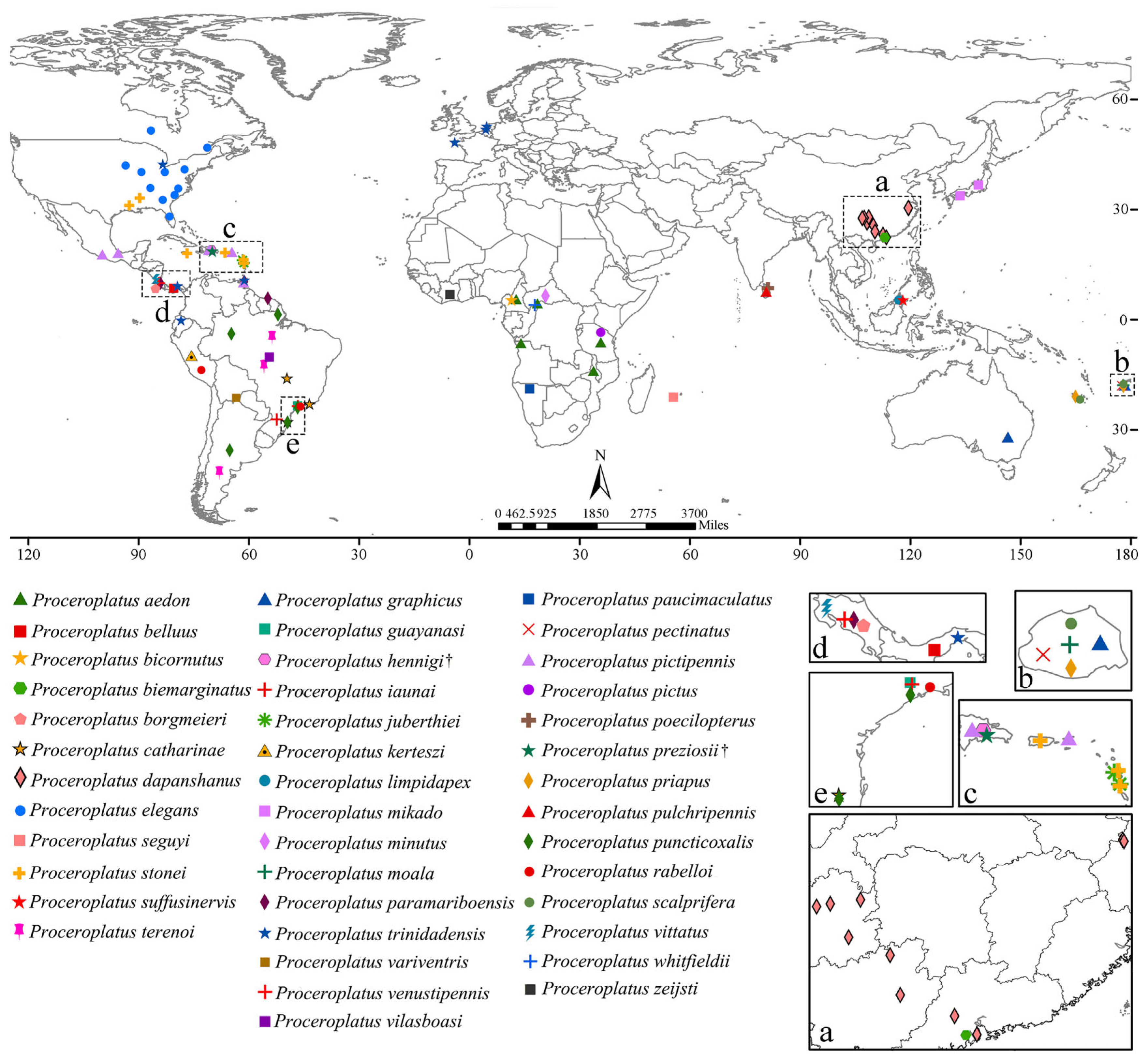
3.2. Species and Distribution
3.3. Key to the Species of Proceroplatus in Asia (Based on Adults)
- 1. Thorax with streaks on scutum ·····················································································································································································································2
- - Thorax without streaks on scutum ·················································································································································································································4
- 2. Scutum with a pair of ill-defined linear streaks; terminalia black ··········································································································P. pulchripennis (Senior-White)
- - Scutum with nonlinear streaks; terminalia brown ·······················································································································································································3
- 3. Two ocelli; scutum with a U-shaped dark streak; Sc reaching Costa; male cercus kidney-shaped ······································································P. dapanshanus sp. n.
- - Three ocelli; scutum without U-shaped streak; Sc not reaching Costa; cercus suboval························································································ P. biemarginatus sp. n.
- 4. Head wholly black; a narrow stripe from middle of R4+5 to middle of M1····································································································· P. suffusinervis (Brunetti)
- - Head brown or other colors; no stripe from middle of R4+5 to middle of M1············································································································································5
- 5. Costa reaching at or near 3/4 distance from R4+5 to M1··············································································································································································6
- - Costa reaching at 1/2 distance from R4+5 to M1; wing tip with patterns·············································································································P. poecilopterus Edwards
- 6. Wings with three brown stripes················································································································································································P. limidapex (Edwards)
- - Wings with a stripe and a rounded spot············································································································································································P. mikado (Okada)
3.4. Description of New Species
3.4.1. Proceroplatus dapanshanus Wang et Huang sp. n.
- urn:lsid:zoobank.org:act:05B6A1C5-2DD1-4DD9-ABAE-FC56EF8CA6D5
3.4.2. Proceroplatus biemarginatus Wang et Huang sp. n.
- urn:lsid:zoobank.org:act:C384DFED-2443-4666-BCFF-A3FA6F7E752E
3.5. Molecular Identification
3.6. Mitogenomic Characteristics
3.6.1. Base Composition
3.6.2. tRNA Structure Prediction
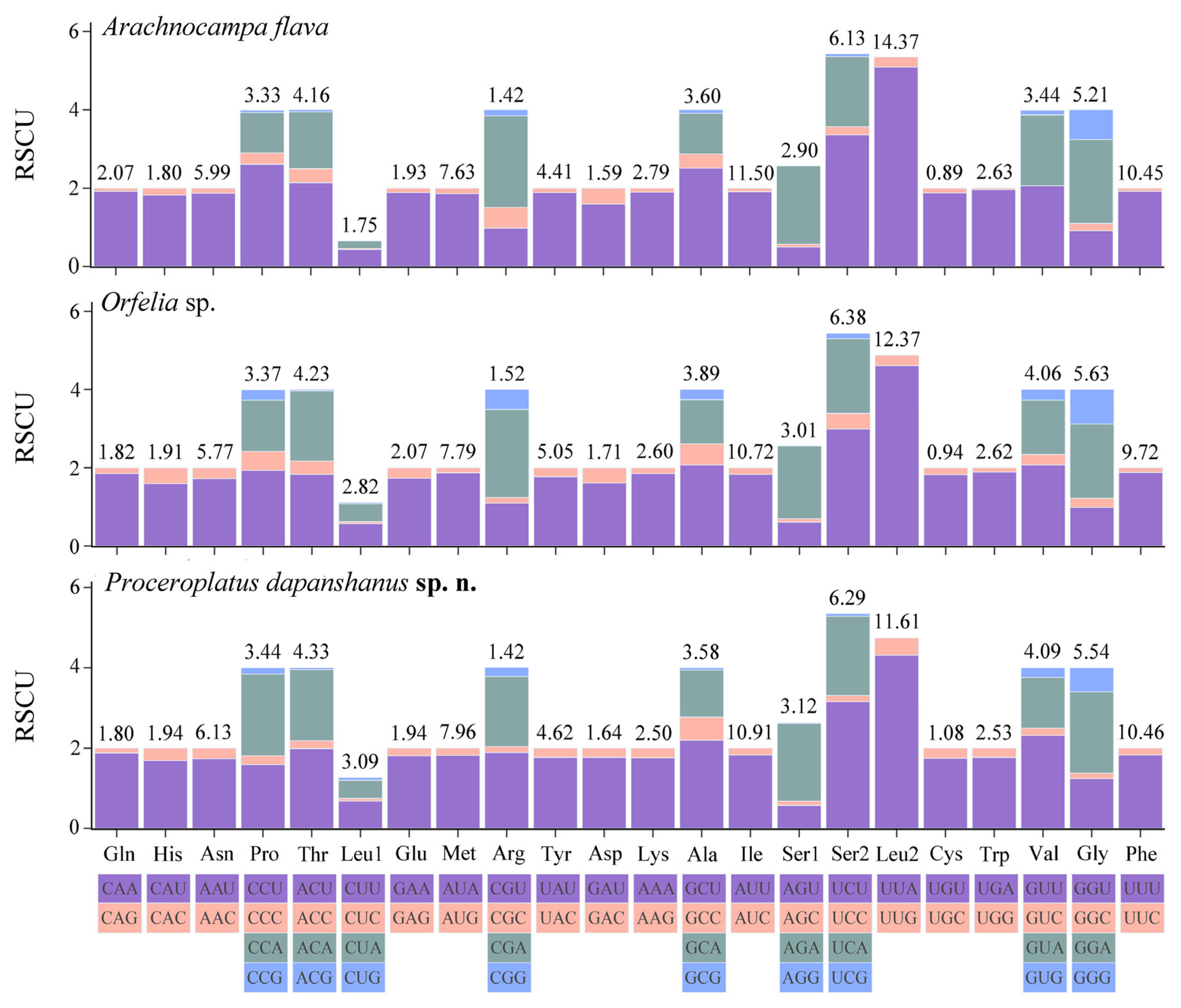
3.6.3. Relative Synonymous Codon Usage (RSCU)
3.6.4. ENc-Plot, PR2-Plot and Neutrality-Plot Analyses
3.7. Phylogeny and Gene Arrangements
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Matile, L. A New Neotropical Fungus Gnat (Diptera: Sciaroidea: Keroplatidae) with Myrmecophagous Larvae. J. N. Y. Entomol. Soc. 1997, 104, 216–220. [Google Scholar]
- Aiello, A.; Jolivet, P. Myrmecophily in Keroplatidae (Diptera: Sciaroidea). J. N. Y. Entomol. Soc. 1996, 104, 226–230. [Google Scholar]
- Evenhuis, N.L. Catalog of the Keroplatidae of the World (Insecta: Diptera); Bishop Museum Bulletin in Entomology: Honolulu, HI, USA; Bishop Museum Press: Honolulu, HI, USA, 2006; ISBN 978-1-58178-054-3. [Google Scholar]
- Chandler, P.J.; Pijnakker, J. Tropical Fungus Gnats Established in Nurseries in the Netherlands (Diptera: Keroplatidae and Mycetophilidae). Br. J. Entomol. Nat. Hist. 2009, 22, 1664.1–1664.13. [Google Scholar]
- Mochizuki, K.; Okamoto, T.; Chen, K.-H.; Wang, C.-N.; Evans, M.; Kramer, A.T.; Kawakita, A. Adaptation to Pollination by Fungus Gnats Underlies the Evolution of Pollination Syndrome in the Genus Euonymus. Ann. Bot. 2023, 132, 319–333. [Google Scholar] [CrossRef]
- Edwards, F.W. British Fungus Gnats (Diptera, Mycetophilidae). With a Revised Generic Classification of the Family. Trans. Entomol. Soc. Lond. 1925, 73, 505–670. [Google Scholar] [CrossRef]
- Edwards, F.W. Notes on the Ceroplatinae, with Descriptions of New Australian Species (Diptera, Mycetophilidae). Proc. Linn. Soc. New South Wales 1929, 54, 162–175. [Google Scholar]
- Laffoon, J.L. Family Mycetophilidae (Fungivoridae). In A Catalog of the Diptera of America North of Mexico; United States Department of Agriculture: Washington, DC, USA, 1965; Volume 276, pp. 196–204. [Google Scholar]
- Matile, L. Diptères Mycetophilidae Du Cameroun et de République Centrafricaine. I. Keroplatinae. Bull. Inst. Française Afr. Noire 1970, 32, 773–816. [Google Scholar]
- Matile, L. Diptères Mycetophilidae de Fernando-Póo. Bull. Muséum Natl. Hist. Nat. 1973, 85, 189–213. [Google Scholar] [CrossRef]
- Papavero, N. Family Keroplatidae (Ceroplatidae, Incl. Macroceridae). Cat. Diptera Am. South United States 1978, 19C, 1–22. [Google Scholar]
- Mantič, M.; Sikora, T.; Burdíková, N.; Blagoderov, V.; Kjærandsen, J.; Kurina, O.; Ševčík, J. Hidden in Plain Sight: Comprehensive Molecular Phylogeny of Keroplatidae and Lygistorrhinidae (Diptera) Reveals Parallel Evolution and Leads to a Revised Family Classification. Insects 2020, 11, 348. [Google Scholar] [CrossRef]
- Penney, D.; Evenhuis, N.L.; Green, D.I. A New Species of Proceroplatus Edwards (Diptera: Keroplatidae) in Miocene Amber from the Dominican Republic. Zootaxa 2013, 3686, 593–599. [Google Scholar] [CrossRef]
- Matile, L. Diptères Mycetophiloidea de La Forêt de Taï (Côte d’Ivoire). I. Keroplatidae. Rev. Française Entomol. (n.s.) 1988, 10, 57–79. [Google Scholar]
- Matile, L. Recherches sur la systématique et l’évolution des Keroplatidae (Diptera, Mycetophiloidea). Mémoires Muséum Natl. Hist. Nat. A 1990, 148, 1–682. [Google Scholar]
- Hebert, P.D.N.; Penton, E.H.; Burns, J.M.; Janzen, D.H.; Hallwachs, W. Ten Species in One: DNA Barcoding Reveals Cryptic Species in the Neotropical Skipper Butterfly Astraptes Fulgerator. Proc. Natl. Acad. Sci. USA 2004, 101, 14812–14817. [Google Scholar] [CrossRef]
- Hebert, P.D.N.; Braukmann, T.W.A.; Prosser, S.W.J.; Ratnasingham, S.; deWaard, J.R.; Ivanova, N.V.; Janzen, D.H.; Hallwachs, W.; Naik, S.; Sones, J.E.; et al. A Sequel to Sanger: Amplicon Sequencing That Scales. BMC Genom. 2018, 19, 219. [Google Scholar] [CrossRef] [PubMed]
- Katoh, K.; Standley, D.M. MAFFT Multiple Sequence Alignment Software Version 7: Improvements in Performance and Usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef] [PubMed]
- Stamatakis, A. RAxML Version 8: A Tool for Phylogenetic Analysis and Post-Analysis of Large Phylogenies. Bioinformatics 2014, 30, 1312–1313. [Google Scholar] [CrossRef]
- Meng, G.; Li, Y.; Yang, C.; Liu, S. MitoZ: A Toolkit for Animal Mitochondrial Genome Assembly, Annotation and Visualization. Nucleic Acids Res. 2019, 47, e63. [Google Scholar] [CrossRef]
- Bernt, M.; Donath, A.; Jühling, F.; Externbrink, F.; Florentz, C.; Fritzsch, G.; Pütz, J.; Middendorf, M.; Stadler, P.F. MITOS: Improved de Novo Metazoan Mitochondrial Genome Annotation. Mol. Phylogenetics Evol. 2013, 69, 313–319. [Google Scholar] [CrossRef]
- Darty, K.; Denise, A.; Ponty, Y. VARNA: Interactive Drawing and Editing of the RNA Secondary Structure. Bioinformatics 2009, 25, 1974–1975. [Google Scholar] [CrossRef] [PubMed]
- Zhang, D.; Gao, F.; Jakovlić, I.; Zou, H.; Zhang, J.; Li, W.X.; Wang, G.T. PhyloSuite: An Integrated and Scalable Desktop Platform for Streamlined Molecular Sequence Data Management and Evolutionary Phylogenetics Studies. Mol. Ecol. Resour. 2020, 20, 348–355. [Google Scholar] [CrossRef] [PubMed]
- Xiang, C.; Gao, F.; Jakovlić, I.; Lei, H.; Hu, Y.; Zhang, H.; Zou, H.; Wang, G.; Zhang, D. Using PhyloSuite for Molecular Phylogeny and Tree-based Analyses. iMeta 2023, 2, e87. [Google Scholar] [CrossRef]
- Perna, N.T.; Kocher, T.D. Patterns of Nucleotide Composition at Fourfold Degenerate Sites of Animal Mitochondrial Genomes. J. Mol. Evol. 1995, 41, 353–358. [Google Scholar] [CrossRef]
- Wright, F. The ‘Effective Number of Codons’ Used in a Gene. Gene 1990, 87, 23–29. [Google Scholar] [CrossRef]
- Fuglsang, A. Impact of Bias Discrepancy and Amino Acid Usage on Estimates of the Effective Number of Codons Used in a Gene, and a Test for Selection on Codon Usage. Gene 2008, 410, 82–88. [Google Scholar] [CrossRef]
- Sueoka, N. Intrastrand Parity Rules of DNA Base Composition and Usage Biases of Synonymous Codons. J. Mol. Evol. 1995, 40, 318–325. [Google Scholar] [CrossRef]
- Sueoka, N. Translation-Coupled Violation of Parity Rule 2 in Human Genes Is Not the Cause of Heterogeneity of the DNA G+ C Content of Third Codon Position. Gene 1999, 238, 53–58. [Google Scholar] [CrossRef]
- Wei, L.; He, J.; Jia, X.; Qi, Q.; Liang, Z.; Zheng, H.; Ping, Y.; Liu, S.; Sun, J. Analysis of Codon Usage Bias of Mitochondrial Genome in Bombyx Mori and Its Relation to Evolution. BMC Evol. Biol. 2014, 14, 262. [Google Scholar] [CrossRef]
- Castresana, J. Selection of Conserved Blocks from Multiple Alignments for Their Use in Phylogenetic Analysis. Mol. Biol. Evol. 2000, 17, 540–552. [Google Scholar] [CrossRef] [PubMed]
- Lanfear, R.; Frandsen, P.B.; Wright, A.M.; Senfeld, T.; Calcott, B. PartitionFinder 2: New Methods for Selecting Partitioned Models of Evolution for Molecular and Morphological Phylogenetic Analyses. Mol. Biol. Evol. 2017, 34, 772–773. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, L.-T.; Schmidt, H.A.; Von Haeseler, A.; Minh, B.Q. IQ-TREE: A Fast and Effective Stochastic Algorithm for Estimating Maximum-Likelihood Phylogenies. Mol. Biol. Evol. 2015, 32, 268–274. [Google Scholar] [CrossRef] [PubMed]
- Lane, J. Neotropical Ceroplatinae (Diptera, Mycetophilidae). Dusenia 1950, 1, 32–69. [Google Scholar]
- Papp, L.; Merz, B.; Földvári, M. A Summary of the Families and Genera with References to the Species Representations. Acta Zool. Acad. Sci. Hung. 2006, 52, 97–269. [Google Scholar]
- Gloaguen, P.-Y. Premier signalement du genre Proceroplatus Edwards, 1925 en France métropolitaine (Diptera Keroplatidae). L’Entomologiste 2022, 78, 161–165. [Google Scholar]
- Matile, L. Keroplatinae Des Iles de l’Océan Indien Occidental (Diptera, Mycetophilidae). Cah. Orstom 1972, 16, 105–123. [Google Scholar]
- Matile, L. Keroplatinae de Madagascar (Diptera: Mycetophilidae). Ann. Natal Mus. 1977, 23, 23–26. [Google Scholar]
- Blasgoderov, V.; Ševčík, J. Keroplatinae (Predaceous Fungus Gnats). Nematocerous Diptera and Lower Brachycera. In Manual of Afrotropical Diptera; Kirk-Spriggs, A.H., Sinclair, B.J., Eds.; South African National Biodiversity Institute: Pretoria, South Africa, 2017; Volume 2, pp. 505–525. [Google Scholar]
- Jia, X.; Liu, S.; Zheng, H.; Li, B.; Qi, Q.; Wei, L.; Zhao, T.; He, J.; Sun, J. Non-Uniqueness of Factors Constraint on the Codon Usage in Bombyx Mori. BMC Genom. 2015, 16, 356. [Google Scholar] [CrossRef]
- Abdoli, R.; Mazumder, T.H.; Nematollahian, S.; Zanjani, R.S.; Mesbah, R.A.; Uddin, A. Gaining Insights into the Compositional Constraints and Molecular Phylogeny of Five Silkworms Mitochondrial Genome. Int. J. Biol. Macromol. 2022, 206, 543–552. [Google Scholar] [CrossRef]
- Chen, H.; Sun, S.; Norenburg, J.L.; Sundberg, P. Mutation and Selection Cause Codon Usage and Bias in Mitochondrial Genomes of Ribbon Worms (Nemertea). PLoS ONE 2014, 9, e85631. [Google Scholar] [CrossRef]
- Shi, A.; Li, C.; Farhan, M.; Xu, C.; Zhang, Y.; Qian, H.; Zhang, S.; Jing, T. Characterization, Codon Usage Pattern and Phylogenetic Implications of the Waterlily Aphid Rhopalosiphum Nymphaeae (Hemiptera: Aphididae) Mitochondrial Genome. Int. J. Mol. Sci. 2024, 25, 11336. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Huang, J.; Wu, H. Mitogenomes Provide Insights into the Phylogeny of Mycetophilidae (Diptera: Sciaroidea). Gene 2021, 783, 145564. [Google Scholar] [CrossRef] [PubMed]
- Cameron, S.L. Insect Mitochondrial Genomics: Implications for Evolution and Phylogeny. Annu. Rev. Entomol. 2014, 59, 95–117. [Google Scholar] [CrossRef] [PubMed]
- Cameron, S.L. Insect Mitochondrial Genomics: A Decade of Progress. Annu. Rev. Entomol. 2025, 70, 83–101. [Google Scholar] [CrossRef] [PubMed]






| Species | A+T% | G+C% | AT-Skew | GC-Skew | Length (bp) |
|---|---|---|---|---|---|
| Mitogenome | |||||
| Arachnocampa flava | 82.013 78.912 78.997 | 17.987 | −0.034 | −0.183 | 16,923 |
| Orfelia sp. | 21.088 | 0.025 | −0.188 | 15,521 | |
| Proceroplatus dapanshanus | 21.003 | 0.005 | −0.185 | 15,688 | |
| Protein coding gene (PCG) | |||||
| Arachnocampa flava | 78.932 76.277 76.816 | 21.068 | −0.164 | 0.006 | 11,202 |
| Orfelia sp. | 23.723 | −0.152 | −0.003 | 10,863 | |
| Proceroplatus dapanshanus | 23.184 | −0.162 | −0.002 | 11,193 | |
| Species | Arachnocampa flava | Orfelia sp. | Proceroplatus dapanshanus | ||||
|---|---|---|---|---|---|---|---|
| PCG | GC3 | ENc | GC3 | ENc | GC3 | ENc | |
| ATP6 | 0.092 | 31.80 | 0.086 | 38.43 | 0.089 | 36.30 | |
| COI | 0.083 | 33.43 | 0.099 | 32.02 | 0.092 | 33.62 | |
| COII | 0.079 | 31.49 | 0.105 | 37.38 | 0.089 | 32.99 | |
| COIII | 0.061 | 31.80 | 0.147 | 38.55 | 0.118 | 35.65 | |
| CYTB | 0.061 | 29.50 | 0.116 | 33.38 | 0.129 | 35.76 | |
| ND1 | 0.063 | 31.25 | 0.113 | 31.26 | 0.111 | 32.09 | |
| ND2 | 0.059 | 26.67 | 0.123 | 32.49 | 0.052 | 32.53 | |
| ND3 | 0.110 | 33.17 | 0.097 | 30.44 | 0.110 | 31.77 | |
| ND4 | 0.077 | 32.32 | 0.101 | 35.09 | 0.099 | 34.37 | |
| ND4L | 0.081 | 27.98 | 0.120 | 36.08 | 0.050 | 25.21 | |
| ND5 | 0.067 | 31.70 | 0.105 | 34.47 | 0.127 | 35.12 | |
| ND6 | 0.017 | 26.44 | 0.088 | 36.90 | 0.081 | 29.73 | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, Q.; Zhu, Y.; Yu, Y.; Liu, L.; Wu, H.; Huang, J. A Review of the Newly Recorded Genus Proceroplatus Edwards, 1925 (Diptera: Keroplatidae) in China with Two New Species, and Its Characterization and Phylogenetic Implication of Mitogenomes. Insects 2025, 16, 883. https://doi.org/10.3390/insects16090883
Wang Q, Zhu Y, Yu Y, Liu L, Wu H, Huang J. A Review of the Newly Recorded Genus Proceroplatus Edwards, 1925 (Diptera: Keroplatidae) in China with Two New Species, and Its Characterization and Phylogenetic Implication of Mitogenomes. Insects. 2025; 16(9):883. https://doi.org/10.3390/insects16090883
Chicago/Turabian StyleWang, Qingyun, Yi Zhu, Yefei Yu, Liwei Liu, Hong Wu, and Junhao Huang. 2025. "A Review of the Newly Recorded Genus Proceroplatus Edwards, 1925 (Diptera: Keroplatidae) in China with Two New Species, and Its Characterization and Phylogenetic Implication of Mitogenomes" Insects 16, no. 9: 883. https://doi.org/10.3390/insects16090883
APA StyleWang, Q., Zhu, Y., Yu, Y., Liu, L., Wu, H., & Huang, J. (2025). A Review of the Newly Recorded Genus Proceroplatus Edwards, 1925 (Diptera: Keroplatidae) in China with Two New Species, and Its Characterization and Phylogenetic Implication of Mitogenomes. Insects, 16(9), 883. https://doi.org/10.3390/insects16090883





