Predicting the Potential Geographical Distribution of Scolytus scolytus in China Using a Biomod2-Based Ensemble Model
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
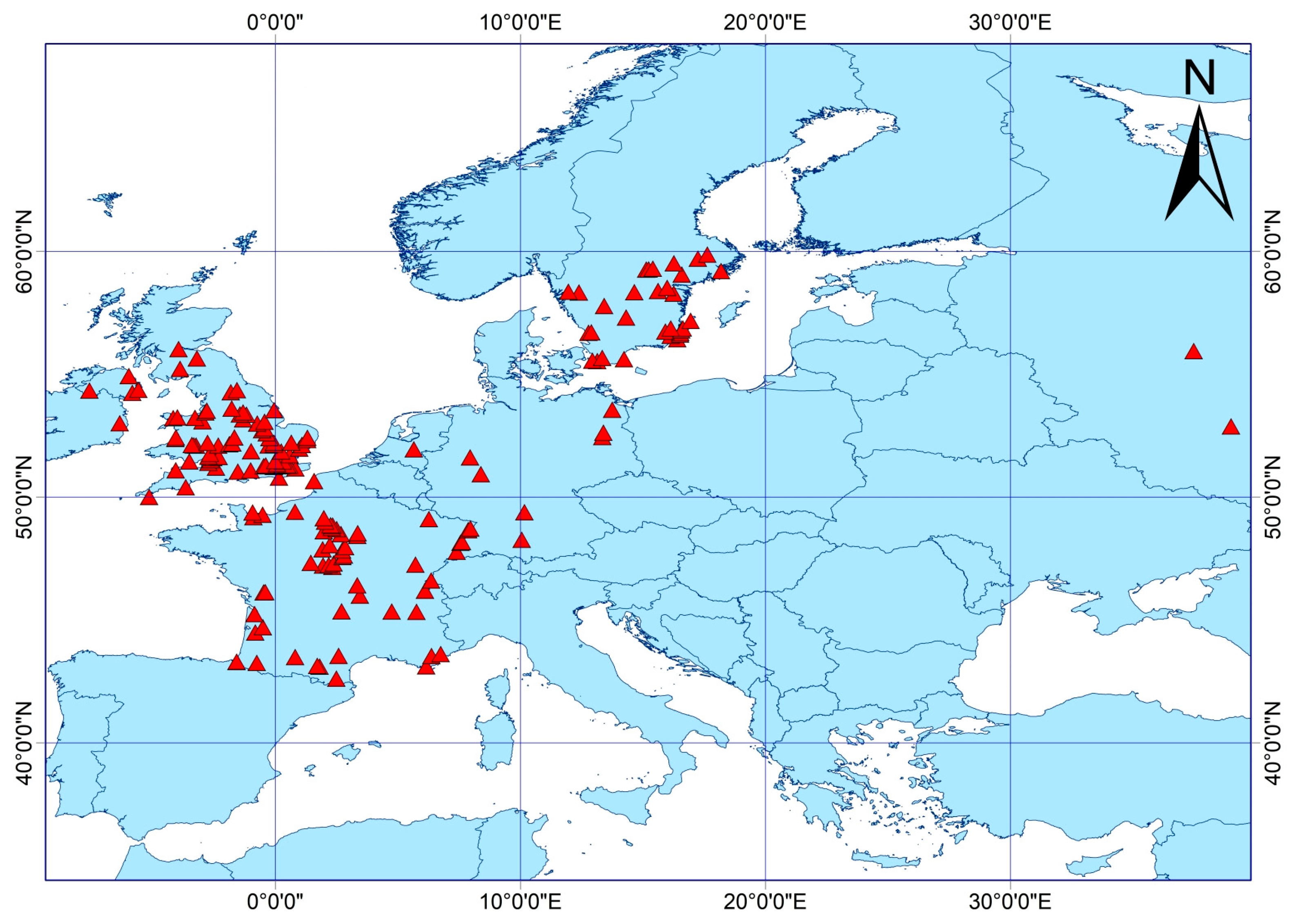
2.1. Environmental and Occurrence Data Collection
2.2. Parameter Setting and Model Ensemble
2.2.1. Environmental Variable Selection
2.2.2. Classification and Threshold Definition of Habitat Suitability
2.2.3. Model Performance Evaluation and Variable Importance
3. Results
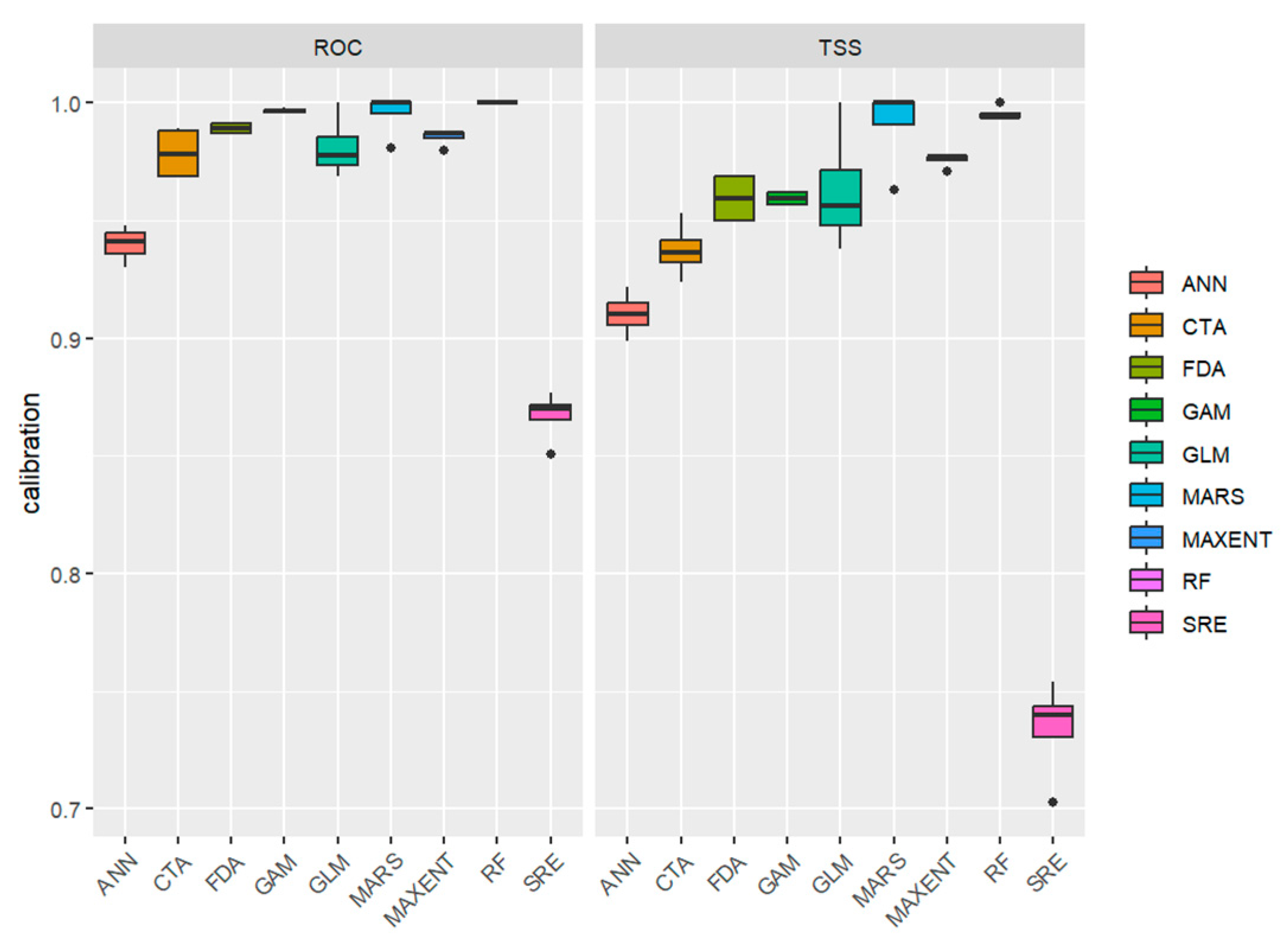
3.1. Evaluation of Model Accuracy
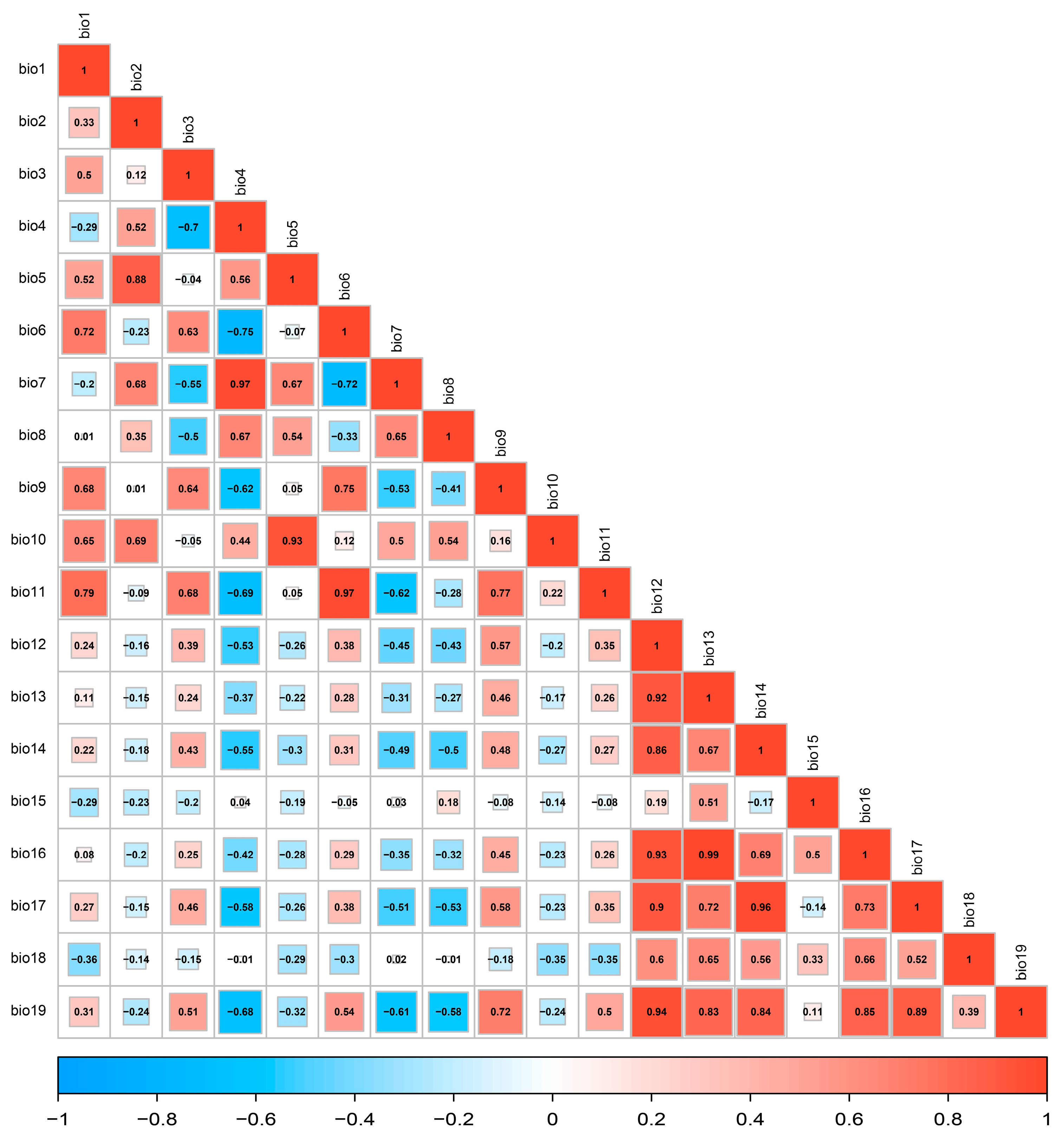
3.2. Screening of Environmental Factors
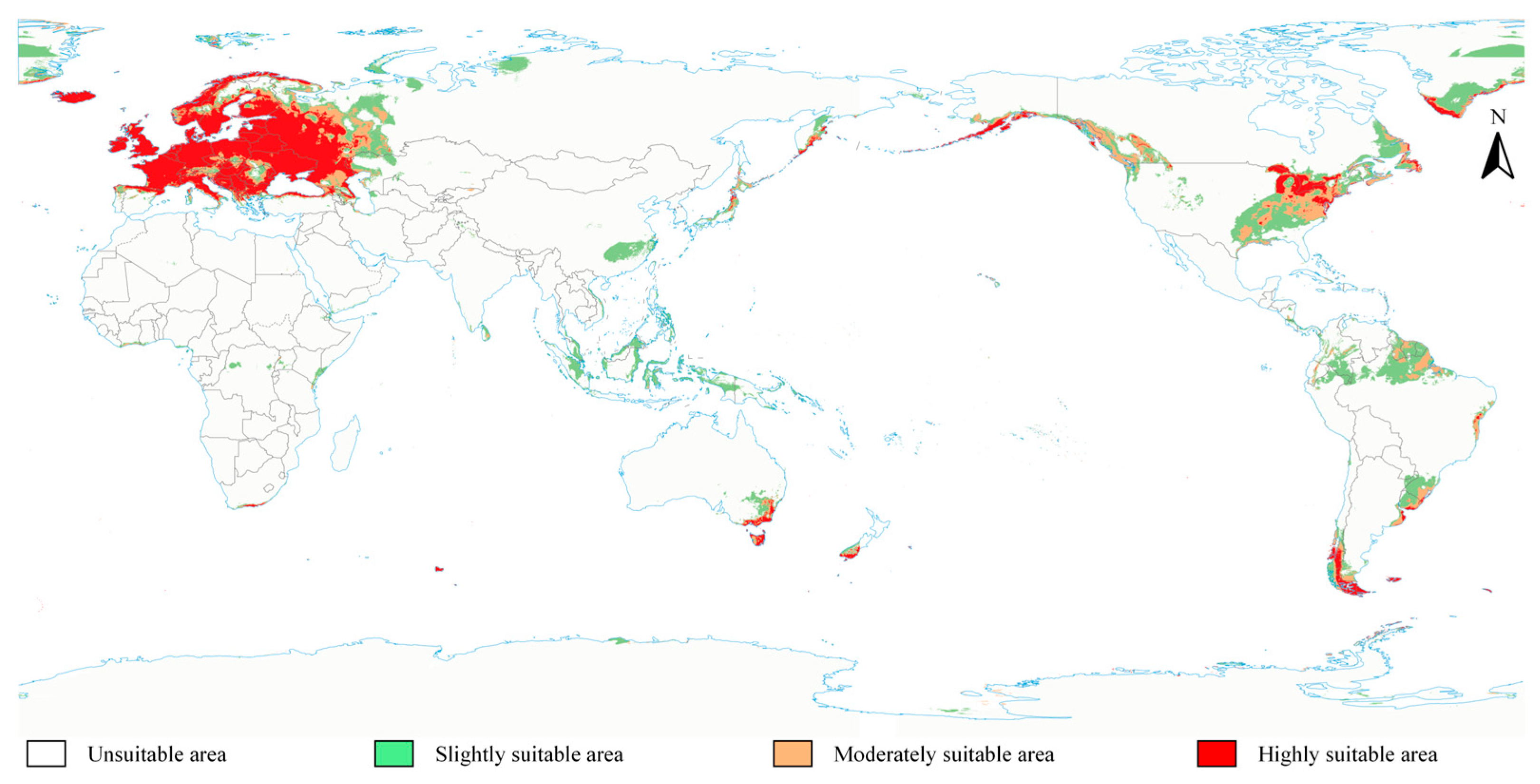
3.3. Global Potential Geographic Distribution of S. scolytus Under Current Climatic Conditions
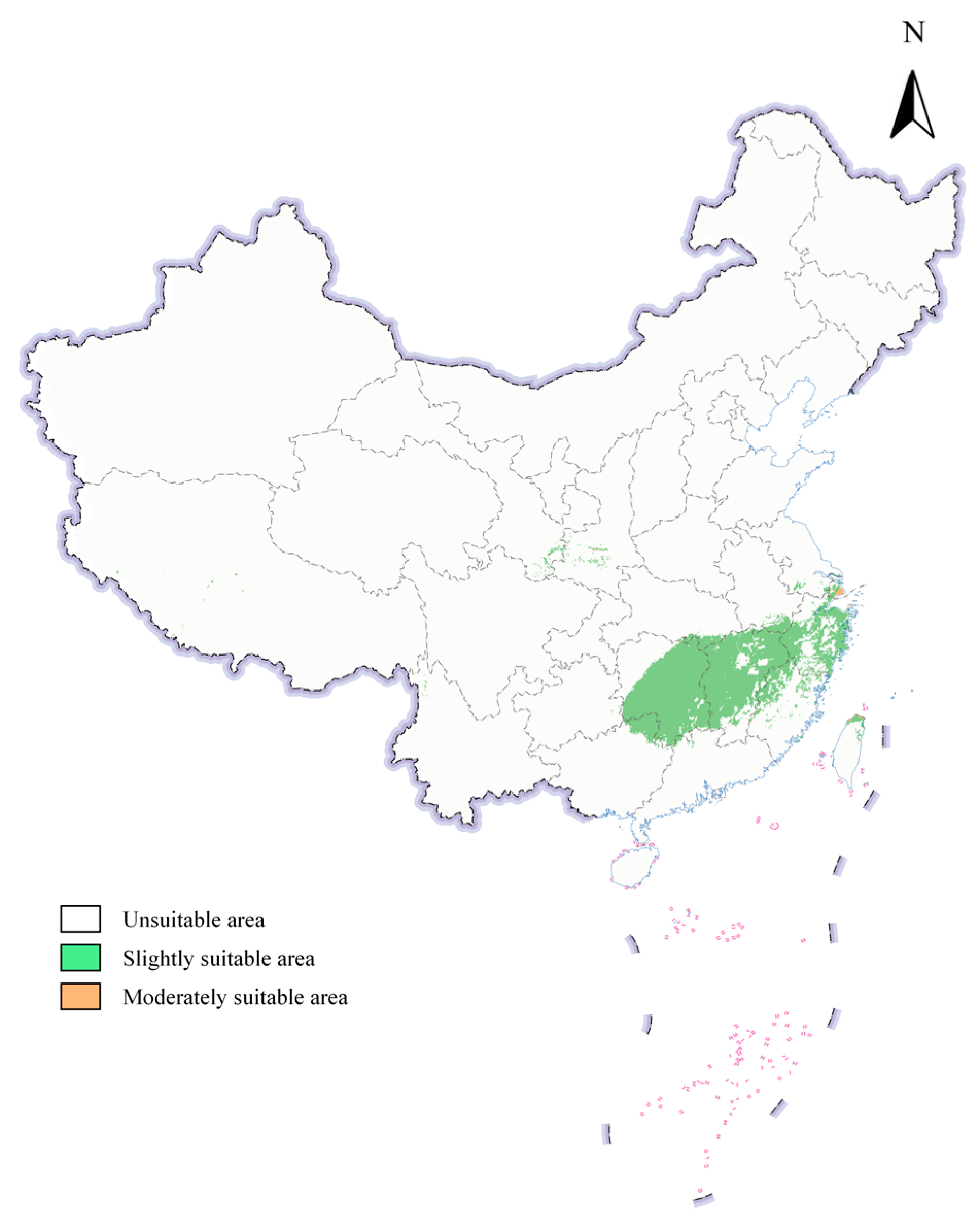
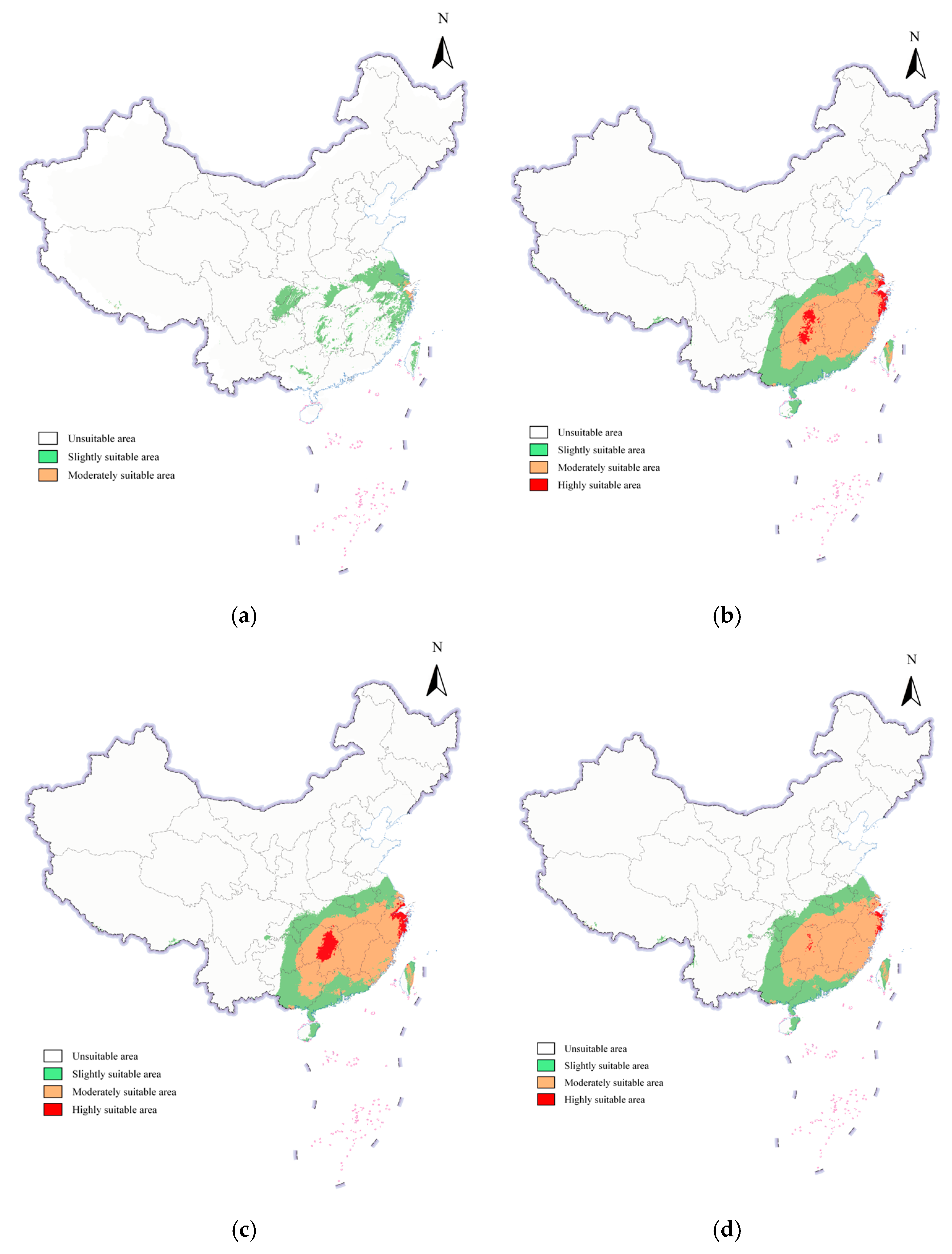
3.4. Potential Geographic Distribution of S. scolytus in China Under Current Climatic Conditions
3.5. Potential Geographic Distribution of S. scolytus in China Under Future Climate Conditions
4. Discussion
4.1. Methodological Discussion
4.2. Invasion Risks and Control Strategies for S. scolytus Under Potential Distribution in China
4.3. Impact of Global Climate Change on the Habitat Suitability of S. scolytus and Its Key Environmental Drivers
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| SDMs | Suitable distribution models |
| BCC-CSM2-MR | Beijing Climate Center climate system model version 2—medium resolution |
| SSP | Shared socioeconomic pathway |
| MaxEnt | Maximum entropy |
| GLM | Generalized linear model |
| GAM | Generalized additive model |
| MARS | Multivariate adaptive regression splines |
| ANN | Artificial neural networks |
| CTA | Classification tree analysis |
| FDA | Flexible discriminant analysis |
| RF | Random forest |
| SRE | Spectral Relaxation Embedding |
| ROC | Receiver operating characteristic |
| TPR | True-positive rate |
| FPR | False-positive rate |
| AUC | Area under the ROC curve |
| TSS | True skill statistic |
References
- Shih, C.; Li, B.; Li, H.; Ju, R. Progress of biological invasions research in China over the last decade. Biodivers. Sci. 2012, 20, 581. [Google Scholar] [CrossRef]
- Wei, F.; Nie, Y.; Miao, H.; Lu, H.; Hu, Y. Advancements of the researches on biodiversity loss mechanisms. Chin. Sci. Bull. 2014, 59, 430–437. [Google Scholar] [CrossRef]
- Wan, F.; Guo, J.; Wang, D. Alien invasive species in China: Their damages and management strategies. Biodivers. Sci. 2002, 10, 119–125. [Google Scholar] [CrossRef]
- Witzell, J.; Sunnerstam, C.; Hansson, T. Vaccination of elms against Dutch Elm Disease—Are the associated epiphytes and endophytes affected? J. Fungi 2023, 9, 297. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Zuo, L.; Liu, Y.; Long, L.; Wu, J.; Yuan, M.; Wang, J.; Yang, M. Comparative transcriptomes of four Elm species provide insights into the genetic features and adaptive evolution of Ulmus spp. For. Ecol. Manag. 2024, 553, 121560. [Google Scholar] [CrossRef]
- Jeong, C.; Lee, C.H.; Seo, J.; Park, J.H.Y.; Lee, K.W. Catechin and flavonoid glycosides from the Ulmus genus: Exploring their nutritional pharmacology and therapeutic potential in osteoporosis and inflammatory conditions. Fitoterapia 2024, 178, 106188. [Google Scholar] [CrossRef] [PubMed]
- Zhang, P.; Ye, J.R.; Zhang, Y.; Wang, L. The research progress of interal and external on pathogens of Dutch Elm Disease. Plant Quar. 2014, 28, 33–38. [Google Scholar]
- López-Almansa, J.C. Ecological factors related to seed germination and early seedling establishment in Ulmus minor Mill., an endangered riparian tree species. For. Int. J. For. Res. 2023, 96, 775–786. [Google Scholar] [CrossRef]
- Macaya-Sanz, D.; Witzell, J.; Collada, C.; Gil, L.; Martín, J.A. Core endophytic mycobiome in Ulmus minor and its relation to Dutch elm disease resistance. Front. Plant Sci. 2023, 14, 1125942. [Google Scholar] [CrossRef] [PubMed]
- Akbari Oghaz, N.; Rahnama, K.; Vatandoost, H.; Afshari, A.; White, J.F.; Hyde, K.D.; Yazdanian, M.; Salari, E.; Hatamzadeh, S.; Taheri, A. Entomopathogenic fungi as guardians of elm trees: A review of dual-action biocontrol agents targeting Scolytus spp. and their associated Ophiostoma species. J. Invertebr. Pathol. 2025, 211, 108361. [Google Scholar] [CrossRef] [PubMed]
- Selikhovkin, A.V.; Nekhaeva, M.Y.; Melnichuk, I.A. Economic and social consequences of invasions of tree pests and pathogens in St. Petersburg. Russ. J. Biol. Invasions 2023, 14, 398–404. [Google Scholar] [CrossRef]
- EPPO. Categorization of Scolytus scolytus. Available online: https://gd.eppo.int/taxon/SCOLSC/categorization (accessed on 17 June 2025).
- Thomas, C.D.; Cameron, A.; Green, R.E.; Bakkenes, M.; Beaumont, L.J.; Collingham, Y.C.; Erasmus, B.F.N.; de Siqueira, M.F.; Grainger, A.; Hannah, L.; et al. Extinction risk from climate change. Nature 2004, 427, 145–148. [Google Scholar] [CrossRef] [PubMed]
- Guillera-Arroita, G.; Lahoz-Monfort, J.J.; Elith, J.; Gordon, A.; Kujala, H.; Lentini, P.E.; McCarthy, M.A.; Tingley, R.; Wintle, B.A. Is my species distribution model fit for purpose? Matching data and models to applications. Glob. Ecol. Biogeogr. 2015, 24, 276–292. [Google Scholar] [CrossRef]
- Elith, J.; Leathwick, J.R. Species distribution models: Ecological explanation and prediction across space and time. Annu. Rev. Ecol. Evol. Syst. 2009, 40, 677–697. [Google Scholar] [CrossRef]
- Kearney, M.; Porter, W. Mechanistic niche modelling: Combining physiological and spatial data to predict species’ ranges. Ecol. Lett. 2009, 12, 334–350. [Google Scholar] [CrossRef] [PubMed]
- Guisan, A.; Thuiller, W. Predicting species distribution: Offering more than simple habitat models. Ecol. Lett. 2005, 8, 993–1009. [Google Scholar] [CrossRef] [PubMed]
- Dormann, C.F.; Calabrese, J.M.; Guillera-Arroita, G.; Matechou, E.; Bahn, V.; Bartoń, K.; Beale, C.M.; Ciuti, S.; Elith, J.; Gerstner, K.; et al. Model averaging in ecology: A review of Bayesian, information-theoretic, and tactical approaches for predictive inference. Ecol. Monogr. 2018, 88, 485–504. [Google Scholar] [CrossRef]
- Hao, T.; Elith, J.; Guillera-Arroita, G.; Lahoz-Monfort, J.J. A review of evidence about use and performance of species distribution modelling ensembles like BIOMOD. Divers. Distrib. 2019, 25, 839–852. [Google Scholar] [CrossRef]
- Zhao, H.; Xian, X.; Yang, N.; Guo, J.; Zhao, L.; Shi, J.; Liu, W.-X. Risk assessment framework for pine wilt disease: Estimating the introduction pathways and multispecies interactions among the pine wood nematode, its insect vectors, and hosts in China. Sci. Total Environ. 2023, 905, 167075. [Google Scholar] [CrossRef] [PubMed]
- Zhao, G.; Cui, X.; Sun, J.; Li, T.; Wang, Q.; Ye, X.; Fan, B. Analysis of the distribution pattern of Chinese Ziziphus jujuba under climate change based on optimized biomod2 and MaxEnt models. Ecol. Indic. 2021, 132, 108256. [Google Scholar] [CrossRef]
- GBIF Occurrence Download. Available online: https://doi.org/10.15468/dl.8zuw3y (accessed on 14 May 2024).
- Bosso, L.; Smeraldo, S.; Rapuzzi, P.; Sama, G.; Garonna, A.P.; Russo, D. Nature protection areas of Europe are insufficient to preserve the threatened beetle Rosalia alpina (Coleoptera: Cerambycidae): Evidence from species distribution models and conservation gap analysis. Ecol. Entomol. 2018, 43, 192–203. [Google Scholar] [CrossRef]
- Yang, T.; Liu, X.; Han, Z. Predicting the effects of climate change on the suitable habitat of Japanese Spanish Mackerel (Scomberomorus niphonius) based on the species distribution model. Front. Mar. Sci. 2022, 9, 927790. [Google Scholar] [CrossRef]
- Xie, C.; Chen, L.; Li, M.; Jim, C.; Liu, D. BIOCLIM modeling for predicting suitable habitat for endangered tree Tapiscia sinensis (Tapisciaceae) in China. Forests 2023, 14, 2275. [Google Scholar] [CrossRef]
- Koo, K.A.; Park, S.U. Data on the predictions of plant redistribution under interplays among climate change, land-use change, and dispersal capacity. Data Brief 2022, 45, 108667. [Google Scholar] [CrossRef] [PubMed]
- Wen, X.; Fang, G.; Chai, S.; He, C.; Sun, S.; Zhao, G.; Lin, X. Can ecological niche models be used to accurately predict the distribution of invasive insects? A case study of Hyphantria cunea in China. Ecol. Evol. 2024, 14, 11159. [Google Scholar] [CrossRef] [PubMed]
- Liang, T.; Wang, Q.; Xin, B.; Wu, Z.; Shi, J. Prediction of the potential global geographical distribution of smaller European elm bark beetle Scolytus multistriatus by using the MaxEnt model. J. Plant Prot. 2023, 50, 1499–1507. [Google Scholar]
- Guo, X.; Xi, S.; Yang, L.; Ji, J.; Ma, X.; Jin, L. Study on prediction of potential suitable distribution areas of Epimedium brevicornu Maxim. in China based on optimal MaxEnt model. Chin. J. Inf. Tradit. Chin. Med. 2024, 31, 1–7. [Google Scholar]
- Phillips, S.J.; Anderson, R.P.; Schapire, R.E. Maximum entropy modeling of species geographic distributions. Ecol. Model. 2006, 190, 231–259. [Google Scholar] [CrossRef]
- Mccaffrey; Daniel, F. Generalized additive models (T. J. Hastie and R. J. Tibshirani). SIAM Rev. 2006, 34, 675–678. [Google Scholar] [CrossRef]
- Jerome, H.F. Multivariate adaptive regression splines. Ann. Stat. 1991, 19, 1–67. [Google Scholar] [PubMed]
- Özesmi, U.; Tan, C.O.; Özesmi, S.L.; Robertson, R.J. Generalizability of artificial neural network models in ecological applications: Predicting nest occurrence and breeding success of the red-winged blackbird Agelaius phoeniceus. Ecol. Model. 2006, 195, 94–104. [Google Scholar] [CrossRef][Green Version]
- Loh, W.Y. Classification and regression trees. WIREs Data Min. Knowl. Discov. 2011, 1, 14–23. [Google Scholar] [CrossRef]
- Hastie, T.; Tibshirani, R.; Buja, A. flexible discriminant analysis by optimal scoring. J. Am. Stat. Assoc. 1994, 89, 1255–1270. [Google Scholar] [CrossRef]
- Breiman, L. Random Forests. Mach. Learn. 2001, 45, 5–32. [Google Scholar] [CrossRef]
- Zha, H.; He, X.; Ding, C.; Simon, H.; Gu, M. Spectral relaxation for K-means clustering. In Proceedings of the 15th International Conference on Neural Information Processing Systems: Natural and Synthetic, Vancouver, BC, Canada, 3–8 December 2001; pp. 1057–1064. [Google Scholar]
- Li, C.; Liu, Y.; Lai, Y.; Shao, H. Comparative study of potential habitats for two endemic grassland caterpillars on the Qinghai-Tibet Plateau based on BIOMOD2 and land use data. Insects 2024, 15, 781. [Google Scholar] [CrossRef] [PubMed]
- Stocker, T.; Plattner, G.K.; Dahe, Q. IPCC Climate Change 2013: The Physical Science Basis-Findings and Lessons Learned. In Proceedings of the EGU General Assembly Conference Abstracts, Vienna, Austria, 27 April–2 May 2014. [Google Scholar]
- Baldwin, M.E.; Kain, J.S. Sensitivity of several performance measures to displacement error, bias, and event frequency. Weather Forecast. 2006, 21, 636–648. [Google Scholar] [CrossRef]
- Landgrebe, T.C.W.; Duin, R.P.W. Efficient multiclass ROC approximation by decomposition via confusion matrix perturbation analysis. IEEE Trans. Pattern Anal. Mach. Intell. 2008, 30, 810–822. [Google Scholar] [CrossRef] [PubMed]
- Santini, A.; Faccoli, M. Dutch elm disease and elm bark beetles: A century of association. iForest—Biogeosciences For. 2014, 8, 126. [Google Scholar] [CrossRef]
- Jürisoo, L.; Süda, I.; Agan, A.; Drenkhan, R. Vectors of Dutch Elm Disease in Northern Europe. Insects 2021, 12, 393. [Google Scholar] [CrossRef] [PubMed]
- Webber, J.F. Relative effectiveness of Scolytus scolytus, S. multistriatus and S. kirschi as vectors of Dutch elm disease. Eur. J. For. Pathol. 1990, 20, 184–192. [Google Scholar] [CrossRef]
- Aoun, M.; Jacobi, V.; Boyle, B.; Bernier, L. Identification and monitoring of Ulmus americana transcripts during in vitro interactions with the Dutch elm disease pathogen Ophiostoma novo-ulmi. Physiol. Mol. Plant Pathol. 2010, 74, 254–266. [Google Scholar] [CrossRef]
- Moser, J.C.; Konrad, H.; Blomquist, S.R.; Kirisits, T. Do mites phoretic on elm bark beetles contribute to the transmission of Dutch elm disease? Naturwissenschaften 2010, 97, 219–227. [Google Scholar] [CrossRef] [PubMed]
- Ryss, A.; Polyanina, K.; Popovichev, B.; Subbotin, S. Description of Bursaphelenchus ulmophilus sp. n. (Nematoda: Parasitaphelenchinae) associated with Dutch elm disease of Ulmus glabra Huds. in the Russian North West. Nematology 2015, 17, 685–703. [Google Scholar] [CrossRef]
- Zhou, Y.; Ge, X.; Zou, Y.; Guo, S.; Wang, T.; Tao, J.; Zong, S. Prediction of the potential geographical distribution of Hylurgus ligniperda at the global scale and in China using the Maxent model. J. Beijing For. Univ. 2022, 44, 90–99. [Google Scholar]
- Huang, D.; Zhang, Y.; Yang, B.; Li, Z. Prediction of potential habitat area of Xyleborinus saxesenii based on optimal MaxEnt model. Sichuan J. Zool. 2024, 43, 251–263. [Google Scholar]
- Bentz, B.J.; Régnière, J.; Fettig, C.J.; Hansen, E.M.; Hayes, J.L.; Hicke, J.A.; Kelsey, R.G.; Negrón, J.F.; Seybold, S.J. Climate change and bark beetles of the western United States and Canada: Direct and indirect effects. BioScience 2010, 60, 602–613. [Google Scholar] [CrossRef]
- González-Hernández, A.; Morales-Villafaña, R.; Romero-Sánchez, M.E.; Islas-Trejo, B.; Pérez-Miranda, R. Modelling potential distribution of a pine bark beetle in Mexican temperate forests using forecast data and spatial analysis tools. J. For. Res. 2018, 31, 649–659. [Google Scholar] [CrossRef]
- Zheng, X.; Wang, Q.; Tian, Y.; Chen, X.; Liu, X. Prediction of potential suitable of tea shot-hole borer Euwallacea fornicatus in China based on MaxEnt model. J. Plant Prot. 2024, 51, 1518–1530. [Google Scholar]
- Zhou, Y.; Guo, S.; Wang, T.; Zong, S.; Ge, X. Modeling the pest-pathogen threats in a warming world for the red turpentine beetle (Dendroctonus valens) and its symbiotic fungus (Leptographium procerum). Pest Manag. Sci. 2024, 80, 3423–3435. [Google Scholar] [CrossRef] [PubMed]
| Model | Overview | Biomod2 Dependent on Packages |
|---|---|---|
| MaxEnt | Uses the maximum entropy principle to model the conditional probability distribution, often used in classification and NLP tasks. Typically optimized using iterative methods like gradient ascent [30]. | maxent, dismo |
| GLM | Fits quadratic response curves with no interactions between covariates, using stepwise backward selection [15]. | glm |
| GAM | Fits smoothed additive response curves through the mgcv package [31]. | gam, mgcv |
| MARS | Fits complex response curves by joining linear segments, using the earth package [32]. | earth |
| ANN | A neural network with one hidden layer optimized using cross validation [33]. | nnet |
| CTA | Decision tree analysis with complex trees, using the rpart package [34]. | rpart |
| FDA | Uses MARS for dimensionality reduction before classification, fitted through mda [35]. | mda |
| RF | Ensembles predictions from 500 random forest trees, using randomForest package [36]. | randomForest |
| SRE | A data dimensionality reduction algorithm applicable to nonlinear data [37]. | gbm |
| Environmental Factors | Average Importance Value | Average Importance Percentage of Total Importance |
|---|---|---|
| bio6 | 0.1460 | 0.3570 |
| bio15 | 0.1140 | 0.2790 |
| bio17 | 0.0444 | 0.1080 |
| bio3 | 0.0295 | 0.0722 |
| bio8 | 0.0254 | 0.0622 |
| bio16 | 0.0231 | 0.0564 |
| bio18 | 0.0212 | 0.0518 |
| bio2 | 0.0053 | 0.0129 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yu, W.; Sun, D.; Ma, J.; Gao, X.; Fang, Y.; Pan, H.; Wang, H.; Shi, J. Predicting the Potential Geographical Distribution of Scolytus scolytus in China Using a Biomod2-Based Ensemble Model. Insects 2025, 16, 742. https://doi.org/10.3390/insects16070742
Yu W, Sun D, Ma J, Gao X, Fang Y, Pan H, Wang H, Shi J. Predicting the Potential Geographical Distribution of Scolytus scolytus in China Using a Biomod2-Based Ensemble Model. Insects. 2025; 16(7):742. https://doi.org/10.3390/insects16070742
Chicago/Turabian StyleYu, Wei, Dongrui Sun, Jiayi Ma, Xinyuan Gao, Yu Fang, Huidong Pan, Huiru Wang, and Juan Shi. 2025. "Predicting the Potential Geographical Distribution of Scolytus scolytus in China Using a Biomod2-Based Ensemble Model" Insects 16, no. 7: 742. https://doi.org/10.3390/insects16070742
APA StyleYu, W., Sun, D., Ma, J., Gao, X., Fang, Y., Pan, H., Wang, H., & Shi, J. (2025). Predicting the Potential Geographical Distribution of Scolytus scolytus in China Using a Biomod2-Based Ensemble Model. Insects, 16(7), 742. https://doi.org/10.3390/insects16070742





