Comprehensive Analysis of the UGT Gene Superfamily in Spodoptera frugiperda
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Identification and Acquisition of the SfUGT Gene Family
2.2. Analysis of Gene Duplication and Ka/Ks Ratios for SfUGT
2.3. Physicochemical Property Analysis of SfUGT Proteins
2.4. Structural Characterization and Motif Landscape of SfUGT Genes
2.5. Phylogenetic Analysis of SfUGT Across Species
2.6. Expression Analysis of the SfUGT Gene Family at Different Developmental Stages
2.7. qPCR Analysis of the SfUGT Gene
3. Results
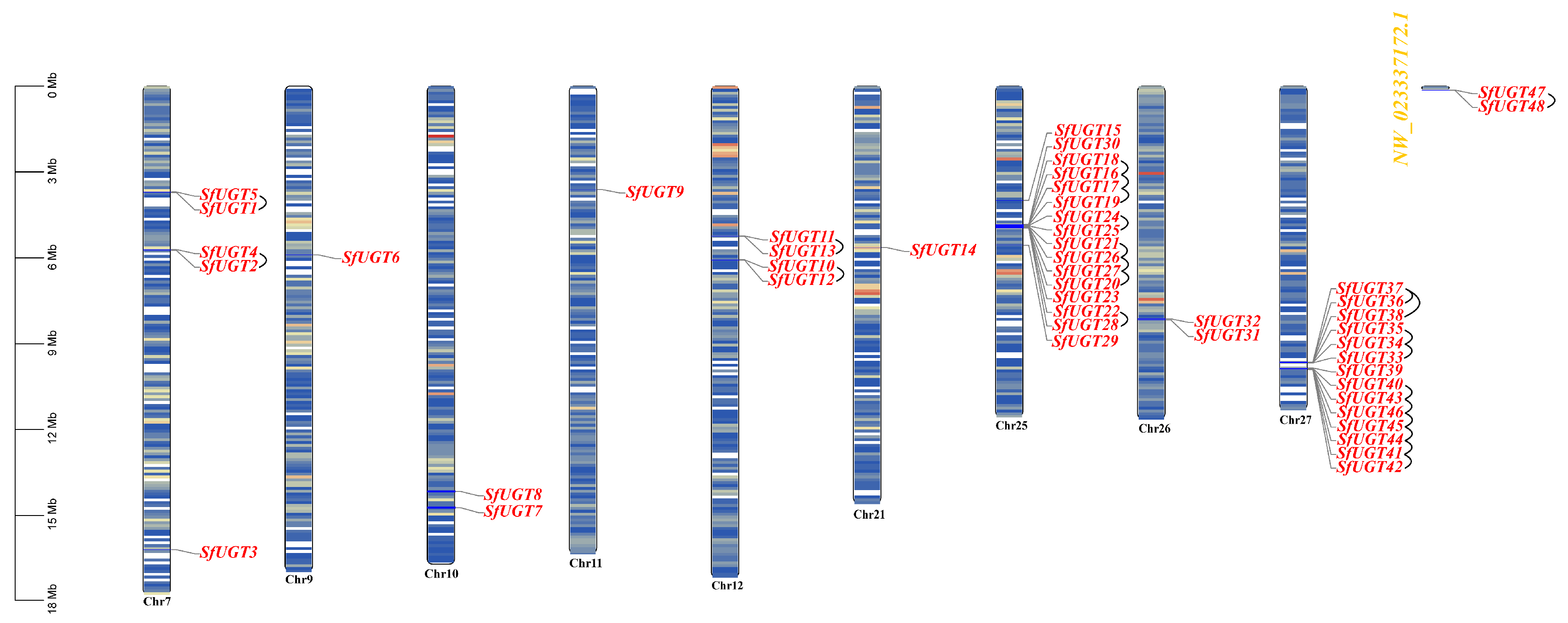
3.1. Discovery and Characterization of the SfUGT Gene Family
3.2. Analysis of SfUGT Gene Duplication and Ka/Ks Ratios
3.3. Construction of SfUGT Phylogenetic Tree and Protein Structure Analysis
3.3.1. Analysis of SfUGT Protein Structure
3.3.2. Protein Secondary Structure Analysis
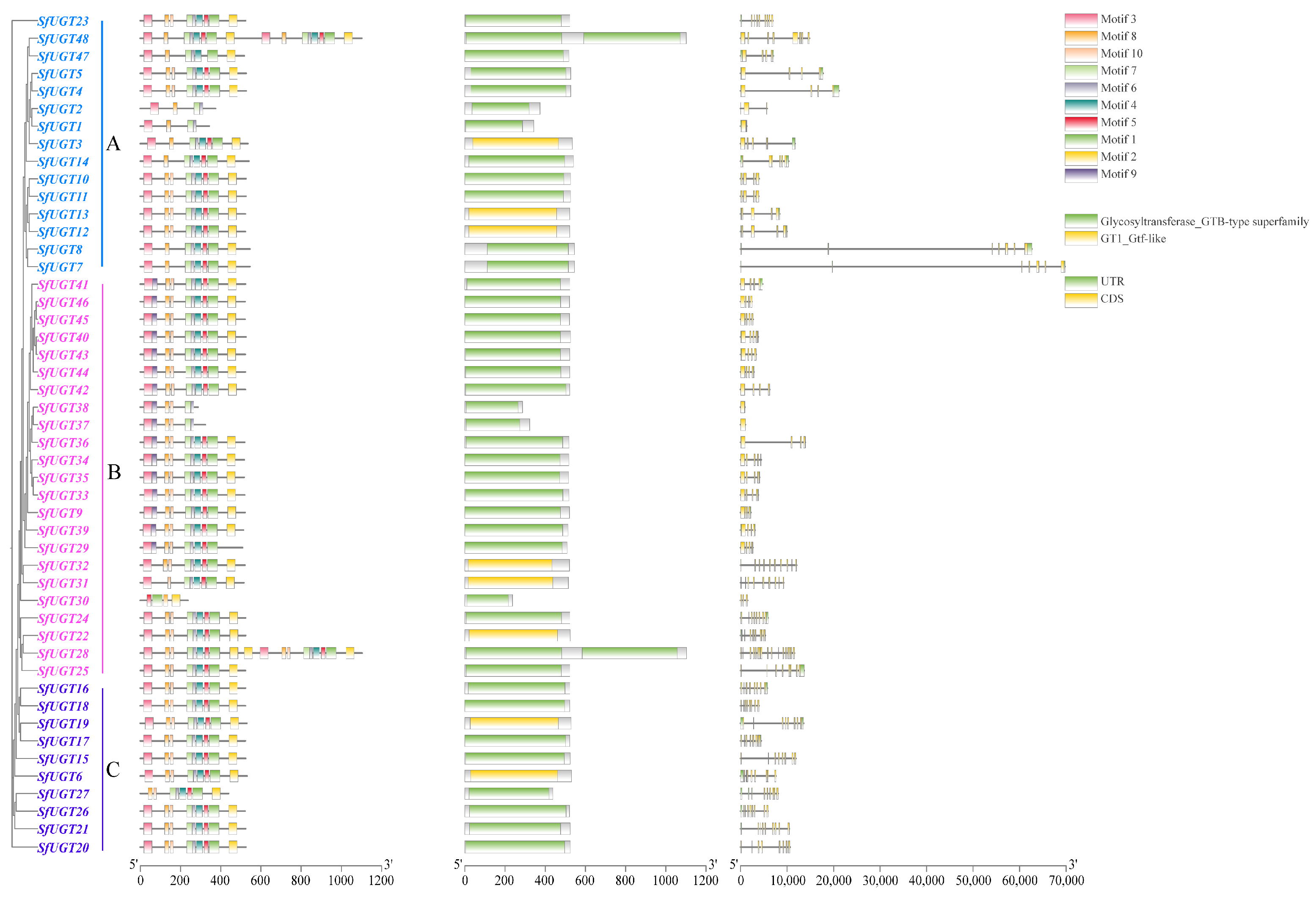
3.4. Analysis of Phylogenetic Relationships, Gene Architecture, and Conserved Motifs in SfUGT Genes
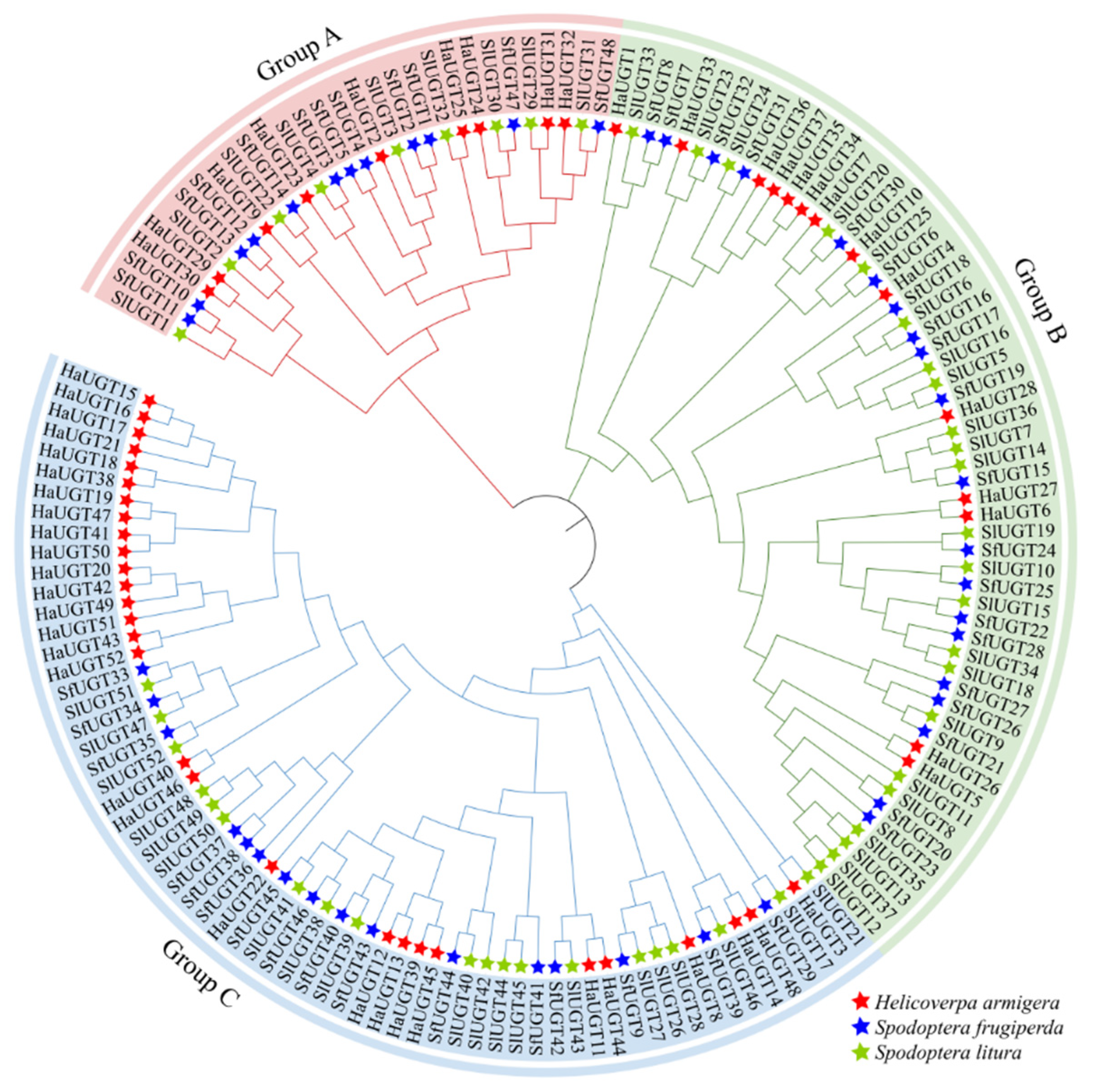
3.5. Phylogenetic Analysis of SfUGT Proteins Across Species
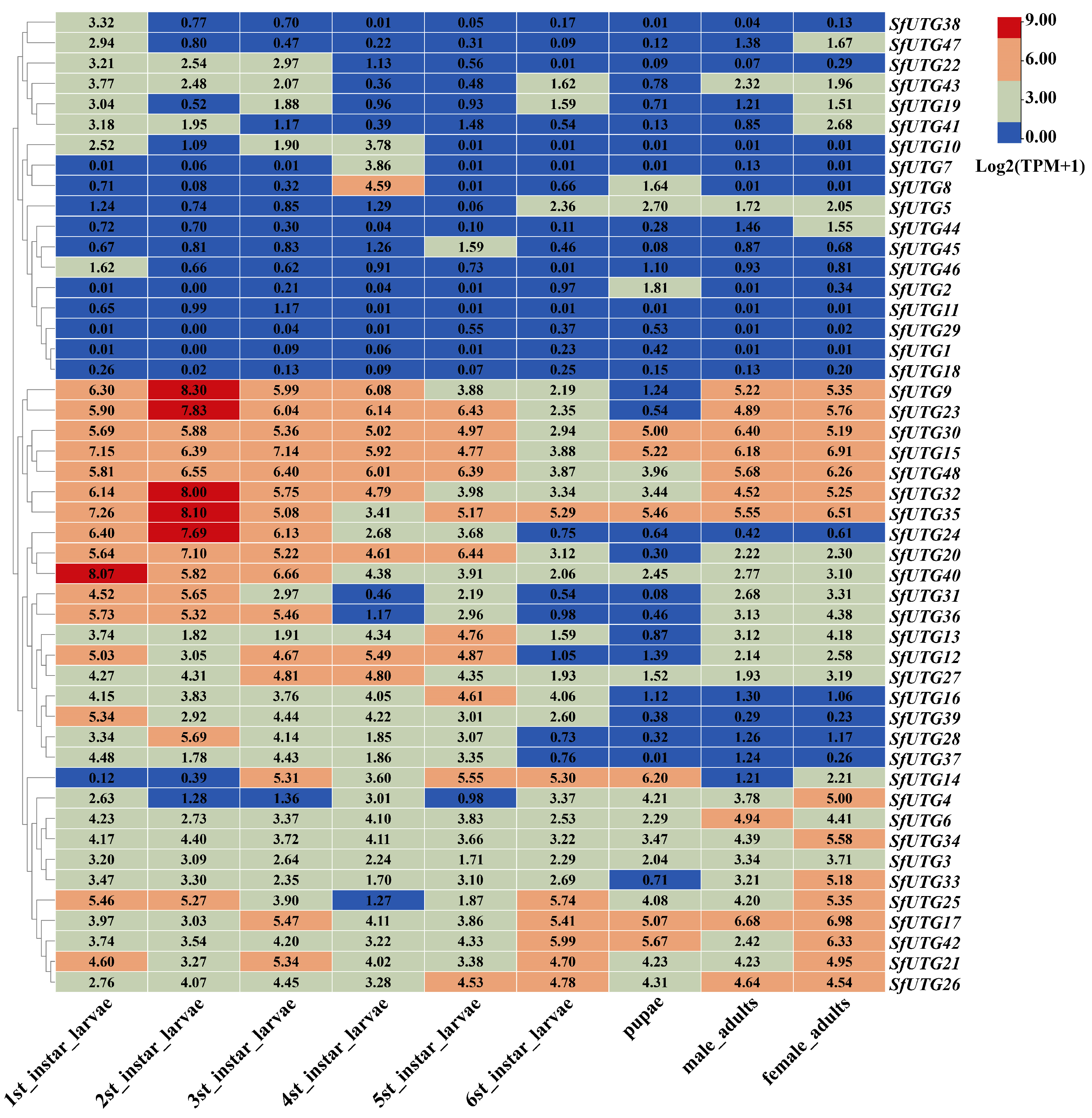
3.6. Analysis of SfUGT Gene Family Expression Across Different Developmental Stages
3.7. Tissue-Specific Expression Analysis of the SfUGT Gene Family
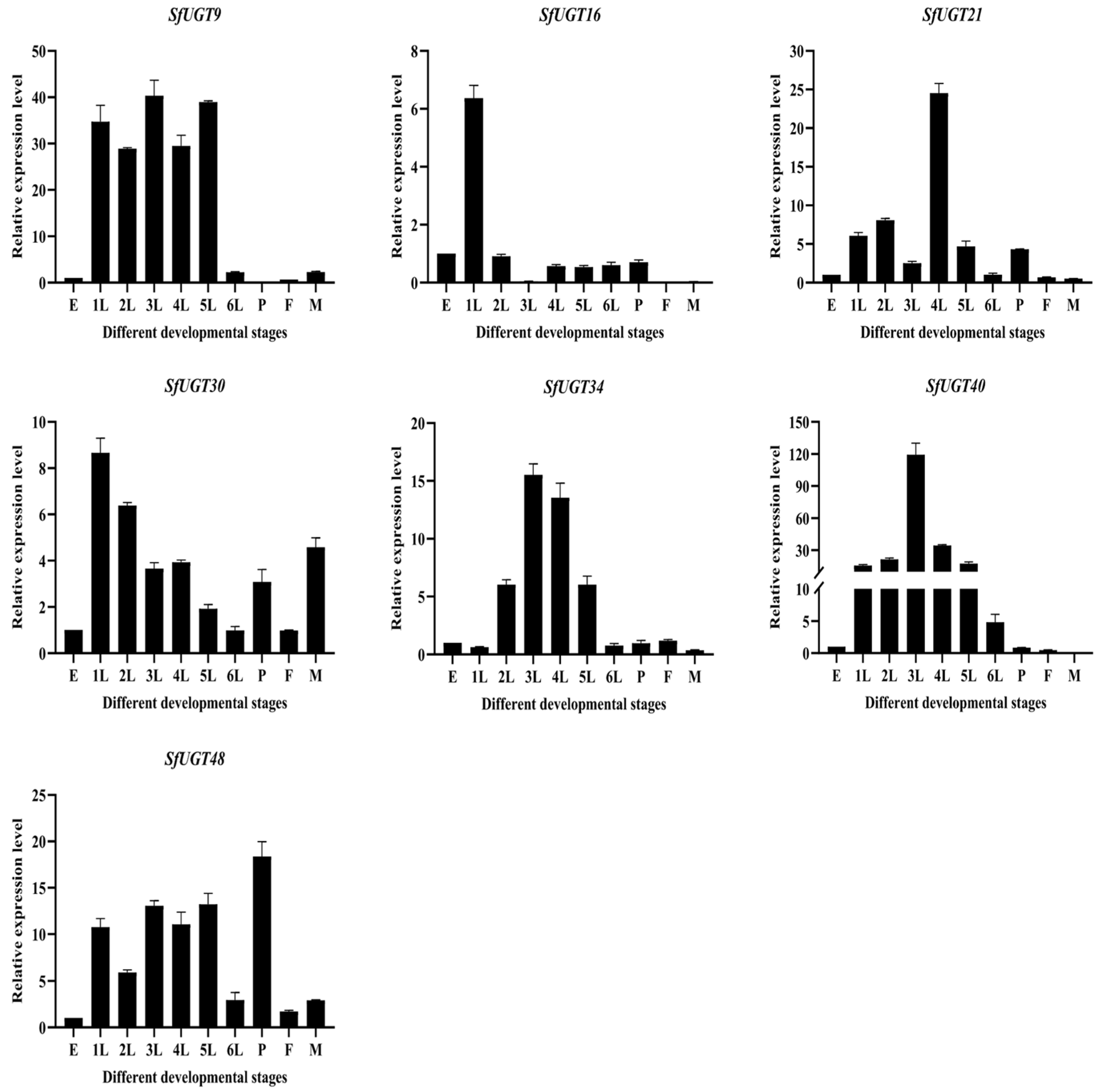
3.8. qPCR Analysis of SfUGT Gene Expression Across Developmental Stages
3.9. qPCR Analysis of SfUGT Gene Expression in Different Tissues
4. Discussion
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Abbas, A.; Ullah, F.; Hafeez, M.; Han, X.; Dara, M.Z.N.; Gul, H.; Zhao, C.R. Biological Control of Fall Armyworm, Spodoptera frugiperda. Agronomy 2022, 12, 2704. [Google Scholar] [CrossRef]
- Koffi, D.; Kyerematen, R.; Eziah, V.Y.; Osei-Mensah, Y.O.; Afreh-Nuamah, K.; Aboagye, E.; Osae, M.; Meagher, R.L. Assessment of impacts of fall armyworm, Spodoptera frugiperda (Lepidoptera: Noctuidae) on maize production in Ghana. J. Integr. Pest Manag. 2020, 11, 20. [Google Scholar] [CrossRef]
- Day, R.; Abrahams, P.; Bateman, M.; Beale, T.; Clottey, V.; Cock, M.; Colmenarez, Y.; Corniani, N.; Early, R.; Godwin, J. Fall armyworm: Impacts and implications for Africa. Outlooks Pest Manag. 2017, 28, 196–201. [Google Scholar] [CrossRef]
- Goergen, G.; Kumar, P.L.; Sankung, S.B.; Togola, A.; Tamò, M. First report of outbreaks of the fall armyworm Spodoptera frugiperda (JE Smith) (Lepidoptera, Noctuidae), a new alien invasive pest in West and Central Africa. PLoS ONE 2016, 11, e0165632. [Google Scholar] [CrossRef]
- Early, R.; González-Moreno, P.; Murphy, S.T.; Day, R. Forecasting the global extent of invasion of the cereal pest Spodoptera frugiperda, the fall armyworm. BioRxiv 2018, 391847. [Google Scholar] [CrossRef]
- Prasanna, B.; Huesing, J.; Eddy, R.; Peschke, V. Fall armyworm in Africa: A Guide for Integrated Pest Management; United States Agency for International Development (USAID): Washington, DC, USA, 2018.
- Nagoshi, R.N.; Meagher, R.L. Review of Fall armyworm (Lepidoptera: Noctuidae) genetic complexity and migration. Fla. Entomol. 2008, 91, 546–554. [Google Scholar]
- Bowles, D.; Isayenkova, J.; Lim, E.-K.; Poppenberger, B. Glycosyltransferases: Managers of small molecules. Curr. Opin. Plant Biol. 2005, 8, 254–263. [Google Scholar] [CrossRef]
- De Bruyn, F.; Maertens, J.; Beauprez, J.; Soetaert, W.; De Mey, M. Biotechnological advances in UDP-sugar based glycosylation of small molecules. Biotechnol. Adv. 2015, 33, 288–302. [Google Scholar] [CrossRef]
- Tephly, T.R.; Burchell, B. UDP-glucuronosyltransferases: A family of detoxifying enzymes. Trends Pharmacol. Sci. 1990, 11, 276–279. [Google Scholar] [CrossRef]
- Ross, J.; Li, Y.; Lim, E.-K.; Bowles, D.J. Higher plant glycosyltransferases. Genome Biol. 2001, 2, 3004.1. [Google Scholar] [CrossRef]
- Howe, G.A.; Herde, M. Interaction of plant defense compounds with the insect gut: New insights from genomic and molecular analyses. Curr. Opin. Insect Sci. 2015, 9, 62–68. [Google Scholar] [CrossRef] [PubMed]
- Mackenzie, P.I.; Bock, K.W.; Burchell, B.; Guillemette, C.; Ikushiro, S.-i.; Iyanagi, T.; Miners, J.O.; Owens, I.S.; Nebert, D.W. Nomenclature update for the mammalian UDP glycosyltransferase (UGT) gene superfamily. Pharmacogenet. Genom. 2005, 15, 677–685. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Yang, X.; Lu, M.; Chen, J.; Shi, T. Gene expression and evolution of Family-1 UDP-glycosyltransferases—Insights from an aquatic flowering plant (Sacred lotus). Aquat. Bot. 2020, 166, 103270. [Google Scholar] [CrossRef]
- Huang, F.-F.; Chai, C.-L.; Zhang, Z.; Liu, Z.-H.; Dai, F.-Y.; Lu, C.; Xiang, Z.-H. The UDP-glucosyltransferase multigene family in Bombyx mori. BMC Genom. 2008, 9, 563. [Google Scholar] [CrossRef]
- Pym, A.; Umina, P.A.; Reidy-Crofts, J.; Troczka, B.J.; Matthews, A.; Gardner, J.; Hunt, B.J.; van Rooyen, A.R.; Edwards, O.R.; Bass, C. Overexpression of UDP-glucuronosyltransferase and cytochrome P450 enzymes confers resistance to sulfoxaflor in field populations of the aphid, Myzus persicae. Insect Biochem. Mol. Biol. 2022, 143, 103743. [Google Scholar] [CrossRef]
- Zheng, S.; Luo, J.; Zhu, X.; Gao, X.; Hua, H.; Cui, J. Transcriptomic analysis of salivary gland and proteomic analysis of oral secretion in Helicoverpa armigera under cotton plant leaves, gossypol, and tannin stresses. Genomics 2022, 114, 110267. [Google Scholar] [CrossRef]
- Yan, M.-W.; Xing, X.-R.; Wu, F.-A.; Wang, J.; Sheng, S. UDP-glycosyltransferases contribute to the tolerance of parasitoid wasps towards insecticides. Pestic. Biochem. Physiol. 2021, 179, 104967. [Google Scholar] [CrossRef]
- Ahn, S.-J.; Vogel, H.; Heckel, D.G. Comparative analysis of the UDP-glycosyltransferase multigene family in insects. Insect Biochem. Mol. Biol. 2012, 42, 133–147. [Google Scholar] [CrossRef]
- Coutinho, P.M.; Deleury, E.; Davies, G.J.; Henrissat, B. An evolving hierarchical family classification for glycosyltransferases. J. Mol. Biol. 2003, 328, 307–317. [Google Scholar] [CrossRef]
- Israni, B.; Wouters, F.C.; Luck, K.; Seibel, E.; Ahn, S.-J.; Paetz, C.; Reinert, M.; Vogel, H.; Erb, M.; Heckel, D.G. The fall armyworm Spodoptera frugiperda utilizes specific UDP-glycosyltransferases to inactivate maize defensive benzoxazinoids. Front. Physiol. 2020, 11, 604754. [Google Scholar] [CrossRef]
- Meech, R.; Mackenzie, P.I. Structure and function of uridine diphosphate glucuronosyltransferases. Clin. Exp. Pharmacol. Physiol. 1997, 24, 907–915. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Zou, K.; Wang, T.; Guan, M.; Duan, H.; Yu, H.; Wu, D.; Du, J. Genome-Wide Identification and Analysis of Family Members with Juvenile Hormone Binding Protein Domains in Spodoptera frugiperda. Insects 2024, 15, 573. [Google Scholar] [CrossRef]
- Yu, J.; Wang, J.; Lin, W.; Li, S.; Li, H.; Zhou, J.; Ni, P.; Dong, W.; Hu, S.; Zeng, C. The genomes of Oryza sativa: A history of duplications. PLoS Biol. 2005, 3, e38. [Google Scholar] [CrossRef]
- Gasteiger, E.; Gattiker, A.; Hoogland, C.; Ivanyi, I.; Appel, R.D.; Bairoch, A. ExPASy: The proteomics server for in-depth protein knowledge and analysis. Nucleic Acids Res. 2003, 31, 3784–3788. [Google Scholar] [CrossRef]
- Horton, P.; Park, K.-J.; Obayashi, T.; Fujita, N.; Harada, H.; Adams-Collier, C.; Nakai, K. WoLF PSORT: Protein localization predictor. Nucleic Acids Res. 2007, 35, W585–W587. [Google Scholar] [CrossRef]
- Geourjon, C.; Deleage, G. SOPMA: Significant improvements in protein secondary structure prediction by consensus prediction from multiple alignments. Bioinformatics 1995, 11, 681–684. [Google Scholar] [CrossRef]
- Bailey, T.L.; Johnson, J.; Grant, C.E.; Noble, W.S. The MEME suite. Nucleic Acids Res. 2015, 43, W39–W49. [Google Scholar] [CrossRef]
- Xiao, H.; Ye, X.; Xu, H.; Mei, Y.; Yang, Y.; Chen, X.; Yang, Y.; Liu, T.; Yu, Y.; Yang, W. The genetic adaptations of fall armyworm Spodoptera frugiperda facilitated its rapid global dispersal and invasion. Mol. Ecol. Resour. 2020, 20, 1050–1068. [Google Scholar] [CrossRef]
- Wang, H.; Zhao, R.; Gao, J.; Xiao, X.; Yin, X.; Hu, S.; Zhang, Y.; Liang, P.; Gu, S. Two cuticle-enriched chemosensory proteins confer multi-insecticide resistance in Spodoptera frugiperda. Int. J. Biol. Macromol. 2024, 266, 130941. [Google Scholar] [CrossRef]
- Zhou, L.; Meng, J.Y.; Ruan, H.Y.; Yang, C.L.; Zhang, C.Y. Expression stability of candidate RT-qPCR housekeeping genes in Spodoptera frugiperda (Lepidoptera: Noctuidae). Arch. Insect Biochem. Physiol. 2021, 108, e21831. [Google Scholar] [CrossRef] [PubMed]
- Thornton, B.; Basu, C. Real-time PCR (qPCR) primer design using free online software. Biochem. Mol. Biol. Educ. 2011, 39, 145–154. [Google Scholar] [CrossRef] [PubMed]
- Jin, Y.; Liu, F.; Huang, W.; Sun, Q.; Huang, X. Identification of reliable reference genes for qRT-PCR in the ephemeral plant Arabidopsis pumila based on full-length transcriptome data. Sci. Rep. 2019, 9, 8408. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Hinton, P.R.; McMurray, I.; Brownlow, C. SPSS Explained; Routledge: London, UK, 2014. [Google Scholar]
- Swift, M.L. GraphPad prism, data analysis, and scientific graphing. J. Chem. Inf. Comput. Sci. 1997, 37, 411–412. [Google Scholar] [CrossRef]
- Innan, H.; Kondrashov, F. The evolution of gene duplications: Classifying and distinguishing between models. Nat. Rev. Genet. 2010, 11, 97–108. [Google Scholar] [CrossRef]
- Yang, M.; Yi, Y.; Xin-Hai, Y.; Hua-Mei, X.; Fei, L. Evolutionary analysis of detoxification gene families of Spodoptera frugiperda. J. Environ. Entomol. 2019, 41, 727–735. [Google Scholar]
- Fitch, W.M. Homology: A personal view on some of the problems. Trends Genet. 2000, 16, 227–231. [Google Scholar] [CrossRef]
- Weiqing, K.; Jinhong, Y. Cloning and RNAi of BmUGT2, a UDP-glucosyltransferase gene in Bombyx mori. J. Anhui Agric. Univ. 2009, 36, 693–698. [Google Scholar]
- Bozzolan, F.; Siaussat, D.; Maria, A.; Durand, N.; Pottier, M.A.; Chertemps, T.; Maïbèche-Coisne, M. Antennal uridine diphosphate (UDP)-glycosyltransferases in a pest insect: Diversity and putative function in odorant and xenobiotics clearance. Insect Mol. Biol. 2014, 23, 539–549. [Google Scholar] [CrossRef]
- Pansopha, P.; Ando, N.; Kanzaki, R. Dynamic use of optic flow during pheromone tracking by the male silkmoth, Bombyx mori. J. Exp. Biol. 2014, 217, 1811–1820. [Google Scholar] [CrossRef]
- Cui, X.; Wang, C.; Wang, X.; Li, G.; Liu, Z.; Wang, H. Molecular mechanism of the UDP-Glucuronosyltransferase 2B20-like Gene (AccUGT2b20-like) in Pesticide Resistance of Apis cerana cerana. Front. Genet. 2020, 11, 592595. [Google Scholar] [CrossRef] [PubMed]
- Kojima, W.; Fujii, T.; Suwa, M.; Miyazawa, M.; Ishikawa, Y. Physiological adaptation of the Asian corn borer Ostrinia furnacalis to chemical defenses of its host plant, maize. J. Insect Physiol. 2010, 56, 1349–1355. [Google Scholar] [CrossRef] [PubMed]
- Lou, L. Effect of Pesticides on UDP Glycosyltransferases Activity in Spodoptera litura; Nanjing Agricultural University: Nanjing, China, 2014. [Google Scholar]
- Tian, F.; Wang, Z.; Li, C.; Liu, J.; Zeng, X. UDP-Glycosyltransferases are involved in imidacloprid resistance in the Asian citrus psyllid, Diaphorina citri (Hemiptera: Lividae). Pestic. Biochem. Physiol. 2019, 154, 23–31. [Google Scholar] [CrossRef] [PubMed]
- Long, G.-Y.; Wang, Z.; Jin, D.-C.; Yang, H.; Yang, S.-J.; Zhou, C.; Zeng, Q.-H. RNAi-mediated held-out wing (HOW) gene knockdown inhibits wing expansion of white-backed planthopper, Sogatella furcifera (Hemiptera: Delphacidae). J. Insect Physiol. 2025, 163, 104810. [Google Scholar] [CrossRef]
- Kumari, R.; Saha, T.; Kumar, P.; Singh, A. CRISPR/Cas9-mediated genome editing technique to control fall armyworm (Spodoptera frugiperda) in crop plants with special reference to maize. Physiol. Mol. Biol. Plants 2024, 30, 1161–1173. [Google Scholar] [CrossRef]
- Shi, Z.; Luo, M.; Yuan, J.; Gao, B.; Yang, M.; Wang, G. CRISPR/Cas9-Based Functional Characterization of SfUGT50A15 Reveals Its Roles in the Resistance of Spodoptera frugiperda to Chlorantraniliprole, Emamectin Benzoate, and Benzoxazinoids. Insects 2024, 15, 314. [Google Scholar] [CrossRef]
- Groen, S.C.; Whiteman, N.K. Ecology and evolution of secondary compound detoxification systems in caterpillars. In Caterpillars in the Middle: Tritrophic Interactions in a Changing World; Springer: Berlin/Heidelberg, Germany, 2022; pp. 115–163. [Google Scholar]


| Primer Name | Sequence of Primers (5′-3′) |
|---|---|
| GADPH-F | CGGTGTCTTCACAACCACAG |
| GADPH-R | TTGACACCAACGACGAACAT |
| SfUGT9-F | ACAATGTCGGTGCTCTGGTT |
| SfUGT9-R | TCTGCGGTGATAACGGTGAC |
| SfUGT16-F | AACTATGGACACGGATTCAATGG |
| SfUGT16-R | CGTACAGCATCATGTTCATTAGC |
| SfUGT21-F | AGGTCCGATACAACGCAACT |
| SfUGT21-R | CAGGTAATGGCTTGACTTCTTCC |
| SfUGT30-F | TGAAGGACTGTGGAAGGTTGT |
| SfUGT30-R | CGGATGAGCGTTAATCAGCATA |
| SfUGT34-F | ACAAGGAGGTCTACAATCAACAG |
| SfUGT34-R | CCTATGAGTGGAACGCCTACTA |
| SfUGT40-F | CTACAGTCCACAGATGAGGCTAT |
| SfUGT40-R | TCTTCCACAGTCAACTTCTCCATA |
| SfUGT48-F | GCTGGTTCCTTGCTCATACTG |
| SfUGT48-R | AACTGCTCCACAACAATCACAT |
| Duplicated Genes | Ka | Ks | Ka/Ks | Divergence Time (Million Years) | Duplicated Type |
|---|---|---|---|---|---|
| SfUGT1-SfUGT5 | 0.40192179 | 1.76697972 | 0.22746259 | 13.6 | Tandem replication |
| SfUGT2-SfUGT4 | 0.37795081 | 3.3103476 | 0.11417254 | 25.5 | Tandem replication |
| SfUGT10-SfUGT12 | 0.27759684 | NaN | NaN | Tandem replication | |
| SfUGT11-SfUGT13 | 0.27068699 | NaN | NaN | Tandem replication | |
| SfUGT16-SfUGT17 | 0.1520578 | 0.91537335 | 0.1661156 | 7 | Tandem replication |
| SfUGT18-SfUGT16 | 0.15772597 | 0.85685295 | 0.18407589 | 6.6 | Tandem replication |
| SfUGT17-SfUGT19 | 0.10253229 | 0.47379352 | 0.21640712 | 3.6 | Tandem replication |
| SfUGT20-SfUGT27 | 0.35321099 | 4.72283199 | 0.07478796 | 36.3 | Tandem replication |
| SfUGT21-SfUGT26 | 0.09656067 | 0.40050911 | 0.24109481 | 3.1 | Tandem replication |
| SfUGT22-SfUGT28 | 0.13243781 | 0.49979133 | 0.2649862 | 3.8 | Tandem replication |
| SfUGT24-SfUGT25 | 0.22994111 | 1.43336234 | 0.16042079 | 11 | Tandem replication |
| SfUGT26-SfUGT27 | 0.14655933 | 0.53908785 | 0.27186538 | 4.1 | Tandem replication |
| SfUGT34-SfUGT33 | 0.21169376 | 1.09658972 | 0.19304737 | 8.4 | Tandem replication |
| SfUGT34-SfUGT35 | 0.21264377 | 1.03035232 | 0.20637967 | 7.9 | Tandem replication |
| SfUGT36-SfUGT37 | 0.07379752 | 0.23438925 | 0.3148503 | 1.8 | Tandem replication |
| SfUGT37-SfUGT38 | 0.01976299 | 0.1246974 | 0.15848757 | 1 | Tandem replication |
| SfUGT40-SfUGT43 | 0.12685392 | 0.57297678 | 0.22139452 | 4.4 | Tandem replication |
| SfUGT41-SfUGT42 | 0.31637091 | 1.07271261 | 0.29492606 | 8.3 | Tandem replication |
| SfUGT41-SfUGT44 | 0.30201086 | 1.72279897 | 0.17530244 | 13.3 | Tandem replication |
| SfUGT43-SfUGT46 | 0.18612883 | 0.98093876 | 0.18974562 | 7.5 | Tandem replication |
| SfUGT44-SfUGT45 | 0.28981858 | 1.44860626 | 0.20006719 | 11.1 | Tandem replication |
| SfUGT45-SfUGT46 | 0.18289155 | 1.01046947 | 0.18099661 | 7.8 | Tandem replication |
| SfUGT47-SfUGT48 | 0.58233839 | 2.35592325 | 0.24718054 | 18.1 | Tandem replication |
| Gene Name | Gene ID | Number of Amino Acid | Molecular Weight | Theoretical pI | Instability Index | Aliphatic | Grand Average of Hydropathicity | Subcellular Localization |
|---|---|---|---|---|---|---|---|---|
| SfUGT1 | 118265011 | 342 | 38,957.63 | 9.16 | 47.02 | 107.43 | 0.111 | mito |
| SfUGT2 | 118265039 | 373 | 41,734.13 | 8.88 | 44.43 | 99.57 | −0.005 | extr |
| SfUGT3 | 118265260 | 534 | 60,369.81 | 7.98 | 53.91 | 104.96 | 0.16 | plas |
| SfUGT4 | 118265297 | 526 | 59,061.97 | 7.78 | 40.81 | 108.42 | 0.134 | plas |
| SfUGT5 | 118265425 | 526 | 59,083.03 | 8.25 | 39.12 | 107.87 | 0.14 | plas |
| SfUGT6 | 118266879 | 530 | 58,835.84 | 7.82 | 40.64 | 103.98 | 0.053 | extr |
| SfUGT7 | 118267687 | 544 | 62,128.39 | 9.07 | 57.92 | 95.18 | −0.042 | plas |
| SfUGT8 | 118267688 | 544 | 61,991.21 | 9.01 | 56.57 | 95.9 | −0.046 | plas |
| SfUGT9 | 118268440 | 520 | 59,548.25 | 6.77 | 48.3 | 108.69 | −0.024 | E.R. |
| SfUGT10 | 118269227 | 525 | 60,827.42 | 6.77 | 47.21 | 100.44 | −0.108 | E.R. |
| SfUGT11 | 118269255 | 525 | 60,668.23 | 6.48 | 45.96 | 103.96 | −0.069 | E.R. |
| SfUGT12 | 118269333 | 521 | 60,644.21 | 8.13 | 44.15 | 96.14 | −0.181 | E.R. |
| SfUGT13 | 118269358 | 521 | 60,690.24 | 8.13 | 43.91 | 95.39 | −0.189 | E.R. |
| SfUGT14 | 118275940 | 539 | 61,284.8 | 9.37 | 40.77 | 102.02 | 0.004 | E.R. |
| SfUGT15 | 118277768 | 523 | 59,649.65 | 8.76 | 39.57 | 102.89 | −0.038 | nucl |
| SfUGT16 | 118277895 | 522 | 59,740.24 | 7 | 39.78 | 101.76 | −0.077 | plas |
| SfUGT17 | 118277896 | 521 | 60,228.23 | 9.15 | 37.79 | 99.5 | −0.14 | E.R. |
| SfUGT18 | 118277897 | 521 | 59,245.87 | 8.75 | 38.97 | 108.89 | 0.024 | plas |
| SfUGT19 | 118277898 | 528 | 60,765.61 | 8.52 | 41.42 | 104.66 | −0.096 | E.R. |
| SfUGT20 | 118277899 | 524 | 59,178.42 | 8.87 | 42.89 | 100.06 | 0.029 | E.R. |
| SfUGT21 | 118277900 | 523 | 59,652.4 | 8.92 | 41.9 | 101.72 | −0.156 | E.R. |
| SfUGT22 | 118277901 | 523 | 59,365.08 | 8.33 | 32.63 | 98.91 | −0.002 | E.R. |
| SfUGT23 | 118277902 | 522 | 59,149.35 | 8.89 | 45.66 | 97.28 | 0.001 | plas |
| SfUGT24 | 118277903 | 522 | 59,049.7 | 6.81 | 40.21 | 102.49 | 0.014 | E.R. |
| SfUGT25 | 118277904 | 522 | 59,434.31 | 8.05 | 40.88 | 99.48 | −0.075 | E.R. |
| SfUGT26 | 118277905 | 520 | 59,257.15 | 9.05 | 41.72 | 104.73 | −0.084 | plas |
| SfUGT27 | 118277906 | 438 | 50,218.57 | 9.06 | 46.38 | 96.96 | −0.118 | pero |
| SfUGT28 | 118278039 | 1102 | 124,725.23 | 8.24 | 35.96 | 107.22 | 0.016 | E.R. |
| SfUGT29 | 118278122 | 508 | 58,469.71 | 5.89 | 55.23 | 107.24 | −0.089 | plas |
| SfUGT30 | 118278231 | 236 | 26,641.16 | 9.35 | 33.12 | 101.14 | −0.1 | cyto |
| SfUGT31 | 118278649 | 514 | 58,243.33 | 8.92 | 56.53 | 105.47 | 0.047 | plas |
| SfUGT32 | 118278849 | 520 | 57,729.49 | 9.26 | 47.01 | 98.85 | 0.018 | pero |
| SfUGT33 | 118279153 | 518 | 59,256.76 | 8.26 | 44.09 | 103.05 | −0.116 | E.R. |
| SfUGT34 | 118279154 | 516 | 59,179.94 | 8.77 | 43.66 | 100.23 | −0.151 | E.R. |
| SfUGT35 | 118279155 | 515 | 58,893.45 | 7.27 | 47.61 | 99.32 | −0.139 | E.R. |
| SfUGT36 | 118279189 | 518 | 58,821.86 | 8.63 | 37.54 | 106.66 | −0.025 | E.R. |
| SfUGT37 | 118279190 | 322 | 37,089.67 | 8.29 | 36.55 | 105.62 | 0.003 | extr |
| SfUGT38 | 118279191 | 286 | 32,873.72 | 6.55 | 36.41 | 109.34 | 0.134 | E.R. |
| SfUGT39 | 118279357 | 512 | 58,603.17 | 6.32 | 42 | 106.58 | −0.026 | E.R. |
| SfUGT40 | 118279411 | 525 | 59,503.47 | 8.22 | 41.82 | 105.24 | 0.011 | E.R. |
| SfUGT41 | 118279412 | 522 | 59,613.42 | 8.34 | 43.32 | 102.3 | −0.056 | E.R. |
| SfUGT42 | 118279413 | 521 | 58,861.73 | 8.83 | 50.4 | 105.47 | −0.012 | plas |
| SfUGT43 | 118279415 | 521 | 59,225.95 | 8.48 | 40.83 | 103.63 | −0.016 | E.R. |
| SfUGT44 | 118279416 | 521 | 59,580.05 | 6.61 | 50.23 | 106.07 | −0.088 | plas |
| SfUGT45 | 118279417 | 520 | 59,555.16 | 7.81 | 41.16 | 106.27 | −0.067 | E.R. |
| SfUGT46 | 118279418 | 520 | 59,370.96 | 7.79 | 47.38 | 109.42 | −0.089 | E.R. |
| SfUGT47 | 118281901 | 516 | 58,246.87 | 8.92 | 42.41 | 104.81 | 0.12 | plas |
| SfUGT48 | 118281915 | 1101 | 125,967.94 | 9.11 | 44.63 | 95.44 | −0.099 | E.R. |
| Gene Name | Alpha Helix | Beta Turn | Random Coil | Extended Strand |
|---|---|---|---|---|
| SfUGT1 | 141 (41.23%) | 9 (2.63%) | 137 (40.06%) | 55 (16.08%) |
| SfUGT2 | 162 (43.43%) | 17 (4.56%) | 140 (37.53%) | 54 (14.48%) |
| SfUGT3 | 253 (47.38%) | 28 (5.24%) | 180 (33.71%) | 73 (13.67%) |
| SfUGT4 | 251 (47.72%) | 26 (4.94%) | 179 (34.03%) | 70 (13.31%) |
| SfUGT5 | 242 (46.01%) | 22 (4.18%) | 194 (36.88%) | 68 (12.93%) |
| SfUGT6 | 259 (48.87%) | 25 (4.72%) | 181 (34.15%) | 65 (12.26%) |
| SfUGT7 | 251 (46.14%) | 27 (4.96%) | 191 (35.11%) | 75 (13.79%) |
| SfUGT8 | 261 (47.98%) | 20 (3.68%) | 192 (35.29%) | 71 (13.05%) |
| SfUGT9 | 232 (44.62%) | 23 (4.42%) | 192 (36.92%) | 73 (14.04%) |
| SfUGT10 | 255 (48.57%) | 21 (4.00%) | 181 (34.48%) | 68 (12.95%) |
| SfUGT11 | 244 (46.48%) | 25 (4.76%) | 179 (34.10%) | 77 (14.67%) |
| SfUGT12 | 244 (46.83%) | 22 (4.22%) | 181 (34.74%) | 74 (14.20%) |
| SfUGT13 | 239 (45.87%) | 25 (4.80%) | 185 (35.51%) | 72 (13.82%) |
| SfUGT14 | 243 (45.08%) | 29 (5.38%) | 191 (35.44%) | 76 (14.10%) |
| SfUGT15 | 239 (45.70%) | 22 (4.21%) | 192 (36.71%) | 70 (13.38%) |
| SfUGT16 | 239 (45.79%) | 30 (5.75%) | 184 (35.25%) | 69 (13.22%) |
| SfUGT17 | 241 (46.26%) | 21 (4.03%) | 187 (35.89%) | 72 (13.82%) |
| SfUGT18 | 233 (44.72%) | 24 (4.61%) | 186 (35.70%) | 78 (14.97%) |
| SfUGT19 | 248 (46.97%) | 24 (4.55%) | 184 (34.85%) | 72 (13.64%) |
| SfUGT20 | 249 (47.52%) | 25 (4.77%) | 180 (34.35%) | 70 (13.36%) |
| SfUGT21 | 243 (46.46%) | 28 (5.35%) | 175 (33.46%) | 77 (14.72%) |
| SfUGT22 | 233 (44.55%) | 23 (4.40%) | 189 (36.14%) | 78 (14.91%) |
| SfUGT23 | 248 (47.51%) | 23 (4.41%) | 184 (35.25%) | 67 (12.84%) |
| SfUGT24 | 234 (44.83%) | 23 (4.41%) | 191 (36.59%) | 74 (14.18%) |
| SfUGT25 | 235 (45.02%) | 24 (4.60%) | 194 (37.16%) | 69 (13.22%) |
| SfUGT26 | 240 (46.15%) | 22 (4.23%) | 189 (36.35%) | 69 (13.27%) |
| SfUGT27 | 200 (45.66%) | 20 (4.57%) | 165 (37.67%) | 53 (12.10%) |
| SfUGT28 | 446 (40.47%) | 90 (8.17%) | 338 (30.67%) | 228 (20.69%) |
| SfUGT29 | 250 (49.21%) | 22 (4.33%) | 165 (32.48%) | 71 (13.98%) |
| SfUGT30 | 104 (44.07%) | 17 (7.20%) | 83 (35.17%) | 32 (13.56%) |
| SfUGT31 | 240 (46.69%) | 26 (5.06%) | 176 (34.24%) | 72 (14.01%) |
| SfUGT32 | 230 (44.23%) | 23 (4.42%) | 194 (37.31%) | 73 (14.04%) |
| SfUGT33 | 231 (44.59%) | 22 (4.25%) | 193 (37.26%) | 72 (13.90%) |
| SfUGT34 | 236 (45.74%) | 26 (5.04%) | 181 (35.08%) | 73 (14.15%) |
| SfUGT35 | 246 (47.77%) | 21 (4.08%) | 179 (34.76%) | 69 (13.40%) |
| SfUGT36 | 245 (47.30%) | 24 (4.63%) | 178 (34.36%) | 71 (13.71%) |
| SfUGT37 | 157 (48.76%) | 11 (3.42%) | 111 (34.47%) | 43 (13.35%) |
| SfUGT38 | 124 (43.36%) | 8 (2.80%) | 104 (36.36%) | 50 (17.48%) |
| SfUGT39 | 234 (45.70%) | 24 (4.69%) | 177 (34.57%) | 77 (15.04%) |
| SfUGT40 | 231 (44.00%) | 23 (4.38%) | 198 (37.71%) | 73 (13.90%) |
| SfUGT41 | 241 (46.17%) | 26 (4.98%) | 179 (34.29%) | 76 (14.56%) |
| SfUGT42 | 236 (45.30%) | 25 (4.80%) | 182 (34.93%) | 78 (14.97%) |
| SfUGT43 | 233 (44.72%) | 22 (4.22%) | 189 (36.28%) | 77 (14.78%) |
| SfUGT44 | 244 (46.83%) | 28 (5.37%) | 181 (34.74%) | 68 (13.05%) |
| SfUGT45 | 231 (44.42%) | 23 (4.42%) | 191 (36.73%) | 75 (14.42%) |
| SfUGT46 | 240 (46.15%) | 23 (4.42%) | 189 (36.35%) | 68 (13.08%) |
| SfUGT47 | 231 (44.77%) | 20 (3.88%) | 189 (36.63%) | 76 (14.73%) |
| SfUGT48 | 474 (43.05%) | 55 (5.00%) | 391 (35.51%) | 181 (16.44%) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, Y.; Guan, M.; Zou, K.; Wang, T.; Wang, H.; Sun, L.; Feng, B.; Ding, J.; Gao, X.; Wang, Y.; et al. Comprehensive Analysis of the UGT Gene Superfamily in Spodoptera frugiperda. Insects 2025, 16, 601. https://doi.org/10.3390/insects16060601
Liu Y, Guan M, Zou K, Wang T, Wang H, Sun L, Feng B, Ding J, Gao X, Wang Y, et al. Comprehensive Analysis of the UGT Gene Superfamily in Spodoptera frugiperda. Insects. 2025; 16(6):601. https://doi.org/10.3390/insects16060601
Chicago/Turabian StyleLiu, Yang, Minghui Guan, Kunliang Zou, Tonghan Wang, Haiyang Wang, Lu Sun, Bo Feng, Jiali Ding, Xiang Gao, Yongfu Wang, and et al. 2025. "Comprehensive Analysis of the UGT Gene Superfamily in Spodoptera frugiperda" Insects 16, no. 6: 601. https://doi.org/10.3390/insects16060601
APA StyleLiu, Y., Guan, M., Zou, K., Wang, T., Wang, H., Sun, L., Feng, B., Ding, J., Gao, X., Wang, Y., Wu, D., & Du, J. (2025). Comprehensive Analysis of the UGT Gene Superfamily in Spodoptera frugiperda. Insects, 16(6), 601. https://doi.org/10.3390/insects16060601





