Intake of Pyriproxyfen Through Contaminated Food by the Predator Ceraeochrysa claveri Navás, 1911 (Neuroptera: Chrysopidae): Evaluation of Long-Term Effects on Testes via Transcriptome Analysis †
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Insect Rearing
2.2. Bioassays
2.3. RNA Extraction, Library Preparation, and Sequencing
2.4. Reads Filtering, De Novo Assembly, and Annotation
2.5. Differentially Expressed Genes (DEGs) and Gene Ontology
2.6. RT-qPCR for DEGs Validation
3. Results
3.1. De Novo Assembly and Annotation
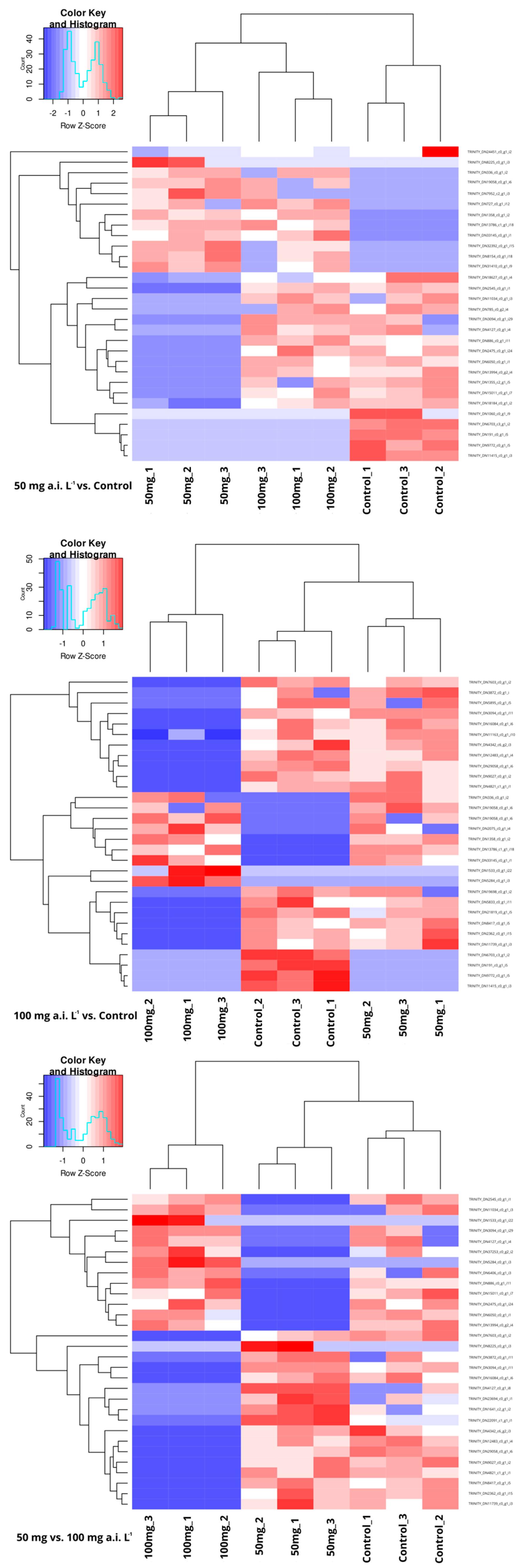
3.2. DEGs Analyses
3.3. RT-qPCR
4. Discussion
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Vasilikopoulos, A.; Misof, B.; Meusemann, K.; Lieberz, D.; Flouri, T.; Beutel, R.G.; Niehuis, O.; Wappler, T.; Rust, J.; Peters, R.S.; et al. An integrative phylogenomic approach to elucidate the evolutionary history and divergence times of Neuropterida (Insecta: Holometabola). BMC Evol. Biol. 2020, 20, 64. [Google Scholar] [CrossRef]
- Neuropterida Species of the World. Catalogue of Life Checklist. Available online: https://www.checklistbank.org/dataset/278910/taxon/3ND (accessed on 25 February 2025).
- Freitas, S.; Penny, N.D. The green lacewings (Neuroptera: Chrysopidae) of Brazilian agro-ecossystems. Proc. Calif. Acad. Sci. 2001, 52, 245–395. [Google Scholar]
- Albuquerque, G.S.; Tauber, C.A.; Tauber, M.J. Green lacewings (Neuroptera: Chrysopidae): Predatory lifestyle. In Insect Bioecology and Nutrition for Integrated Pest Management, 1st ed.; Panizzi, A.R., Parra, J.R.P., Eds.; CRC Press: Boca Raton, FL, USA, 2012; pp. 593–631. [Google Scholar]
- Pappas, M.L.; Broufas, G.D.; Koveos, D.S. Chrysopid predators and their role in biological control. J. Entomol. 2011, 8, 301–326. [Google Scholar] [CrossRef]
- Machado, R.J.P.; Martins, C.C.; Freitas, S.D.; Penny, N.D. Capítulo 29: Neuroptera Linnaeus, 1758. In Insetos do Brasil: Diversidade e Taxonomia, 2nd ed.; Rafael, J.A., Melo, G.A.R., Carvalho, C.J.B.D., Eds.; Editora INPA: Manaus, Brazil, 2024; pp. 552–567. [Google Scholar] [CrossRef]
- Fogel, M.N.; Schneider, M.I.; Rimoldi, F.; Ladux, L.S.; Desneux, N.; Ronco, A.E. Toxicity assessment of four insecticides with different modes of action on pupae and adults of Eriopis connexa (Coleoptera: Coccinellidae), a relevant predator of the Neotropical region. Environ. Sci. Pollut. Res. 2016, 23, 14918–14926. [Google Scholar] [CrossRef]
- Ono, É.K.; Zanardi, O.Z.; Aguiar Santos, K.F.; Yamamoto, P.T. Susceptibility of Ceraeochrysa cubana larvae and adults to six insect growth-regulator insecticides. Chemosphere 2017, 168, 49–57. [Google Scholar] [CrossRef]
- Sohail, M.; Nasar, M.H.; Muhammad, R.; Soomro, Q.A.; Asif, M.U.; Maari, J.M. Resistance potential of Chrysoperla carnea (Stephens) to insecticides used against sucking complex of cotton. Int. J. Ecotoxicol. Ecobiol. 2019, 4, 1–7. [Google Scholar] [CrossRef]
- Jindra, M.; Bittova, L. The juvenile hormone receptor as a target of juvenoid “Insect growth regulators”. Arch. Insect Biochem. Physiol. 2020, 103, e21615. [Google Scholar] [CrossRef]
- The IRAC Mode of Action Classification Online. Insecticide Resistance Action Committee. Available online: https://irac-online.org/mode-of-action/classification-online/ (accessed on 28 January 2024).
- Boina, D.R.; Rogers, M.E.; Wang, N.; Stelinski, L.L. Effect of pyriproxyfen, a juvenile hormone mimic, on egg hatch, nymph development, adult emergence and reproduction of the Asian citrus psyllid, Diaphorina citri Kuwayama. Pest Manag. Sci. 2010, 66, 349–357. [Google Scholar] [CrossRef]
- Duan, D.; Zheng, R.; Lin, S.; Chen, Y.; Tian, H.; Zhao, J.; Tian, S.; Wei, H.; Gu, X. Modulation of juvenile hormone esterase gene expression against development of Plutella xylostella (Lepidoptera: Plutellidae). J. Econ. Entomol. 2016, 109, 865–872. [Google Scholar] [CrossRef]
- Kancharlapalli, S.J.; Brelsfoard, C.L. The impact of non-lethal doses of pyriproxyfen on male and female Aedes albopictus reproductive fitness. Front. Insect Sci. 2024, 4, 1430422. [Google Scholar] [CrossRef]
- Xu, Q.; Tang, B.; Zou, Q.; Zheng, H.; Liu, X.; Wang, S. Effects of pyriproxyfen on female reproduction in the common cutworm, Spodoptera litura (F.) (Lepidoptera: Noctuidae). PLoS ONE 2015, 10, e0138171. [Google Scholar] [CrossRef] [PubMed]
- Litsey, E.M.; Fine, J.D. Developmental exposure to hormone-mimicking insect growth disruptors alters expression of endocrine-related genes in worker honey bee (Hymenoptera: Apidae) brains and hypopharyngeal glands. J. Econ. Entomol. 2024, 117, 377–387. [Google Scholar] [CrossRef] [PubMed]
- Qian, H.-Y.; Zhang, X.; Zhao, G.-D.; Guo, H.-M.; Li, G.; Xu, A.-Y. Effects of pyriproxyfen exposure on reproduction and gene expressions in silkworm, Bombyx mori. Insects 2020, 11, 467. [Google Scholar] [CrossRef] [PubMed]
- Guo, H.; Zhang, X.; Sun, J.; Zheng, X.; Zhao, G.; Li, G.; Qian, H. Effects of pyriproxyfen exposure on damage to midgut and related gene expressions in the Bombyx mori silkworm. Sci. Asia 2021, 47, 733. [Google Scholar] [CrossRef]
- Zhao, G.; Zhang, X.; Wang, C.; Zhang, H.; Guo, H.; Qian, H.; Li, G.; Xu, A. Effect of pyriproxyfen exposure on cocooning and gene expression in the silk gland of Bombyx mori (Linnaeus, 1758). Ecotoxicol. Environ. Saf. 2020, 202, 110914. [Google Scholar] [CrossRef]
- Wang, P.; Cui, Q.; Wang, X.; Liu, Y.; Zhang, Y.; Huang, X.; Jiang, S.; Jiang, M.; Bi, L.; Li, B.; et al. The inhibition of ecdysone signal pathway was the key of pyriproxyfen poisoning for silkworm, Bombyx mori. Pestic. Biochem. Physiol. 2023, 189, 105307. [Google Scholar] [CrossRef]
- Luo, J.; Liu, S.; Hou, J.; Chen, L.; Li, H.; Liao, S.; Tan, Q.; Yang, T.; Yi, G.; Zhang, F.; et al. The comparison of juvenile hormone and transcriptional changes between three different juvenile hormone analogs insecticides on honey bee worker larval’s development. Agronomy 2021, 11, 2497. [Google Scholar] [CrossRef]
- Garcia, A.S.G.; Scudeler, E.L.; Santos, D.C. Ultrastructure of testes of Ceraeochrysa claveri (Neuroptera: Chrysopidae) after exposure to azadirachtin and pyriproxyfen. Microsc. Microanal. 2020, 26, 205–206. [Google Scholar] [CrossRef]
- Scudeler, E.L.; Carvalho, S.F.D.; Garcia, A.S.G.; Santorum, M.; Padovani, C.R.; Santos, D.C.D. Midgut and fat body: Multisystemic action of pyriproxyfen on non-target organism Ceraeochrysa claveri (Neuroptera: Chrysopidae). Environ. Pollut. 2022, 293, 118580. [Google Scholar] [CrossRef]
- Venkatesan, T.; Jalali, S.K.; Murthy, K.S.; Rabindra, R.J.; Lalitha, Y. Occurrence of insecticide resistance in field populations of Chrysoperla zastrowi arabica (Neuroptera: Chrysopidae) in India. Indian J. Agric. Sci. 2009, 79, 910–912. [Google Scholar]
- Pathan, A.K.; Sayyed, A.H.; Aslam, M.; Liu, T.-X.; Razzaq, M.; Gillani, W.A. Resistance to pyrethroids and organophosphates increased fitness and predation potential of Chrysoperla carnea (Neuroptera: Chrysopidae). J. Econ. Entomol. 2010, 103, 823–834. [Google Scholar] [CrossRef]
- Fátima, T.A.; Andrade, C.G.; Costa, L.V.; Fonseca, M.V. Selectivity of seven insecticides against pupae and adults of Chrysoperla externa (Neuroptera: Chrysopidae). Rev. Colom. Entomol. 2013, 39, 34–39. [Google Scholar]
- Mansoor, M.M.; Abbas, N.; Shad, S.A.; Pathan, A.K.; Razaq, M. Increased fitness and realized heritability in emamectin benzoate-resistant Chrysoperla carnea (Neuroptera: Chrysopidae). Ecotoxicology 2013, 22, 1232–1240. [Google Scholar] [CrossRef]
- Mansoor, M.M.; Shad, S.A. Genetics, cross-resistance and realized heritability of resistance to acetamiprid in generalist predator, Chrysoperla carnea (Steph.) (Neuroptera: Chrysopidae). Egypt. J. Biol. Pest Control. 2020, 30, 23. [Google Scholar] [CrossRef]
- Pathak, J.; Ramasamy, G.G.; Agrawal, A.; Srivastava, S.; Basavaarya, B.R.; Muthugounder, M.; Muniyappa, V.K.; Maria, P.; Rai, A.; Venkatesan, T. Comparative transcriptome analysis to reveal differentially expressed cytochrome P450 in response to imidacloprid in the aphid lion, Chrysoperla zastrowi sillemi (Esben-Petersen). Insects 2022, 13, 900. [Google Scholar] [CrossRef]
- Graveley, B.R.; Brooks, A.N.; Carlson, J.W.; Duff, M.O.; Landolin, J.M.; Yang, L.; Artieri, C.G.; Van Baren, M.J.; Boley, N.; Booth, B.W.; et al. The developmental transcriptome of Drosophila melanogaster. Nature 2011, 471, 473–479. [Google Scholar] [CrossRef]
- Oppenheim, S.J.; Baker, R.H.; Simon, S.; DeSalle, R. We can’t all be supermodels: The value of comparative transcriptomics to the study of non-model insects. Insect Mol. Biol. 2015, 24, 139–154. [Google Scholar] [CrossRef]
- Wang, Y.; Zhou, X.; Wang, L.; Liu, X.; Yang, D.; Rokas, A. Gene selection and evolutionary modeling affect phylogenomic inference of Neuropterida based on transcriptome data. Int. J. Mol. Sci. 2019, 20, 1072. [Google Scholar] [CrossRef]
- Liu, Z.-F.; Liang, Y.; Sun, X.; Yang, J.; Zhang, P.-J.; Gao, Y.; Fan, J.-B.; Fan, R.-J. Analysis of differentially expressed genes of Chrysoperla sinica related to flight capacity by transcriptome. J. Insect Sci. 2021, 21, 18. [Google Scholar] [CrossRef]
- Li, Z.-Q.; Zhang, S.; Luo, J.-Y.; Wang, S.-B.; Wang, C.-Y.; Lv, L.-M.; Dong, S.-L.; Cui, J.-J. Identification and expression pattern of candidate olfactory genes in Chrysoperla sinica by antennal transcriptome analysis. Comp. Biochem. Physiol. Part D Genom. Proteom. 2015, 15, 28–38. [Google Scholar] [CrossRef]
- Du, Q.; Shan, Y.; Hu, H.; Wu, C.; Wang, D.; Song, X.; Ma, Y.; Xi, J.; Ren, X.; Ma, X.; et al. Fitness effect and transcription profile reveal sublethal effect of nitenpyram on the predator Chrysopa pallens (Neuroptera: Chrysopidae). Arch. Insect Biochem. Physiol. 2024, 115, e22073. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.-Q.; Zhang, S.; Ma, Y.; Luo, J.-Y.; Wang, C.-Y.; Lv, L.-M.; Dong, S.-L.; Cui, J.-J. First transcriptome and digital gene expression analysis in Neuroptera with an emphasis on chemoreception genes in Chrysopa pallens (Rambur). PLoS ONE 2013, 8, e67151. [Google Scholar] [CrossRef]
- Shi, Y.; Pandit, A.; Nachman, R.J.; Christiaens, O.; Davies, S.A.; Dow, J.A.T.; Smagghe, G. Transcriptome analysis of neuropeptides in the beneficial insect lacewing (Chrysoperla carnea) identifies kinins as a selective pesticide target: A biostable kinin analogue with activity against the peach potato aphid Myzus persicae. J. Pest Sci. 2023, 96, 253–264. [Google Scholar] [CrossRef]
- Scudeler, E.L.; Barroso, G.; Daquila, B.V.; Carvalho, S.F.; Conte, H.; Santos, D.C. Pyriproxyfen exposure compromises cocoon spinning and damages the Malpighian tubules of the nontarget predator Ceraeochrysa claveri (Neuroptera: Chrysopidae). Environ. Pollut. 2024, 363, 125255. [Google Scholar] [CrossRef]
- MAPA. Ministério da Agricultura, Pecuária e Abastecimento. Agrofit: Sistema de Agrotóxicos Fitossanitários. Available online: https://agrofit.agricultura.gov.br/agrofit_cons/principal_agrofit_cons (accessed on 11 January 2024).
- Andrews, S. FastQC: A Quality Control Tool for High Throughput Sequence Data. Available online: https://www.bioinformatics.babraham.ac.uk/projects/fastqc (accessed on 2 March 2023).
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef]
- Bushnell, B. BBMap: A Fast, Accurate, Splice-Aware Aligner. Available online: https://escholarship.org/uc/item/1h3515gn#main (accessed on 13 March 2023).
- Grabherr, M.G.; Haas, B.J.; Yassour, M.; Levin, J.Z.; Thompson, D.A.; Amit, I.; Adiconis, X.; Fan, L.; Raychowdhury, R.; Zeng, Q.; et al. Full-length transcriptome assembly from RNA-seq data without a reference genome. Nat. Biotechnol. 2011, 29, 644–652. [Google Scholar] [CrossRef]
- Haas, B.J.; Papanicolaou, A.; Yassour, M.; Grabherr, M.; Blood, P.D.; Bowden, J.; Couger, M.B.; Eccles, D.; Li, B.; Lieber, M.; et al. De novo transcript sequence reconstruction from RNA-seq using the Trinity platform for reference generation and analysis. Nat. Protoc. 2013, 8, 1494–1512. [Google Scholar] [CrossRef]
- Li, W.; Godzik, A. Cd-Hit: A fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics 2006, 22, 1658–1659. [Google Scholar] [CrossRef]
- Fu, L.; Niu, B.; Zhu, Z.; Wu, S.; Li, W. CD-HIT: Accelerated for clustering the next-generation sequencing data. Bioinformatics 2012, 28, 3150–3152. [Google Scholar] [CrossRef]
- Kim, D.; Paggi, J.M.; Park, C.; Bennett, C.; Salzberg, S.L. Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat. Biotechnol. 2019, 37, 907–915. [Google Scholar] [CrossRef]
- Manni, M.; Berkeley, M.R.; Seppey, M.; Zdobnov, E.M. BUSCO: Assessing genomic data quality and beyond. Curr. Protoc. 2021, 1, e323. [Google Scholar] [CrossRef] [PubMed]
- Manni, M.; Berkeley, M.R.; Seppey, M.; Simão, F.A.; Zdobnov, E.M. BUSCO update: Novel and streamlined workflows along with broader and deeper phylogenetic coverage for scoring of eukaryotic, prokaryotic, and viral genomes. Mol. Biol. Evol. 2021, 38, 4647–4654. [Google Scholar] [CrossRef] [PubMed]
- TransDecoder. Available online: https://github.com/TransDecoder/TransDecoder (accessed on 20 March 2023).
- Casimiro-Soriguer, C.S.; Muñoz-Mérida, A.; Pérez-Pulido, A.J. Sma3s: A universal tool for easy functional annotation of proteomes and transcriptomes. Proteomics 2017, 17, 1700071. [Google Scholar] [CrossRef]
- Patro, R.; Duggal, G.; Love, M.I.; Irizarry, R.A.; Kingsford, C. Salmon provides fast and bias-aware quantification of transcript expression. Nat. Methods 2017, 14, 417–419. [Google Scholar] [CrossRef]
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010, 26, 139–140. [Google Scholar] [CrossRef]
- McCarthy, D.J.; Chen, Y.; Smyth, G.K. Differential expression analysis of multifactor RNA-seq experiments with respect to biological variation. Nucleic Acids Res. 2012, 40, 4288–4297. [Google Scholar] [CrossRef]
- Chen, Y.; Lun, A.T.L.; Smyth, G.K. From reads to genes to pathways: Differential expression analysis of RNA-seq experiments using Rsubread and the edgeR quasi-likelihood pipeline. F1000Research 2016, 5, 1438. [Google Scholar] [CrossRef]
- The R Project for Statistical Computing. Available online: https://www.r-project.org/ (accessed on 18 April 2023).
- Benjamini, Y.; Yekutieli, D. The control of the false discovery rate in multiple testing under dependency. Ann. Stat. 2001, 29, 1165–1188. [Google Scholar] [CrossRef]
- Carbon, S.; Ireland, A.; Mungall, C.J.; Shu, S.; Marshall, B.; Lewis, S. AmiGO: Online access to ontology and annotation data. Bioinformatics 2009, 25, 288–289. [Google Scholar] [CrossRef]
- Untergasser, A.; Nijveen, H.; Rao, X.; Bisseling, T.; Geurts, R.; Leunissen, J.A.M. Primer3Plus, an enhanced web interface to Primer3. Nucleic Acids Res. 2007, 35, W71–W74. [Google Scholar] [CrossRef]
- Stothard, P. The sequence manipulation suite: JavaScript programs for analyzing and formatting protein and DNA sequences. BioTechniques 2000, 28, 1102–1104. [Google Scholar] [CrossRef] [PubMed]
- Kibbe, W.A. OligoCalc: An online oligonucleotide properties calculator. Nucleic Acids Res. 2007, 35, W43–W46. [Google Scholar] [CrossRef] [PubMed]
- Lü, J.; Yang, C.; Zhang, Y.; Pan, H. Selection of reference genes for the normalization of RT-qPCR data in gene expression studies in insects: A systematic review. Front. Physiol. 2018, 9, 1560. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Bustin, S.A.; Benes, V.; Garson, J.A.; Hellemans, J.; Huggett, J.; Kubista, M.; Mueller, R.; Nolan, T.; Pfaffl, M.W.; Shipley, G.L.; et al. The MIQE guidelines: Minimum information for publication of quantitative real-time PCR experiments. Clin. Chem. 2009, 55, 611–622. [Google Scholar] [CrossRef]
- Puente, X.S.; López-Otn, C. Cloning and expression analysis of a novel human serine hydrolase with sequence similarity to prokaryotic enzymes involved in the degradation of aromatic compounds. J. Biol. Chem. 1995, 270, 12926–12932. [Google Scholar] [CrossRef]
- Derewenda, Z.S.; Derewenda, U. Relationships among serine hydrolases: Evidence for a common structural motif in triacylglyceride lipases and esterases. Biochem. Cell Biol. 1991, 69, 842–851. [Google Scholar] [CrossRef]
- Cygler, M.; Schrag, J.D.; Sussman, J.L.; Harel, M.; Silman, I.; Gentry, M.K.; Doctor, B.P. Relationship between sequence conservation and three-dimensional structure in a large family of esterases, lipases, and related proteins. Protein Sci. 1993, 2, 366–382. [Google Scholar] [CrossRef]
- Zhao, S.; Shao, C.; Goropashnaya, A.V.; Stewart, N.C.; Xu, Y.; Tøien, Ø.; Barnes, B.M.; Fedorov, V.B.; Yan, J. Genomic analysis of expressed sequence tags in American black bear Ursus americanus. BMC Genom. 2010, 11, 201. [Google Scholar] [CrossRef]
- Gao, K.; Deng, X.Y.; Shang, M.K.; Qian, H.Y.; Guo, X.J. Molecular cloning and characterization of biphenyl hydrolase-like (BPHL) protein gene from silkworm, Bombyx mori. J. Asia Pac. Entomol. 2016, 19, 611–617. [Google Scholar] [CrossRef]
- Garcia, A.S.G.; Scudeler, E.L.; Pinheiro, P.F.F.; Dos Santos, D.C. Can Exposure to neem oil affect the spermatogenesis of predator Ceraeochrysa claveri? Protoplasma 2019, 256, 693–701. [Google Scholar] [CrossRef] [PubMed]
- Grewal, P.K.; McLaughlan, J.M.; Moore, C.J.; Browning, C.A.; Hewitt, J.E. Characterization of the LARGE family of putative glycosyltransferases associated with dystroglycanopathies. Glycobiology 2005, 15, 912–923. [Google Scholar] [CrossRef]
- Ashikov, A.; Buettner, F.F.; Tiemann, B.; Gerardy-Schahn, R.; Bakker, H. LARGE2 Generates the same xylose- and glucuronic acid-containing glycan structures as LARGE. Glycobiology 2013, 23, 303–309. [Google Scholar] [CrossRef]
- Liang, D.-M.; Liu, J.-H.; Wu, H.; Wang, B.-B.; Zhu, H.-J.; Qiao, J.-J. Glycosyltransferases: Mechanisms and applications in natural product development. Chem. Soc. Rev. 2015, 44, 8350–8374. [Google Scholar] [CrossRef]
- Biswas, A.; Thattai, M. Promiscuity and specificity of eukaryotic glycosyltransferases. Biochem. Soc. Trans. 2020, 48, 891–900. [Google Scholar] [CrossRef]
- Nagare, M.; Ayachit, M.; Agnihotri, A.; Schwab, W.; Joshi, R. Glycosyltransferases: The multifaceted enzymatic regulator in insects. Insect Mol. Biol. 2021, 30, 123–137. [Google Scholar] [CrossRef]
- Ahn, S.-J.; Vogel, H.; Heckel, D.G. Comparative analysis of the UDP-glycosyltransferase multigene family in insects. Insect Biochem. Mol. Biol. 2012, 42, 133–147. [Google Scholar] [CrossRef]
- Huang, F.-F.; Chai, C.-L.; Zhang, Z.; Liu, Z.-H.; Dai, F.-Y.; Lu, C.; Xiang, Z.-H. The UDP-glucosyltransferase multigene family in Bombyx mori. BMC Genom. 2008, 9, 563. [Google Scholar] [CrossRef]
- Li, X.; Zhu, B.; Gao, X.; Liang, P. Over-expression of UDP-glycosyltransferase gene UGT2B17 is involved in chlorantraniliprole resistance in Plutella xylostella (L.): UGT2B17 gene and chlorantraniliprole resistance. Pest. Manag. Sci. 2017, 73, 1402–1409. [Google Scholar] [CrossRef]
- Zhang, Y.-N.; Ma, J.-F.; Xu, L.; Dong, Z.-P.; Xu, J.-W.; Li, M.-Y.; Zhu, X.-Y. Identification and expression patterns of UDP-glycosyltransferase (UGT) genes from insect pest Athetis lepigone (Lepidoptera: Noctuidae). J. Asia Pac. Entomol. 2017, 20, 253–259. [Google Scholar] [CrossRef]
- Kaplanoglu, E.; Chapman, P.; Scott, I.M.; Donly, C. Overexpression of a cytochrome P450 and a UDP-glycosyltransferase is associated with imidacloprid resistance in the Colorado potato beetle, Leptinotarsa decemlineata. Sci. Rep. 2017, 7, 1762. [Google Scholar] [CrossRef]
- Reid, W.R.; Sun, H.; Becnel, J.J.; Clark, A.G.; Scott, J.G. Overexpression of a Glutathione S-transferase (Mdgst) and a Galactosyltransferase—Like gene (Mdgt1) is responsible for imidacloprid resistance in house flies. Pest Manag. Sci. 2019, 75, 37–44. [Google Scholar] [CrossRef]
- Barabási, A.-L.; Oltvai, Z.N. Network biology: Understanding the cell’s functional organization. Nat. Rev. Genet. 2004, 5, 101–113. [Google Scholar] [CrossRef] [PubMed]
- Alberghina, L.; Höfer, T.; Vanoni, M. Molecular networks and system-level properties. J. Biotechnol. 2009, 144, 224–233. [Google Scholar] [CrossRef] [PubMed]
- Atchley, W.R.; Fitch, W.M. A natural classification of the basic helix–loop–helix class of transcription factors. Proc. Natl. Acad. Sci. USA 1997, 94, 5172–5176. [Google Scholar] [CrossRef]
- Jones, S. An Overview of the Basic Helix-Loop-Helix Proteins. Genome Biol. 2004, 5, 226. [Google Scholar] [CrossRef]
- Billin, A.N.; Eilers, A.L.; Queva, C.; Ayer, D.E. Mlx, a novel Max-like BHLHZip protein that interacts with the Max network of transcription factors. J. Biol. Chem. 1999, 274, 36344–36350. [Google Scholar] [CrossRef]
- Lüscher, B. Function and regulation of the transcription factors of the Myc/Max/Mad network. Gene 2001, 277, 1–14. [Google Scholar] [CrossRef]
- Yuan, J.; Tirabassi, R.S.; Bush, A.B.; Cole, M.D. The C. Elegans MDL-1 and MXL-1 proteins can functionally substitute for vertebrate MAD and MAX. Oncogene 1998, 17, 1109–1118. [Google Scholar] [CrossRef]
- Steiger, D.; Furrer, M.; Schwinkendorf, D.; Gallant, P. Max-independent functions of Myc in Drosophila melanogaster. Nat. Genet. 2008, 40, 1084–1091. [Google Scholar] [CrossRef]
- McFerrin, L.G.; Atchley, W.R. Evolution of the Max and Mlx networks in animals. Genome Biol. Evol. 2011, 3, 915–937. [Google Scholar] [CrossRef]
- Carroll, P.A.; Freie, B.W.; Cheng, P.F.; Kasinathan, S.; Gu, H.; Hedrich, T.; Dowdle, J.A.; Venkataramani, V.; Ramani, V.; Wu, X.; et al. The glucose-sensing transcription factor MLX balances metabolism and stress to suppress apoptosis and maintain spermatogenesis. PLoS Biol. 2021, 19, e3001085. [Google Scholar] [CrossRef] [PubMed]
- Tadokoro, S.; Shattil, S.J.; Eto, K.; Tai, V.; Liddington, R.C.; De Pereda, J.M.; Ginsberg, M.H.; Calderwood, D.A. Talin binding to Integrin ß tails: A final common step in Integrin activation. Science 2003, 302, 103–106. [Google Scholar] [CrossRef] [PubMed]
- Ye, F.; Snider, A.K.; Ginsberg, M.H. Talin and Kindlin: The one-two punch in Integrin activation. Front. Med. 2014, 8, 6–16. [Google Scholar] [CrossRef]
- Tanentzapf, G.; Brown, N.H. An interaction between Integrin and the Talin FERM domain mediates Integrin activation but not linkage to the cytoskeleton. Nat. Cell Biol. 2006, 8, 601–606. [Google Scholar] [CrossRef]
- Franco-Cea, A.; Ellis, S.J.; Fairchild, M.J.; Yuan, L.; Cheung, T.Y.S.; Tanentzapf, G. Distinct developmental roles for direct and indirect Talin-mediated linkage to actin. Dev. Biol. 2010, 345, 64–77. [Google Scholar] [CrossRef]
- Bogatan, S.; Cevik, D.; Demidov, V.; Vanderploeg, J.; Panchbhaya, A.; Vitkin, A.; Jacobs, J.R. Talin is required continuously for cardiomyocyte remodeling during heart growth in Drosophila. PLoS ONE 2015, 10, e0131238. [Google Scholar] [CrossRef]
- Lemke, S.B.; Weidemann, T.; Cost, A.-L.; Grashoff, C.; Schnorrer, F. A small proportion of Talin molecules transmit forces at developing muscle attachments in vivo. PLoS Biol. 2019, 17, e3000057. [Google Scholar] [CrossRef] [PubMed]
- Tomacheski, J.F.; Scudeler, E.L.; Santos, D.C. Evaluation of Talin expression on testes from adults of Ceraeochrysa claveri Navás, 1911 (Neuroptera: Chrysopidae) exposed to pyriproxyfen during larval phase. In Proceedings of the 2nd International Electronic Conference on Entomology, Basel, Switzerland, 19–21 May 2025. [Google Scholar]
- Ziesche, T.M.; Ordon, F.; Schliephake, E.; Will, T. Long-term data in agricultural landscapes indicate that insect decline promotes pests well adapted to environmental changes. J. Pest Sci. 2024, 97, 1281–1297. [Google Scholar] [CrossRef]
- Serrão, J.E.; Plata-Rueda, A.; Martínez, L.C.; Zanuncio, J.C. Side-effects of pesticides on non-target insects in agriculture: A mini-review. Sci. Nat. 2022, 109, 17. [Google Scholar] [CrossRef]
- Quandahor, P.; Kim, L.; Kim, M.; Lee, K.; Kusi, F.; Jeong, I. Effects of agricultural pesticides on decline in insect species and individual numbers. Environments 2024, 11, 182. [Google Scholar] [CrossRef]
- Stark, J.D.; Vargas, R.; Banks, J.E. Incorporating ecologically relevant measures of pesticide effect for estimating the compatibility of pesticides and biocontrol agents. J. Econ. Entomol. 2007, 100, 1027–1032. [Google Scholar] [CrossRef] [PubMed]
- Godoy, M.S.D.; Lira, A.G.D.; Lima, A.G.D.; Molina-Rugama, A.J.; Pastori, P.L.; Loureiro, E.D.S.; Fonseca Junior, E.A.D.; Mesquita, S.C.S. Residual toxicity of insecticides applied to first-stage lacewing larvae. Rev. Caatinga 2025, 38, e12569. [Google Scholar] [CrossRef]
- Amarasekare, K.G.; Shearer, P.W. Comparing effects of insecticides on two green lacewings species, Chrysoperla johnsoni and Chrysoperla carnea (Neuroptera: Chrysopidae). J. Econ. Entomol. 2013, 106, 1126–1133. [Google Scholar] [CrossRef]
- Golmohammadi, G.R.; Trshizi, H.R.R.; Shoshtari, R.V.; Faravardeh, L.; Rafei-Karehroudi, Z. Lethal and sublethal effects of three insecticides on green lacewing, Chrysoperla carnea (Neuroptera: Chrysopidae) under laboratory conditions. J. Entomol. Soc. Iran 2021, 41, 105–121. [Google Scholar] [CrossRef]
- Armas, F.S.D.; Rakes, M.; Pasini, R.A.; Araújo, M.B.; Nava, D.E.; Grützmacher, A.D. Residual toxicity of four insecticides on larvae and adults of the predator Chrysoperla externa (Hagen, 1861) (Neuroptera: Chrysopidae). Rev. Bras. Frutic. 2023, 45, e-926. [Google Scholar] [CrossRef]
- Van Leeuwen, T.; Dermauw, W.; Mavridis, K.; Vontas, J. Significance and interpretation of molecular diagnostics for insecticide resistance management of agricultural pests. Curr. Opin. Insect Sci. 2020, 39, 69–76. [Google Scholar] [CrossRef]
- EPA. United Stated Environmental Protection Agency. Exposure Assessment Tools by Chemical Classes—Pesticides. Available online: https://www.epa.gov/expobox/exposure-assessment-tools-chemical-classes-pesticides (accessed on 1 May 2025).
- Khaliq, A.; Javed, M.; Sohail, M.; Sagheer, M. Environmental effects on insects and their population dynamics. J. Entomol. Zool. Stud. 2014, 2, 1–7. [Google Scholar]
- Terblanche, J.S. Physiological performance of field-released insects. Curr. Opin. Insect Sci. 2014, 4, 60–66. [Google Scholar] [CrossRef]
- Azpiazu, C.; Bosch, J.; Viñuela, E.; Medrzycki, P.; Teper, D.; Sgolastra, F. Chronic oral exposure to field-realistic pesticide combinations via pollen and nectar: Effects on feeding and thermal performance in a solitary bee. Sci. Rep. 2019, 9, 13770. [Google Scholar] [CrossRef]
- Enserink, M. Lab v. Field: The case for studying real-life bugs. Science 2002, 298, 92–93. [Google Scholar] [CrossRef] [PubMed]
- Rejesus, R.M.; Jones, M.S. Perspective: Enhancing economic evaluations and impacts of integrated pest management farmer field schools (IPM-FFS) in low-income countries. Pest Manag. Sci. 2020, 76, 3527–3536. [Google Scholar] [CrossRef] [PubMed]
- Mulungu, K.; Abro, Z.; Niassy, S.; Muriithi, B.; Picthar, J.; Kidoido, M.; Subramanian, S.; Mohamed, S.; Khan, Z.; Hailu, G.; et al. The economic, social, and environmental impact of ecologically centered integrated pest management practices in east Africa. J. Environ. Manag. 2024, 371, 123241. [Google Scholar] [CrossRef] [PubMed]


| Gene Name | Forward Primer (5′-3′) | Reverse Primer (5′-3′) |
|---|---|---|
| BPHL | CCCCACCACTCCATCCTACT | GTTTTCTTGTTGCCGGGTGC |
| GAPDH | GAACGGGGTCAAGGTAGTGG | AAGTGGTGAAGACTCCGGTT |
| Glycosyltransferase-like protein large2 | GGACTGTATTTGTCATTAAAAAGG | GTTCGTGACCTTTGGTCCATA |
| MLX-interacting | TAACATGGCTGCTTTGCTTA | ACCTTTGTCACCCGCTGAAT |
| Talin-1 | AACTTCTCGTCCTGCACCT | AATGAAACCGGTCCACTTTG |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tomacheski, J.F.; Garcia, A.S.G.; Nakajima, R.T.; Patroni, F.M.d.S.; Scudeler, E.L.; Nóbrega, R.H.; Santos, D.C.d. Intake of Pyriproxyfen Through Contaminated Food by the Predator Ceraeochrysa claveri Navás, 1911 (Neuroptera: Chrysopidae): Evaluation of Long-Term Effects on Testes via Transcriptome Analysis. Insects 2025, 16, 567. https://doi.org/10.3390/insects16060567
Tomacheski JF, Garcia ASG, Nakajima RT, Patroni FMdS, Scudeler EL, Nóbrega RH, Santos DCd. Intake of Pyriproxyfen Through Contaminated Food by the Predator Ceraeochrysa claveri Navás, 1911 (Neuroptera: Chrysopidae): Evaluation of Long-Term Effects on Testes via Transcriptome Analysis. Insects. 2025; 16(6):567. https://doi.org/10.3390/insects16060567
Chicago/Turabian StyleTomacheski, Jefferson Fogaça, Ana Silvia Gimenes Garcia, Rafael Takahiro Nakajima, Fábio Malta de Sá Patroni, Elton Luiz Scudeler, Rafael Henrique Nóbrega, and Daniela Carvalho dos Santos. 2025. "Intake of Pyriproxyfen Through Contaminated Food by the Predator Ceraeochrysa claveri Navás, 1911 (Neuroptera: Chrysopidae): Evaluation of Long-Term Effects on Testes via Transcriptome Analysis" Insects 16, no. 6: 567. https://doi.org/10.3390/insects16060567
APA StyleTomacheski, J. F., Garcia, A. S. G., Nakajima, R. T., Patroni, F. M. d. S., Scudeler, E. L., Nóbrega, R. H., & Santos, D. C. d. (2025). Intake of Pyriproxyfen Through Contaminated Food by the Predator Ceraeochrysa claveri Navás, 1911 (Neuroptera: Chrysopidae): Evaluation of Long-Term Effects on Testes via Transcriptome Analysis. Insects, 16(6), 567. https://doi.org/10.3390/insects16060567







