Conservation and Variability in Mitochondrial Genomes of Perlodidae: Insights from Comparative Mitogenomics
Simple Summary
Abstract
1. Introduction
2. Material and Methods
2.1. Sample Preparation and DNA Extraction
2.2. PCR Amplification and Sequencing
2.3. Mitogenome Assembly and Annotation
3. Results and Discussion
3.1. Mitogenome Organization and Base Composition
3.2. Protein-Coding Genes
3.3. Transfer RNA Genes
3.4. Ribosomal RNA Genes
3.5. The Control Region
3.6. Mitochondrial Genome Analysis of Perlodidae
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Zwick, P. Phylogenetic system and zoogeography of the Plecoptera. Annu. Rev. Entomol. 2000, 45, 709–746. [Google Scholar] [CrossRef]
- DeWalt, R.E.; Maehr, M.D.; Neu-Becker, U.; Stueber, G. Plecoptera Species File Online. Available online: http://Plecoptera.SpeciesFile.org (accessed on 26 January 2024).
- DeWalt, R.E.; Kondratieff, B.C.; Sandberg, J.B. Order Plecoptera. In Thorp and Covich’s Freshwater Invertebrates; Academic Press: Cambridge, MA, USA, 2015; pp. 933–949. [Google Scholar]
- Stewart, K.D.; Nelson, C.H.; Dufeld, R.M. Occurrence of stonefies (Plecoptera) in the diet of the red-spotted newt, Notophthalmus viridescens. Entomol. News. 2001, 112, 225–229. [Google Scholar]
- Baumann, R.W. Nearctic stonefly genera as indicators of ecological parameters (Plecoptera: Insecta). Great Basin Nat. 1979, 39, 241–244. [Google Scholar]
- El Yaagoubi, S.; Edegbene, A.O.; El Alami, M.; Errochdi, S.; Harrak, R. Ephemeroptera, Plecoptera and Trichoptera (EPT) trait-based biomonitoring of rivers within the northwestern Rif of Morocco: Implications for determining riverine ecosystems ecological health in Africa. Aquat. Sci. 2024, 86, 54. [Google Scholar] [CrossRef]
- Ding, S.; Li, W.; Wang, Y.; Cameron, S.L.; Murányi, D.; Yang, D. The phylogeny and evolutionary timescale of stoneflies (Insecta: Plecoptera) inferred from mitochondrial genomes. Mol. Phylogenet. Evol. 2019, 135, 123–135. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Cao, J.J.; Li, W.H. Complete mitochondrial genome of Suwallia teleckojensis (Plecoptera: Chloroperlidae) and implications for the higher phylogeny of stoneflies. Int. J. Mol. Sci. 2018, 19, 680. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Cao, J.J.; Murányi, D.; Guo, X.; Guo, C.Y.; Li, W.H. Family–level phylogeny of infraorder Systellognatha (Insecta: Plecoptera) inferred from mitochondrial genomes. Zool. Scr. 2022, 51, 589–602. [Google Scholar] [CrossRef]
- Wang, Y.; Cao, J.J.; Li, N.; Ma, G.-Y.; Li, W.-H. The first mitochondrial genome from Scopuridae (Insecta: Plecoptera) reveals structural features and phylogenetic implications. Int. J. Biol. Macromol. 2019, 122, 893–902. [Google Scholar] [CrossRef] [PubMed]
- Mo, R.R.; Wang, Y.; Cao, J.J.; Wang, G.Q.; Li, W.H.; Murányi, D. Two complete mitochondrial genomes of the subfamily Chloroperlinae (Plecoptera: Chloroperlidae) and their phylogenetic implications. Arthropod Syst. Phylogeny 2022, 80, 155–168. [Google Scholar] [CrossRef]
- Rehman, A.; Huo, Q.B.; Du, Y.Z. The First Complete Mitochondrial Genome of Genus Isocapnia (Plecoptera: Capniidae) and Phylogenetic Assignment of Superfamily Nemouroidea. Genes 2023, 14, 965. [Google Scholar] [CrossRef]
- Cao, J.J.; Wang, Y.; Chen, M.D.; Yuan, M.Y.; Li, W.H. The complete mitochondrial genome of a stonefly species, Suwallia bimaculata (Plecoptera: Chloroperlidae). Mitochondrial DNA Part B 2019, 4, 2828–2829. [Google Scholar] [CrossRef]
- Cao, J.J.; Wang, Y.; Guo, X.; Wang, G.Q.; Li, W.H.; Murányi, D. Two complete mitochondrial genomes from Leuctridae (Plecoptera: Nemouroidea): Implications for the phylogenetic relationships among stoneflies. J. Insect Sci. 2021, 21, 16. [Google Scholar] [CrossRef] [PubMed]
- Morgan, C.C.; Foster, P.G.; Webb, A.E.; Pisani, D.; McInerney, J.O.; O’Connell, M.J. Heterogeneous models place the root of the placental mammal phylogeny. Mol. Biol. Evol. 2013, 30, 2145–2156. [Google Scholar] [CrossRef]
- Stewart, J.B.; Beckenbach, A.T. Insect mitochondrial genomics 2: The complete mitochondrial genome sequence of a giant stonefly, Pteronarcys princeps, asymmetric directional mutation bias, and conserved plecopteran A+T-region elements. Genome 2006, 49, 815–824. [Google Scholar] [CrossRef] [PubMed]
- Wang, K.; Ding, S.; Yang, D. The complete mitochondrial genome of a stonefly species, Kamimuria chungnanshana Wu, 1948 (Plecoptera: Perlidae). Mitochondrial DNA Part A 2015, 27, 3810–3811. [Google Scholar] [CrossRef] [PubMed]
- Qian, Y.H.; Wu, H.Y.; Ji, X.Y.; Yu, W.W.; Du, Y.Z. Mitochondrial genome of the stonefly Kamimuria wangi (Plecoptera: Perlidae) and phylogenetic position of Plecoptera based on mitogenomes. PLoS ONE 2014, 9, e86328. [Google Scholar]
- Wang, K.; Wang, Y.; Yang, D. The complete mitochondrial genome of a stonefly species, Togoperla sp. (Plecoptera: Perlidae). Mitochondrial DNA Part A 2016, 27, 1703–1704. [Google Scholar] [CrossRef] [PubMed]
- Wu, H.Y.; Ji, X.Y.; Yu, W.W.; Du, Y.Z. Complete mitochondrial genome of the stonefly Cryptoperla stilifera Sivec (Plecoptera: Peltoperlidae) and the phylogeny of Polyneopteran insects. Gene 2014, 537, 177–183. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.T.; Du, Y.Z. First Mitochondrial Genome from Nemouridae (Plecoptera) Reveals Novel Features of the Elongated Control Region and Phylogenetic Implications. Int. J. Mol. Sci. 2017, 18, 996. [Google Scholar] [CrossRef]
- Huang, M.C.; Wang, Y.Y.; Liu, X.Y.; Li, W.H.; Kang, Z.H.; Wang, K.; Li, X.K.; Yang, D. The complete mitochondrial genome and its remarkable secondary structure for a stonefly Acroneuria hainana Wu (Insecta: Plecoptera, Perlidae). Gene 2015, 557, 52–60. [Google Scholar] [CrossRef]
- Chen, Z.T.; Wu, H.Y.; Du, Y.Z. The nearly complete mitochondrial genome of a stonefly species, Styloperla sp. (Plecoptera: Styloperlidae). Mitochondrial DNA Part A 2016, 27, 2728–2729. [Google Scholar] [CrossRef] [PubMed]
- Li, X.T.; Chen, Z.T. Complete mitochondrial genome and phylogenetic position of Filchneria songi in Perlodidae (Insecta: Plecoptera). Mitochondrial DNA Part B Resour. 2021, 6, 3400–3401. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Guo, C.Y.; Yue, X.X.; Fan, X.; Fan, Y.Y.; Cao, J.J. Mitochondrial genomes of Nemourinae species (Plecoptera: Nemouridae) and the phylogenetic implications. J. Insect Sci. 2024, 24, 4. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Cao, J.J.; Guo, X.; Guo, C.Y.; Li, W.H.; Murányi, D. Comparative analysis of mitochondrial genomes among the family Peltoperlidae (Plecoptera: Systellognatha) and phylogenetic implications. Front. Ecol. Evol. 2022, 10, 979847. [Google Scholar] [CrossRef]
- Chen, Z.T.; Du, Y.Z. Comparison of the complete mitochondrial genome of the stonefly Sweltsa longistyla (Plecoptera: Chloroperlidae) with mitogenomes of three other stoneflies. Gene 2015, 558, 82–87. [Google Scholar] [CrossRef]
- Zhao, M.Y.; Huo, Q.B.; Du, Y.Z. Molecular phylogeny inferred from the mitochondrial genomes of Plecoptera with Oyamia nigribasis (Plecoptera: Perlidae). Sci. Rep. 2020, 10, 20955. [Google Scholar] [CrossRef] [PubMed]
- Xiang, Y.; Zhao, M.; Huo, Q.; Du, Y. Mitochondrial Genomes of the Genus Claassenia (Plecoptera: Perlidae) and Phylogenetic Assignment to Subfamily Perlinae. Genes 2021, 12, 1986. [Google Scholar] [CrossRef]
- Grant, J.R.; Stothard, P. The CGView server: A comparative genomics tool for circular genomes. Nucleic Acids Res. 2008, 36, 181–184. [Google Scholar] [CrossRef]
- Laslett, D.; Canbäck, B. ARWEN: A program to detect tRNA genes in metazoan mitochondrial nucleotide sequences. Bioinformatics 2008, 24, 172–175. [Google Scholar] [CrossRef] [PubMed]
- Zhang, D.; Gao, F.; Jakovlić, I.; Zou, H.; Zhang, J.; Li, W.X.; Wang, G.T. PhyloSuite: An integrated and scalable desktop platform for streamlined molecular sequence data management and evolutionary phylogenetics studies. Mol. Ecol. Resour. 2020, 20, 348–355. [Google Scholar] [CrossRef]
- Perna, N.T.; Kocher, T.D. Patterns of nucleotide composition at fourfold degenerate sites of animal mitogenome. J. Mol. Evol. 1995, 41, 353–358. [Google Scholar] [CrossRef] [PubMed]
- Clary, D.O.; Wolstenholme, D.R. The mitochondrial DNA molecular of Drosophila yakuba: Nucleotide sequence, gene organization, and genetic code. J. Mol. Evol. 1985, 22, 252–271. [Google Scholar] [CrossRef] [PubMed]
- Shen, Y.; Du, Y.Z. The complete mitochondrial genome of Flavoperla biocellata Chu, 1929 (Plecoptera: Perlidae) and the phylogenetic analyses of Plecoptera. PeerJ 2020, 8, e8762. [Google Scholar] [CrossRef]
- Zhang, D.X.; Hewitt, G.M. Insect mitochondrial control region: A review of its structure, evolution and usefulness in evolutionary studies. Biochem. Syst. Ecol. 1997, 25, 99–120. [Google Scholar] [CrossRef]
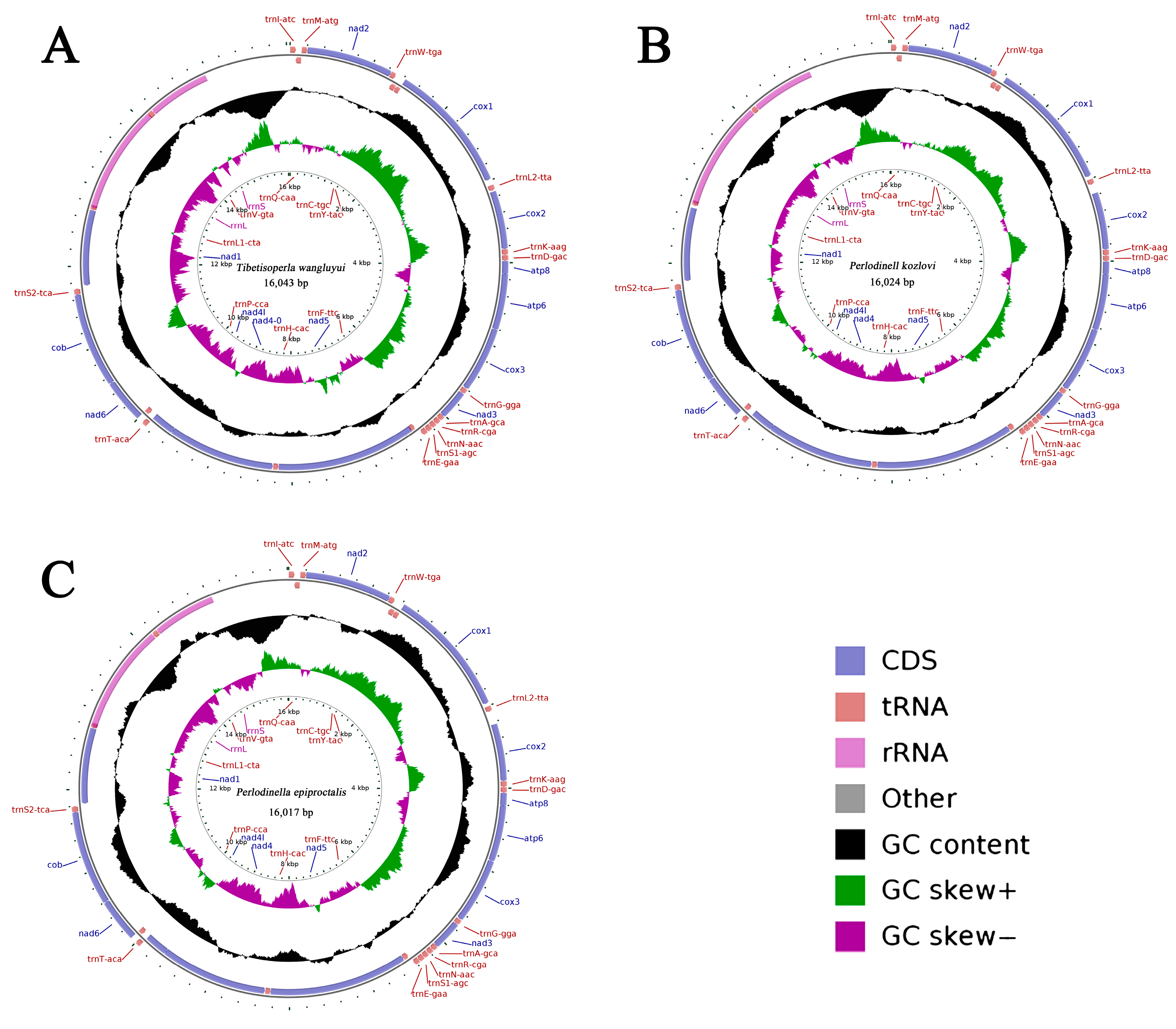
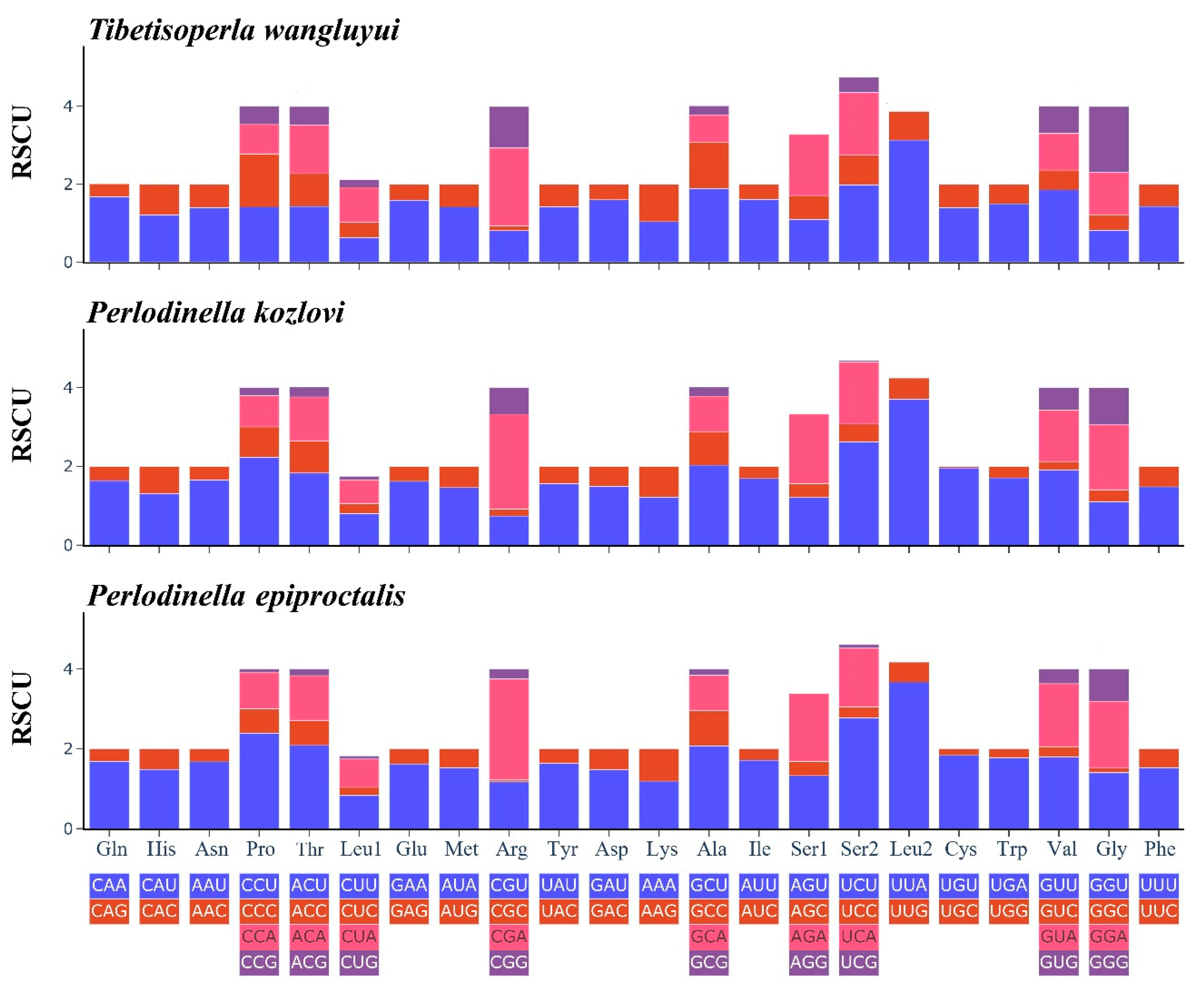
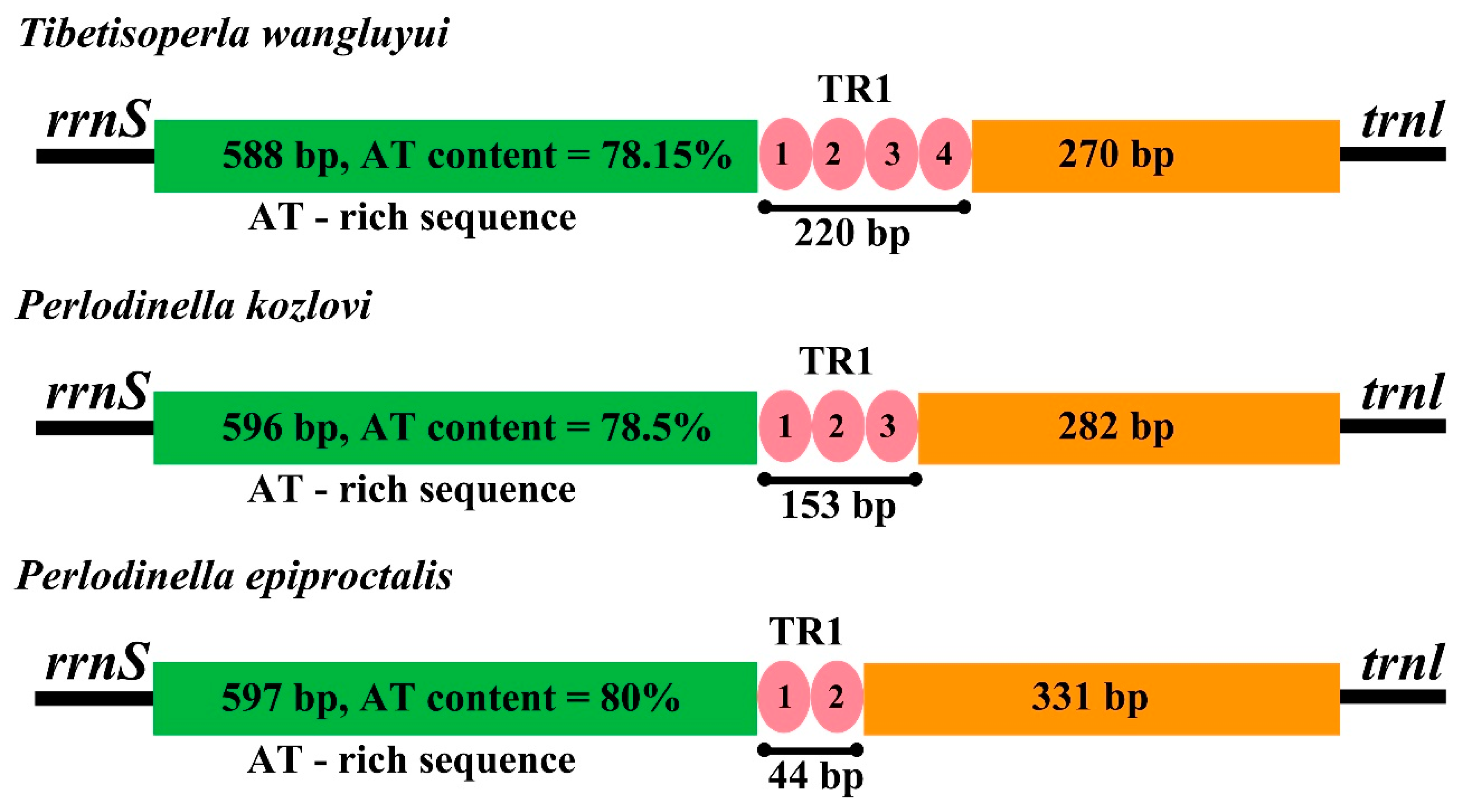
| Family | Organism | Number | AT% | GenBank Number |
|---|---|---|---|---|
| Perlodidae | Isoperla bilineata | 15,048 | 67.8 | NC_038190.1 |
| Isoperla eximia | 16,034 | 67.8 | NC_038167.1 | |
| Arcynopteryx dichroa | 16,215 | 69.3 | NC_059845.1 | |
| Filchneria songi | 16,028 | 70.1 | MZ475123.1 | |
| Filchneria zhouchangfai | 16,032 | 69.8 | NC_086967.1 | |
| Perlodes sp. | 16,039 | 70.4 | MF197377.1 | |
| Perlodinella shennongjia | 17,612 | 70.7 | NC_086966.1 | |
| Pseudomegarcys japonica | 16,067 | 66.5 | NC_038168.1 | |
| Stavsolus manchuricus | 16,130 | 66.7 | PQ616052 | |
| Neowuia wuyishana | 16,672 | 70 | PQ616053 | |
| Perlodinella kozlovi | 16,024 | 69.4 | PQ616054 | |
| Perlodinella epiproctalis | 16,017 | 70.2 | PQ616056 | |
| Isoperla qinlinga | 16,195 | 68.5 | PQ616055 | |
| Tibetisoperla wangluyui | 16,043 | 67.1 | PQ644302 | |
| Megarcys ochracea | 16,239 | 67.4 | PQ222379 | |
| Skwala compacta | 16,418 | 69.2 | PP997962 | |
| Leuctridae | Rhopalopsole subnigra | 15,562 | 69.7 | OQ612622.1 |
| Rhopalopsole bulbifera | 15,599 | 70.7 | NC_042207.1 |
| Subfamily | Species | Length (bp) | GC Skew | A (%) | T (%) | C (%) | G (%) | A + T (%) | AT Skew (+) | AT Skew (−) |
|---|---|---|---|---|---|---|---|---|---|---|
| Perlodinae | Neowuia wuyishana | 16,672 | −0.241 | 36.2 | 33.8 | 18.6 | 11.3 | 70 | 0.034 | −0.034 |
| Megarcys ochracea | 16,239 | 0.266 | 31.9 | 35.5 | 11.9 | 20.7 | 67.4 | −0.053 | 0.053 | |
| Stavsolus manchuricus | 16,130 | −0.248 | 34.6 | 32.1 | 20.8 | 12.5 | 66.7 | 0.039 | −0.039 | |
| Skwala compacta | 16,418 | 0.238 | 32.6 | 36.6 | 11.7 | 19.1 | 69.2 | −0.057 | 0.057 | |
| Pseudomegarcys japonica | 16,067 | −0.279 | 35.4 | 31.1 | 21.5 | 12 | 66.5 | 0.066 | −0.066 | |
| Perlodinella shennongjia | 17,612 | −0.212 | 37.1 | 33.6 | 17.9 | 11.4 | 70.7 | 0.049 | −0.049 | |
| Perlodinella kozlovi | 16,024 | −0.218 | 35.9 | 33.5 | 18.7 | 12 | 69.4 | 0.035 | −0.035 | |
| Perlodinella epiproctalis | 16,017 | −0.214 | 36.2 | 34 | 18.1 | 11.7 | 70.2 | 0.031 | −0.031 | |
| Filchneria zhouchangfai | 16,032 | −0.228 | 36.5 | 33.3 | 18.5 | 11.7 | 69.8 | 0.047 | −0.047 | |
| Filchneria songi | 16,028 | −0.199 | 36.5 | 33.6 | 18 | 12 | 70.1 | 0.04 | −0.04 | |
| Arcynopteryx dichroa | 16,215 | −0.238 | 36.5 | 32.8 | 19 | 11.7 | 69.3 | 0.053 | −0.053 | |
| Isoperlinae | Tibetisoperla wangluyui | 16,043 | −0.233 | 34.7 | 32.4 | 20.3 | 12.6 | 67.1 | 0.035 | −0.035 |
| Isoperla qinlinga | 16,195 | −0.234 | 35.3 | 33.2 | 19.4 | 12.1 | 68.5 | 0.031 | −0.031 | |
| Isoperla eximia | 16,034 | −0.226 | 35 | 32.8 | 19.8 | 12.4 | 67.8 | 0.031 | −0.031 | |
| Isoperla bilineata | 15,048 | −0.229 | 34.6 | 33.2 | 19.8 | 12.4 | 67.8 | 0.022 | −0.022 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yang, X.; Huo, Q.-B.; Rehman, A.; Zhu, Y.-F.; Du, Y.-Z. Conservation and Variability in Mitochondrial Genomes of Perlodidae: Insights from Comparative Mitogenomics. Insects 2025, 16, 245. https://doi.org/10.3390/insects16030245
Yang X, Huo Q-B, Rehman A, Zhu Y-F, Du Y-Z. Conservation and Variability in Mitochondrial Genomes of Perlodidae: Insights from Comparative Mitogenomics. Insects. 2025; 16(3):245. https://doi.org/10.3390/insects16030245
Chicago/Turabian StyleYang, Xiao, Qing-Bo Huo, Abdur Rehman, Ya-Fei Zhu, and Yu-Zhou Du. 2025. "Conservation and Variability in Mitochondrial Genomes of Perlodidae: Insights from Comparative Mitogenomics" Insects 16, no. 3: 245. https://doi.org/10.3390/insects16030245
APA StyleYang, X., Huo, Q.-B., Rehman, A., Zhu, Y.-F., & Du, Y.-Z. (2025). Conservation and Variability in Mitochondrial Genomes of Perlodidae: Insights from Comparative Mitogenomics. Insects, 16(3), 245. https://doi.org/10.3390/insects16030245







