Influence of Host’s Plant Diet on Gut Microbial Communities and Metabolic Potential in Spodoptera frugiperda
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Insect Rearing
2.2. Sample Collection and DNA Extraction
2.3. 16S rRNA Gene Sequencing
2.4. Data Analysis
3. Results
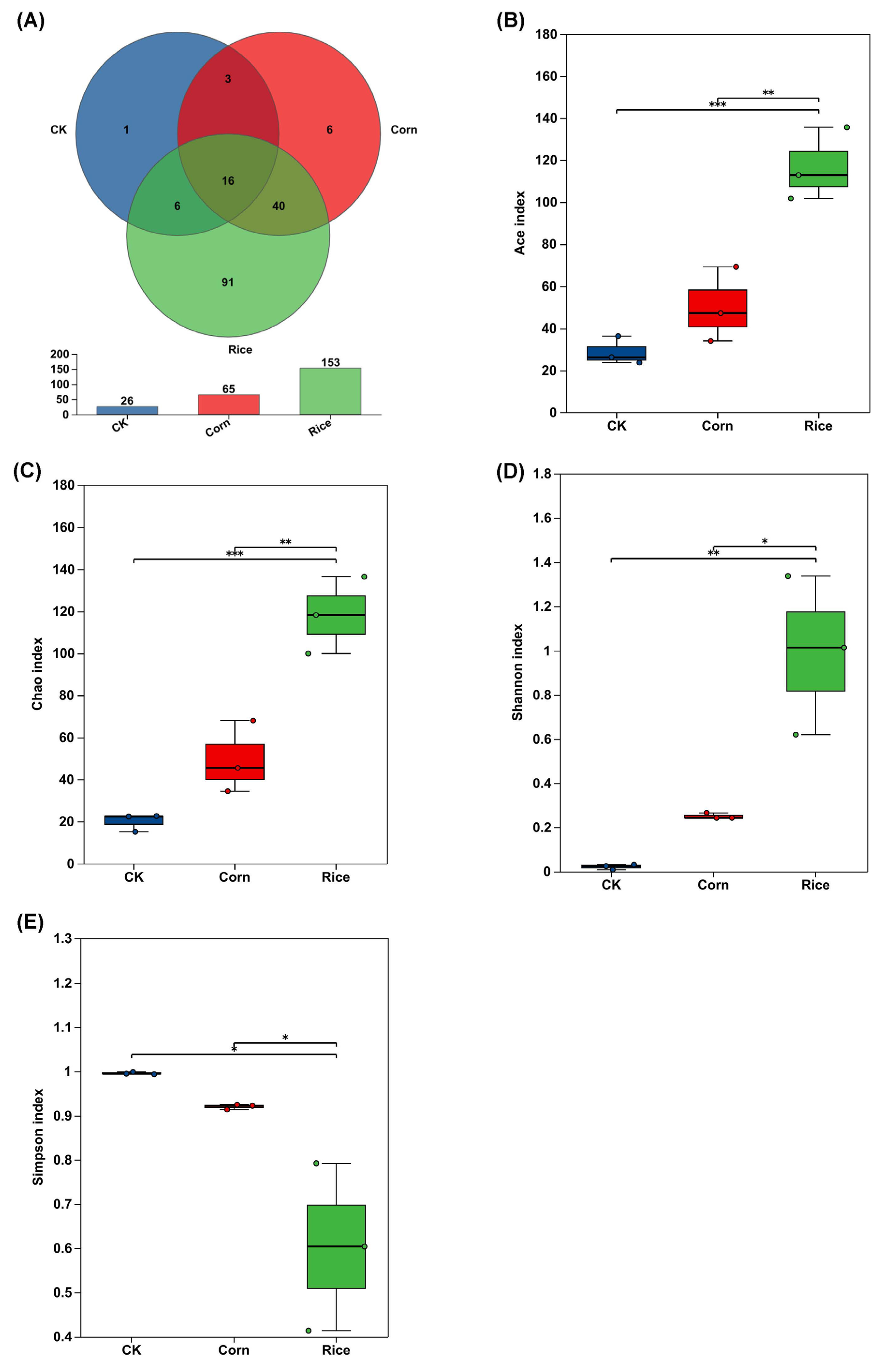
3.1. Gut Microbial Diversity in S. frugiperda
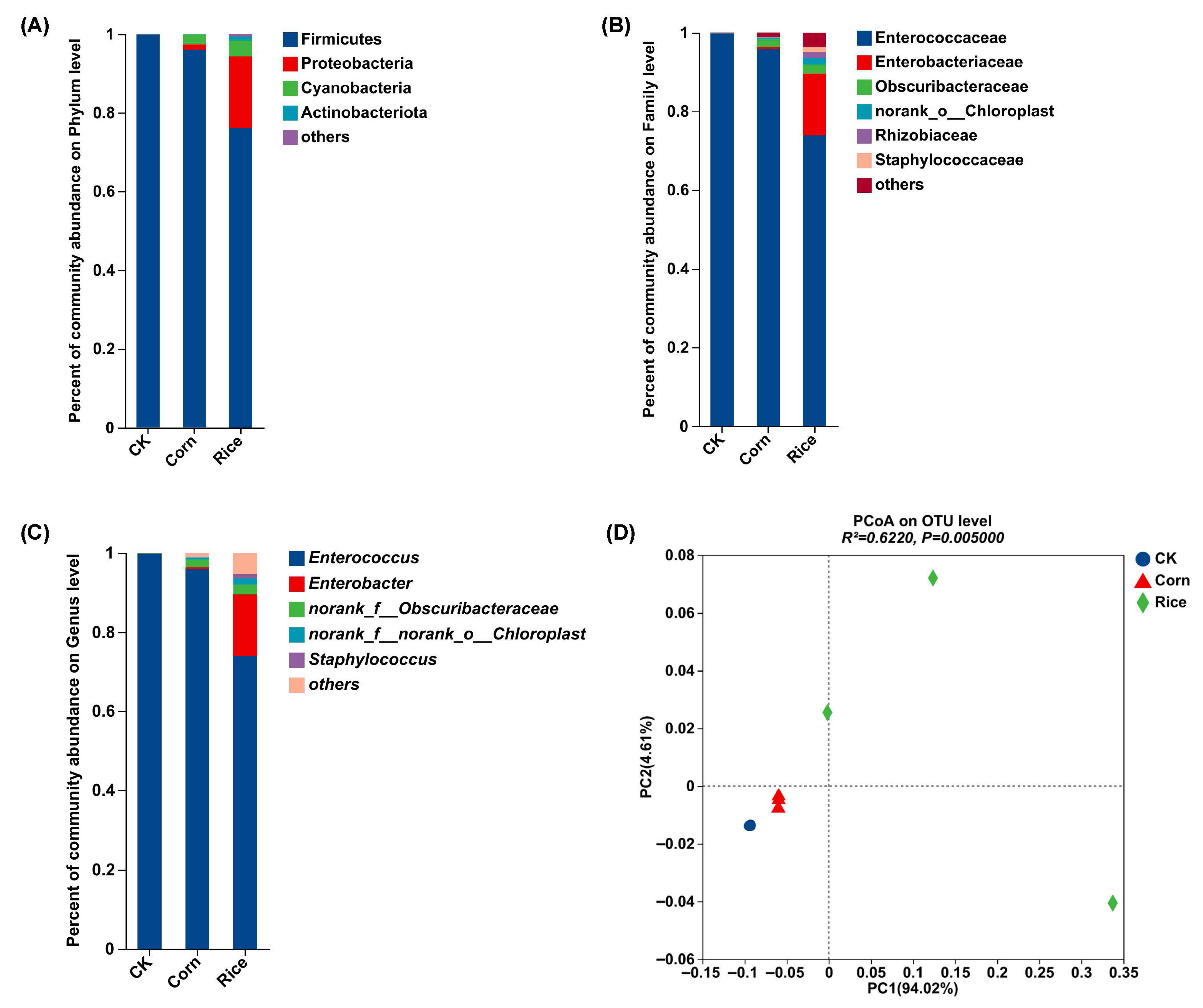
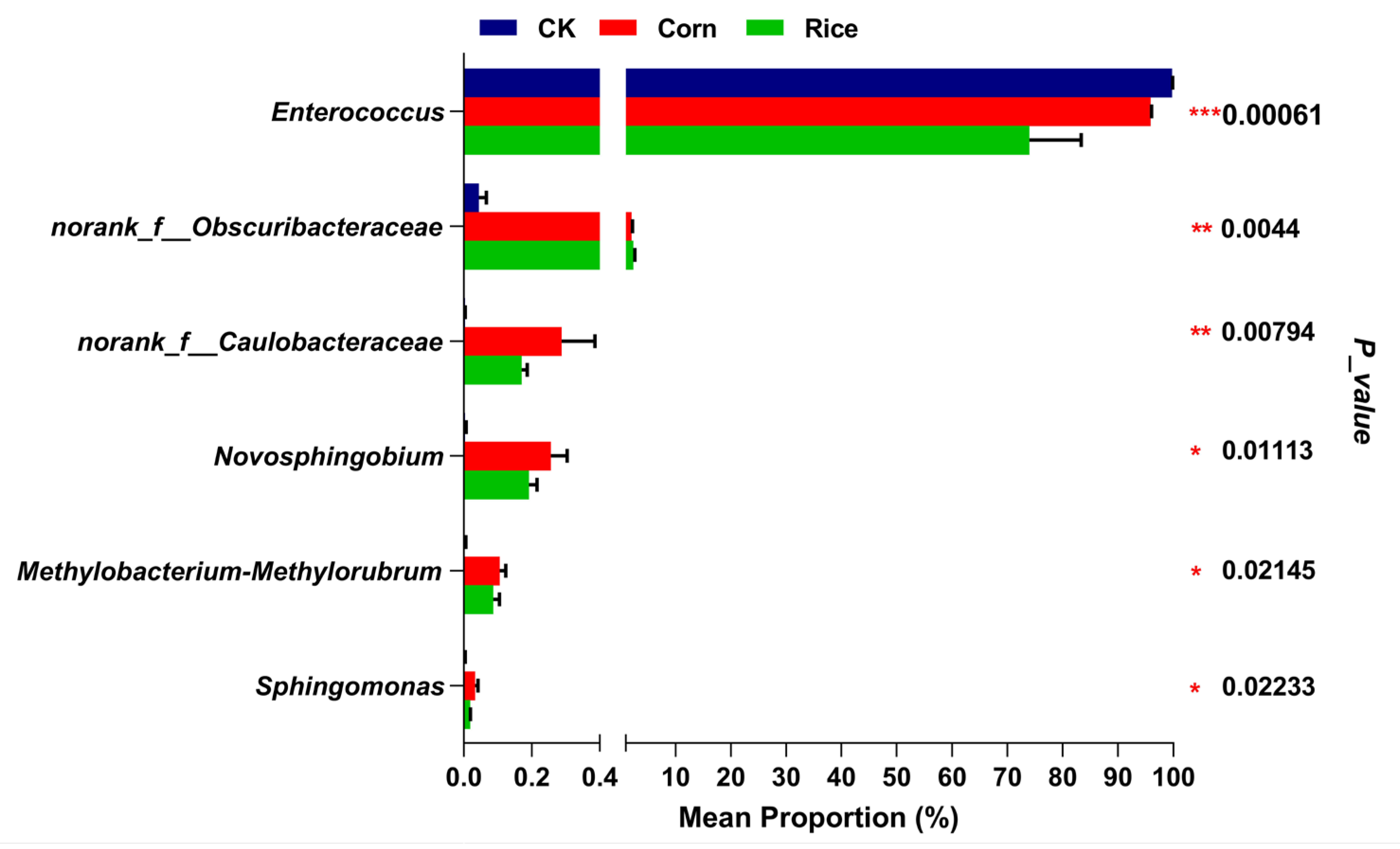
3.2. Gut Microbial Composition in S. frugiperda
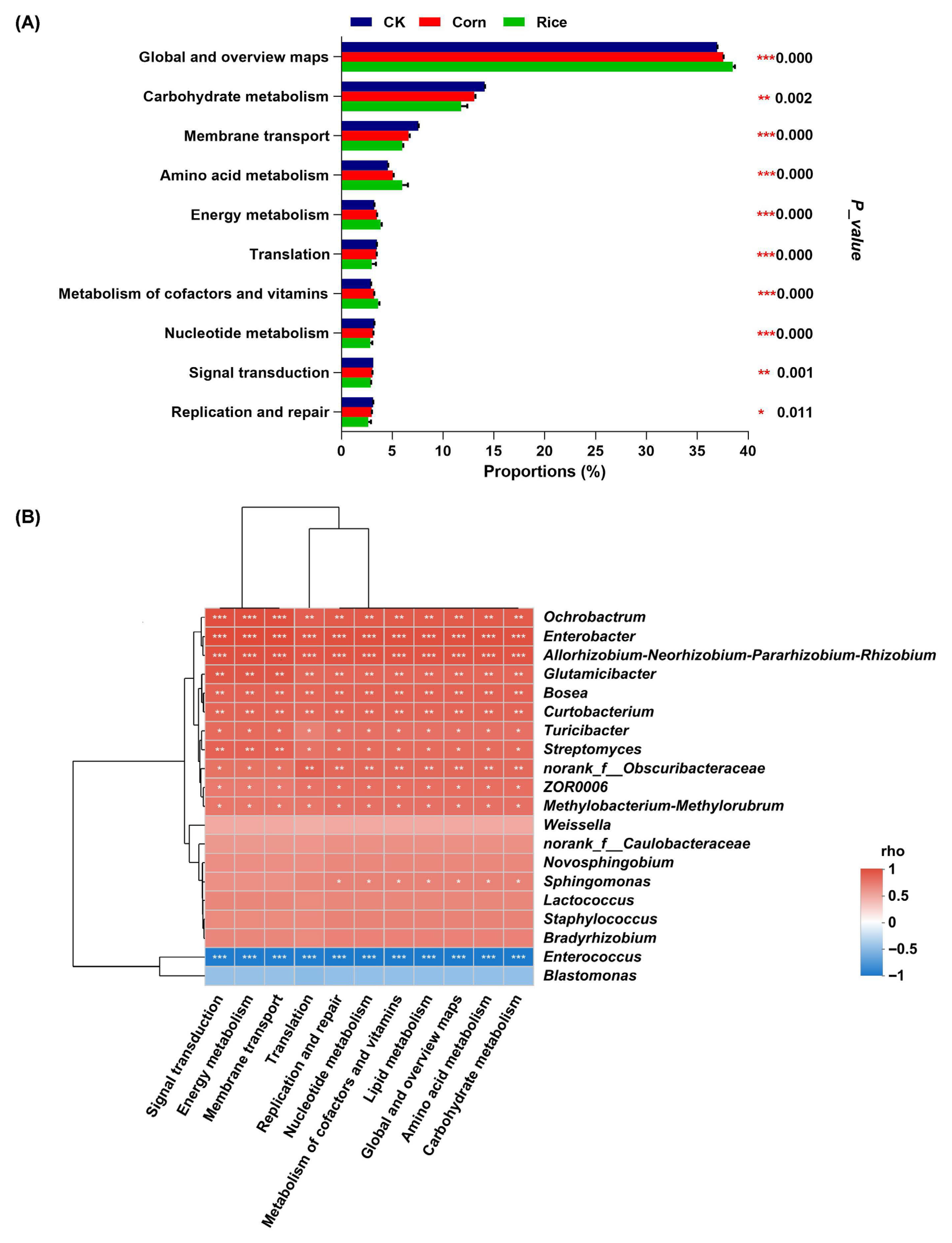
3.3. Functional Predictions of Gut Microbial Groups in S. frugiperda
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Tay, W.T.; Meagher, R., Jr.; Czepak, C.; Groot, A.T. Spodoptera frugiperda: Ecology, evolution, and management options of an invasive species. Annu. Rev. Entomol. 2023, 68, 299–317. [Google Scholar] [CrossRef] [PubMed]
- Yuning, L.; Luyang, L.; Xueming, C.; Xianmei, Y.; Jintian, L.; Benshui, S. The bacterial and fungal communities of the larval midgut of Spodoptera frugiperda (Lepidoptera: Noctuidae) varied by feeding on two cruciferous vegetables. Sci. Rep. 2022, 12, 13063. [Google Scholar] [CrossRef]
- Makgoba, M.C.; Tshikhudo, P.P.; Nnzeru, L.R.; Makhado, R.A. Impact of fall armyworm (Spodoptera frugiperda) (J.E. Smith) on small-scale maize farmers and its control strategies in the Limpopo province, South Africa. Jàmbá J. Disaster Risk Stud. 2021, 13, a1016. [Google Scholar] [CrossRef]
- Soumia, P.S.; Shirsat, D.V.; Chitra, N.; Guru-Pirasanna-Pandi, G.; Karuppaiah, V.; Gadge, A.S.; Thangasamy, A.; Mahajan, V. Invasion of fall armyworm, (Spodoptera frugiperda, J E Smith) (Lepidoptera, Noctuidae) on onion in the maize–onion crop sequence from Maharashtra, India. Front. Ecol. Evol. 2023, 11, 1279640. [Google Scholar] [CrossRef]
- Mlambo, S.; Mubayiwa, M.; Tarusikirwa, V.L.; Machekano, H.; Mvumi, B.M.; Nyamukondiwa, C. The fall armyworm and larger grain borer pest invasions in Africa: Drivers, impacts and implications for food systems. Biology 2024, 13, 160. [Google Scholar] [CrossRef]
- Ngegba, P.M.; Khalid, M.Z.; Jiang, W.; Zhong, G. An overview of insecticide resistance mechanisms, challenges, and management strategies in Spodoptera frugiperda. Crop Prot. 2025, 197, 107322. [Google Scholar] [CrossRef]
- Zheng, R.; Cheng, L.; Peng, J.; Li, Q.; Yang, F.; Yang, D.; Xia, Y.; Tang, Q. Comparative analysis of gut microbiota and immune genes linked with the immune system of wild and captive Spodoptera frugiperda (Lepidoptera: Noctuidae). Dev. Comp. Immunol. 2023, 138, 104530. [Google Scholar] [CrossRef] [PubMed]
- Zhang, N.; Qian, Z.; He, J.; Shen, X.; Lei, X.; Sun, C.; Fan, J.; Felton, G.W.; Shao, Y. Gut bacteria of lepidopteran herbivores facilitate digestion of plant toxins. Proc. Natl. Acad. Sci. USA 2024, 121, e2412165121. [Google Scholar] [CrossRef]
- Basit, A.; Haq, I.U.; Hyder, M.; Humza, M.; Younas, M.; Akhtar, M.R.; Ghafar, M.A.; Liu, T.X.; Hou, Y. Microbial symbiosis in lepidoptera: Analyzing the gut microbiota for sustainable pest management. Biology 2025, 14, 937. [Google Scholar] [CrossRef] [PubMed]
- Su, C.; Xie, T.; Jiang, L.; Wang, Y.; Wang, Y.; Nie, R.; Zhao, Y.; He, B.; Ma, J.; Yang, Q.; et al. Host genetics and larval host plant modulate microbiome structure and evolution underlying the intimate insect-microbe-plant interactions in Parnassius species on the Qinghai-Tibet Plateau. Ecol. Evol. 2024, 14, e11218. [Google Scholar] [CrossRef] [PubMed]
- Fu, Y.; Zhang, L.; Zhao, Q.; Fu, D.; Yu, H.; Xu, J.; Yang, S. Antibiotics ingestion altered the composition of gut microbes and affected the development and reproduction of the fall armyworm. J. Pest Sci. 2024, 97, 2187–2201. [Google Scholar] [CrossRef]
- Mugo-Kamiri, L.; Querejeta, M.; Raymond, B.; Herniou, E.A. The effect of diet composition on the diversity of active gut bacteria and on the growth of Spodoptera exigua (Lepidoptera: Noctuidae). J. Insect Sci. 2024, 24, ieae031. [Google Scholar] [CrossRef]
- Zhang, M.; Yan, J.; Zavala, J.A.; Palli, S.R.; Smagghe, G.; Gao, Y. Gut bacteria in potato tuberworm (Phthorimaea operculella) populations are dominated by Enterococcus spp. and these play a significant role in carbohydrate metabolism and host growth. J. Pest Sci. 2025, 98, 1583–1598. [Google Scholar] [CrossRef]
- Zhang, X.; Wu, Y.G.; Zhang, J.L.; Li, P.; Tang, Y.; Mu, Y.P.; Wang, M.Y.; Wang, W.; Mao, Y.B. Gut microbiota facilitates the adaptation of Apolygus lucorum (Hemiptera: Miridae) to its host plant. J. Econ. Entomol. 2025, 118, 1553–1564. [Google Scholar] [CrossRef]
- Chen, C.; Hao, Y.; Yang, J.; Zhang, J.; Wang, H.; Liu, Y. Influences of rearing season, host plant, and silkworm species on gut bacterial community. Insects 2025, 16, 47. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; He, Z.; Zhang, D.; Zhao, F.; Zhang, Y.; Ding, R. The role of gut microbiota at different developmental stages in the adaptation of the Etiella zinckenella to a plant host. Sci. Rep. 2025, 15, 4971. [Google Scholar] [CrossRef]
- Han, S.; Zhou, Y.; Wang, D.; Qin, Q.; Song, P.; He, Y. Effect of different host plants on the diversity of gut bacterial communities of Spodoptera frugiperda (J. E. Smith, 1797). Insects 2023, 14, 264. [Google Scholar] [CrossRef] [PubMed]
- Wu, L.H.; Hu, C.X.; Liu, T.X. Metagenomic profiling of gut microbiota in Fall Armyworm (Spodoptera frugiperda) larvae fed on different host plants. BMC Microbiol. 2024, 24, 337. [Google Scholar] [CrossRef]
- Chen, Y.P.; Li, Y.H.; Sun, Z.X.; Du, E.W.; Lu, Z.H.; Li, H.; Gui, F.R. Effects of host plants on bacterial community structure in larvae midgut of Spodoptera frugiperda. Insects 2022, 13, 373. [Google Scholar] [CrossRef]
- Chen, J.; Ma, Y.; Huang, S.; Li, J.; Zhang, Y.; Wang, H.; Qi, G.; Shi, Q.; Zhang, Z.; Yang, M.; et al. The dynamics of the microbial community in fall armyworm Spodoptera frugiperda during a life cycle. Entomol. Exp. Appl. 2023, 171, 502–513. [Google Scholar] [CrossRef]
- Marulanda-Moreno, S.M.; Saldamando-Benjumea, C.; Vivero Gomez, R.; Cadavid-Restrepo, G.; Moreno-Herrera, C.X. Comparative analysis of Spodoptera frugiperda (J. E. Smith) (Lepidoptera, Noctuidae) corn and rice strains microbiota revealed minor changes across life cycle and strain endosymbiont association. PeerJ 2024, 12, e17087. [Google Scholar] [CrossRef]
- Hafeez, M.; Li, X.W.; Zhang, J.M.; Zhang, Z.J.; Huang, J.; Wang, L.K.; Khan, M.M.; Shah, S.; Fernández-Grandon, G.M.; Lu, Y.B. Role of digestive protease enzymes and related genes in host plant adaptation of a polyphagous pest, Spodoptera frugiperda. Insect Sci. 2021, 28, 611–626. [Google Scholar] [CrossRef] [PubMed]
- Huang, Q.; Shan, H.W.; Chen, J.P.; Wu, W. Diversity and dynamics of bacterial communities in the digestive and excretory systems across the life cycle of leafhopper, Recilia dorsalis. Insects 2023, 14, 545. [Google Scholar] [CrossRef]
- Peng, H.; Zhang, Z.; Kang, X.; Zhang, Y.; Zhang, H.; Wang, Y.; Yang, D.; Zhang, J.; Wang, Y.; Cui, L.; et al. Unveiling gut microbiota and metabolic functions contributed to polyvinyl chloride degradation in Spodoptera frugiperda larvae. J. Hazard. Mater. 2025, 492, 138209. [Google Scholar] [CrossRef]
- Kasten, J.P.; Precetti, A.A.C.M.; Parra, J.R.P. Dados biológicos comparativos de Spodoptera frugiperda (J. E. Smith, 1797) em duas dietas artificiais e substrato natural. Rev. Agric. 1978, 53, 68–78. [Google Scholar]
- Liu, C.; Zhao, D.; Ma, W.; Guo, Y.; Wang, A.; Wang, Q.; Lee, D.J. Denitrifying sulfide removal process on high-salinity wastewaters in the presence of Halomonas sp. Appl. Microbiol. Biotechnol. 2016, 100, 1421–1426. [Google Scholar] [CrossRef]
- Chen, S.; Zhou, Y.; Chen, Y.; Gu, J. fastp: An ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 2018, 34, i884–i890. [Google Scholar] [CrossRef]
- Magoč, T.; Salzberg, S.L. FLASH: Fast length adjustment of short reads to improve genome assemblies. Bioinformatics 2011, 27, 2957–2963. [Google Scholar] [CrossRef] [PubMed]
- Edgar, R.C. UPARSE: Highly accurate OTU sequences from microbial amplicon reads. Nat. Methods 2013, 10, 996–998. [Google Scholar] [CrossRef] [PubMed]
- Edgar, R.C.; Haas, B.J.; Clemente, J.C.; Quince, C.; Knight, R. UCHIME improves sensitivity and speed of chimera detection. Bioinformatics 2011, 27, 2194–2200. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Garrity, G.M.; Tiedje, J.M.; Cole, J.R. Naïve Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl. Environ. Microb. 2007, 73, 5261–5267. [Google Scholar] [CrossRef]
- Bolyen, E.; Rideout, J.R.; Dillon, M.R.; Bokulich, N.A.; Abnet, C.C.; Al-Ghalith, G.A.; Alexander, H.; Alm, E.J.; Arumugam, M.; Asnicar, F.; et al. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat. Biotechnol. 2019, 37, 852–857. [Google Scholar] [CrossRef] [PubMed]
- Schloss, P.D.; Westcott, S.L.; Ryabin, T.; Hall, J.R.; Hartmann, M.; Hollister, E.B.; Lesniewski, R.A.; Oakley, B.B.; Parks, D.H.; Robinson, C.J.; et al. Introducing mothur: Open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl. Environ. Microb. 2009, 75, 7537–7541. [Google Scholar] [CrossRef]
- Douglas, G.M.; Maffei, V.J.; Zaneveld, J.R.; Yurgel, S.N.; Brown, J.R.; Taylor, C.M.; Huttenhower, C.; Langille, M.G.I. PICRUSt2 for prediction of metagenome functions. Nat. Biotechnol. 2020, 38, 685–688. [Google Scholar] [CrossRef]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2016; Available online: http://www.r-project.org (accessed on 28 August 2024).
- Chen, Y.; Chen, Y.; Li, Y.; Du, E.; Sun, Z.; Lu, Z.; Gui, F. Comparative study of the gut microbial community structure of Spodoptera frugiperda and Spodoptera literal (Lepidoptera). PeerJ 2024, 12, e17450. [Google Scholar] [CrossRef] [PubMed]
- Paniagua Voirol, L.R.; Frago, E.; Kaltenpoth, M.; Hilker, M.; Fatouros, N.E. Bacterial symbionts in lepidoptera: Their diversity, transmission, and impact on the host. Front. Microbiol. 2018, 9, 556. [Google Scholar] [CrossRef] [PubMed]
- Xia, X.; Lan, B.; Tao, X.; Lin, J.; You, M. Characterization of Spodoptera litura gut bacteria and their role in feeding and growth of the host. Front. Microbiol. 2020, 11, 1492. [Google Scholar] [CrossRef]
- Yang, F.Y.; Ahmed Saqib, H.; Chen, J.H.; Ruan, Q.Q.; Vasseur, L.; He, W.Y.; You, M.S. Differential profiles of gut microbiota and metabolites associated with host shift of Plutella xylostella. Int. J. Mol. Sci. 2020, 21, 6283. [Google Scholar] [CrossRef]
- Rowland, I.; Gibson, G.; Heinken, A.; Scott, K.; Swann, J.; Thiele, I.; Tuohy, K. Gut microbiota functions: Metabolism of nutrients and other food components. Eur. J. Nutr. 2018, 57, 1–24. [Google Scholar] [CrossRef]
- Kong, H.G.; Son, J.S.; Chung, J.H.; Lee, S.; Kim, J.S.; Ryu, C.M. Population dynamics of intestinal Enterococcus modulate Galleria mellonella metamorphosis. Microbiol. Spectr. 2023, 11, e278022. [Google Scholar] [CrossRef]
- Frias, J.; Garriga, A.; Peñalver, A.; Teixeira, M.; Beltrí, R.; Toubarro, D.; Simões, N. Exploring gut microbiome variations between Popillia japonica populations of Azores. Microorganisms 2023, 11, 1972. [Google Scholar] [CrossRef]
- Mondal, S.; Somani, J.; Roy, S.; Babu, A.; Pandey, A.K. Insect microbial symbionts: Ecology, interactions, and biological significance. Microorganisms 2023, 11, 2665. [Google Scholar] [CrossRef]
- Rozadilla, G.; Cabrera, N.A.; Virla, E.G.; Greco, N.M.; McCarthy, C.B. Gut microbiota of Spodoptera frugiperda (J.E. Smith) larvae as revealed by metatranscriptomic analysis. J. Appl. Entomol. 2020, 144, 351–363. [Google Scholar] [CrossRef]
- Chang, H.; Guo, J.; Qi, G.; Gao, Y.; Wang, S.; Wang, X.; Liu, Y. Comparative analyses of the effects of sublethal doses of emamectin benzoate and tetrachlorantraniliprole on the gut microbiota of Spodoptera frugiperda (Lepidoptera: Noctuidae). J. Insect Sci. 2023, 23, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Coolen, S.; van der Molen, M.R.; Welte, C.U. The secret life of insect-associated microbes and how they shape insect-plant interactions. FEMS Microbiol. Ecol. 2022, 98, fiac083. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dong, W.-Y.; Hafeez, M.; Zhao, S.-Y.; Zhang, J.-M.; Imran, M.; Ullah, F.; Li, X.-W.; Lu, Y.-B. Influence of Host’s Plant Diet on Gut Microbial Communities and Metabolic Potential in Spodoptera frugiperda. Insects 2025, 16, 1042. https://doi.org/10.3390/insects16101042
Dong W-Y, Hafeez M, Zhao S-Y, Zhang J-M, Imran M, Ullah F, Li X-W, Lu Y-B. Influence of Host’s Plant Diet on Gut Microbial Communities and Metabolic Potential in Spodoptera frugiperda. Insects. 2025; 16(10):1042. https://doi.org/10.3390/insects16101042
Chicago/Turabian StyleDong, Wan-Ying, Muhammad Hafeez, Sheng-Yuan Zhao, Jin-Ming Zhang, Muhammad Imran, Farman Ullah, Xiao-Wei Li, and Yao-Bin Lu. 2025. "Influence of Host’s Plant Diet on Gut Microbial Communities and Metabolic Potential in Spodoptera frugiperda" Insects 16, no. 10: 1042. https://doi.org/10.3390/insects16101042
APA StyleDong, W.-Y., Hafeez, M., Zhao, S.-Y., Zhang, J.-M., Imran, M., Ullah, F., Li, X.-W., & Lu, Y.-B. (2025). Influence of Host’s Plant Diet on Gut Microbial Communities and Metabolic Potential in Spodoptera frugiperda. Insects, 16(10), 1042. https://doi.org/10.3390/insects16101042








