Exploring Potential Distribution and Environmental Preferences of Three Species of Dicranomyia (Diptera: Limoniidae: Limoniinae) Across the Western Palaearctic Realm Using Maxent
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
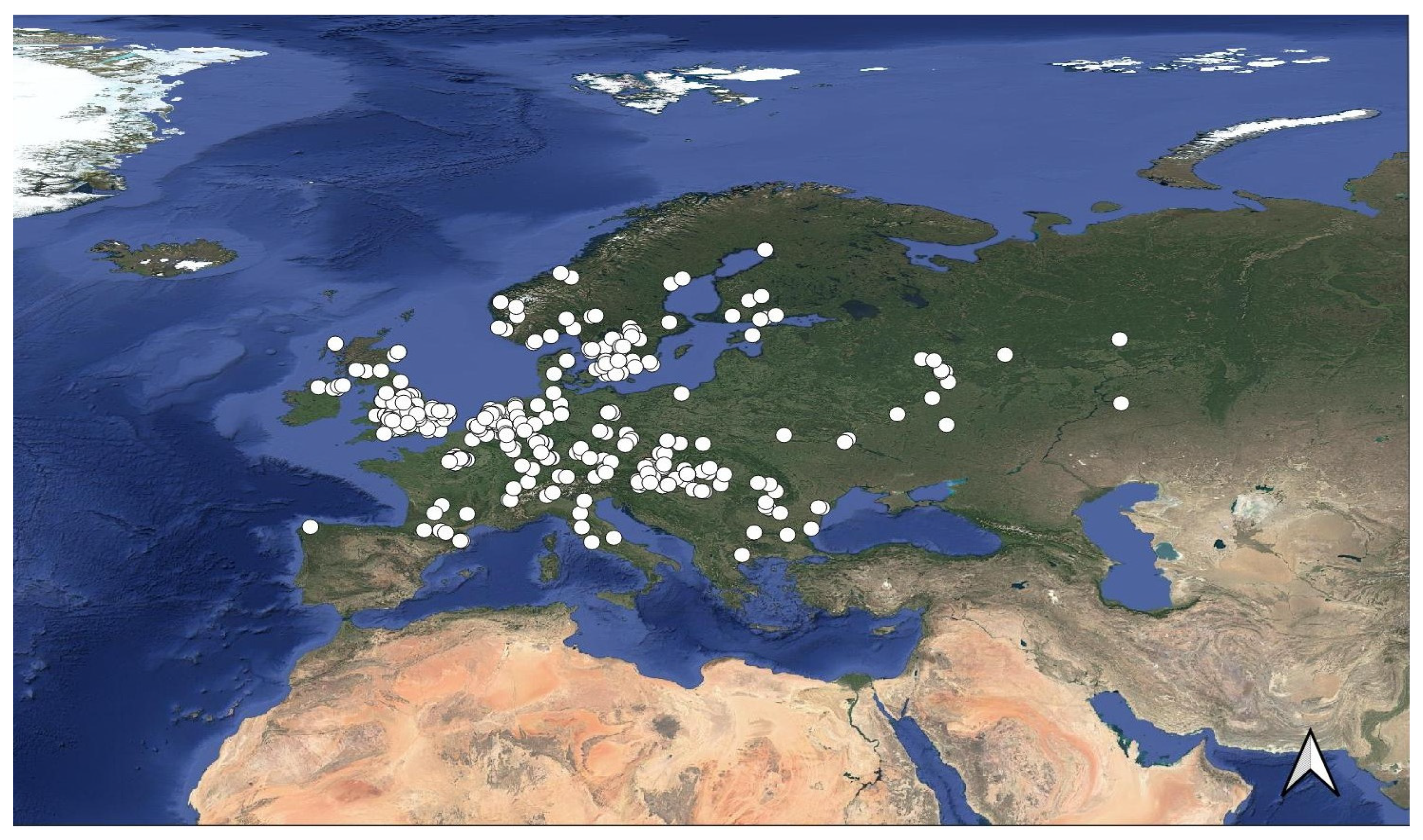
2.1. Occurrence Points and Study Area
2.2. Environmental Variables
2.3. Modeling Approach and Evaluation
2.4. Most Influential Variables
2.5. Sampling Bias
3. Results
3.1. Evaluation of the Models
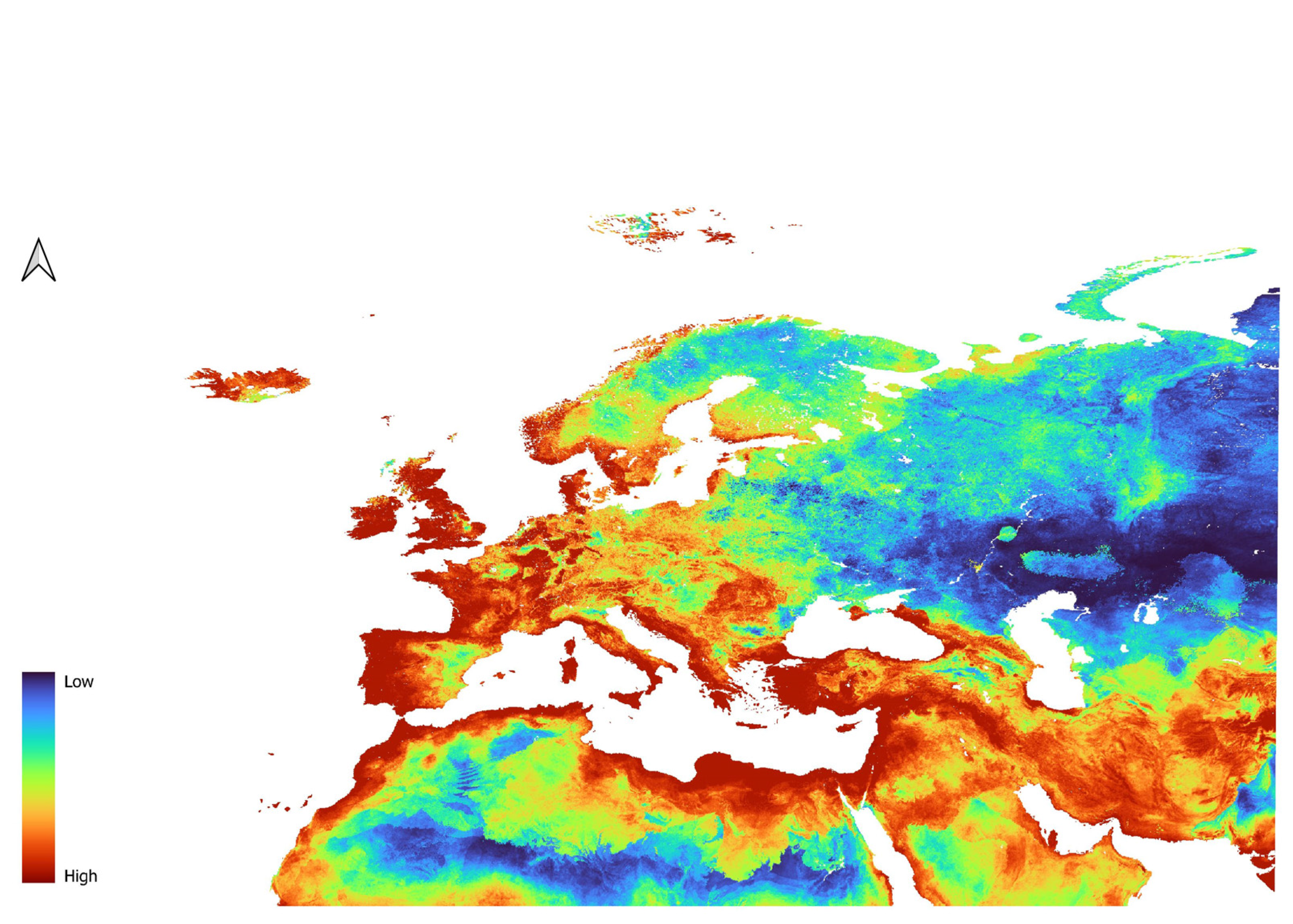
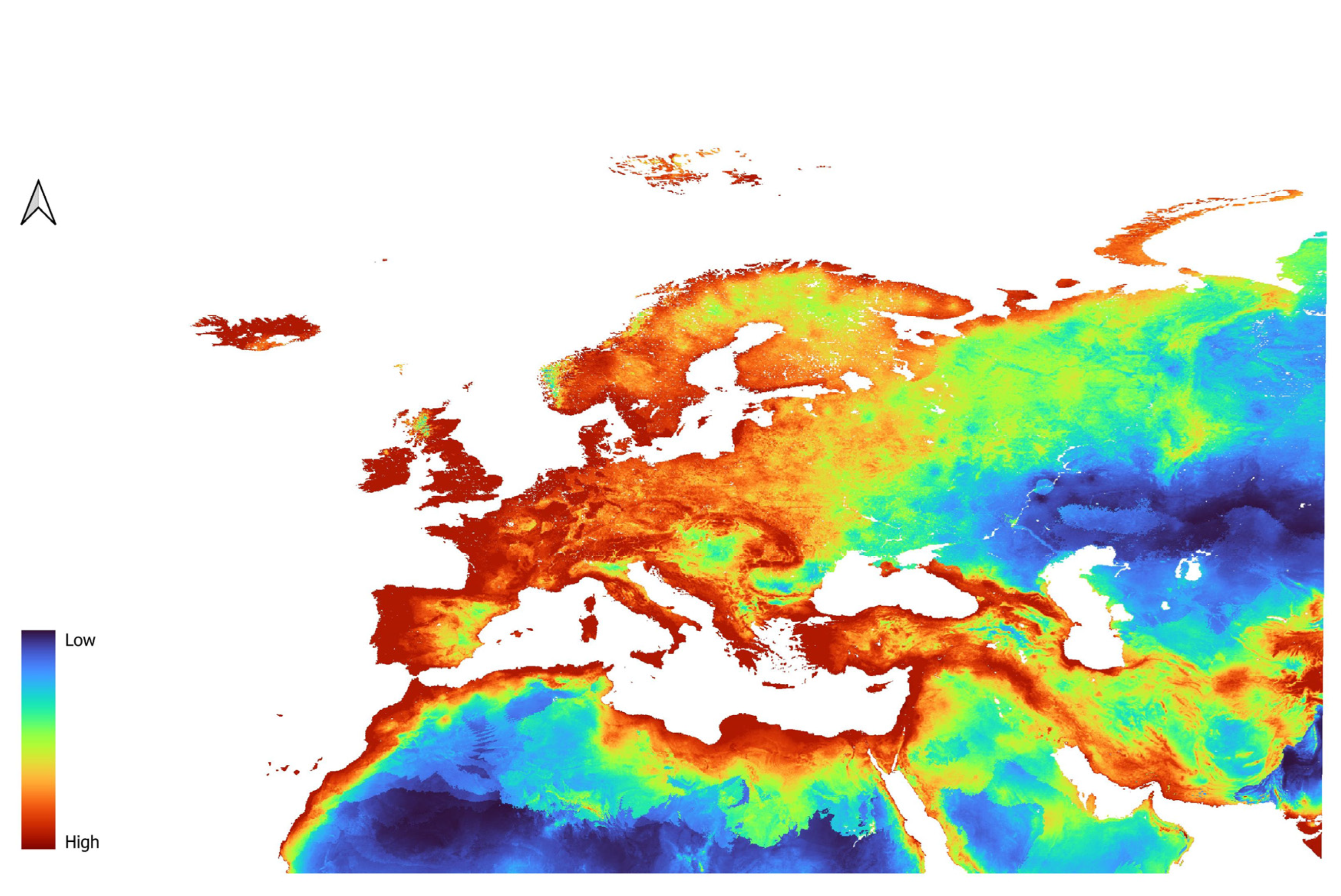
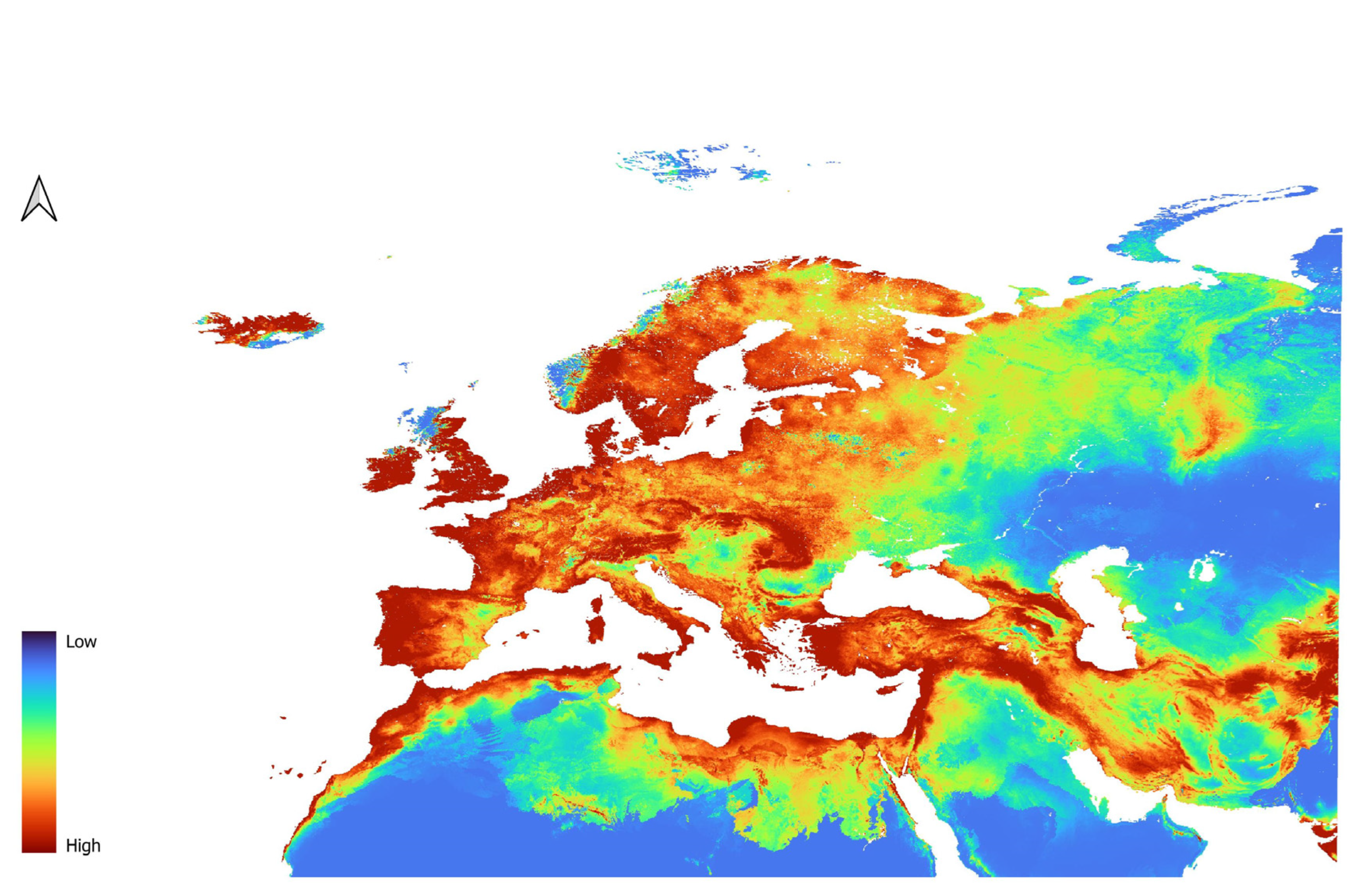
3.2. Dicranomyia affinis Models
3.3. Dicranomyia chorea Models
3.4. Dicranomyia mitis Models
3.5. Variable Importance
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| Akaike Information Criterion corrected for small sample size | AICc |
| Area Under the Curve of the Receiver Operating Characteristic | AUC |
| Ecological Niche Modeling | ENM |
| Global Biodiversity Information Facility | GBIF |
| Linear feature of Maxent | L |
| Quadratic feature of Maxent | Q |
| Linear Product features of Maxent | LP |
| Linear Quadratic features of Maxent | LQ |
| Principal Component Analysis | PCA |
| Quadratic Product features of Maxent | QP |
| Naturalis Biodiversity Center, Leiden, The Netherlands | RMNH |
| Spatial Distribution Modeling | SDM |
| 10,000 background points | 10k |
| 500,000 background points | 500k |
References
- Wigginton, N.S. Evidence of an Anthropocene Epoch. Science 2016, 351, 134–136. [Google Scholar] [CrossRef]
- Janekovi, F.; Novak, T. PCA—A Powerful Method for Analyze Ecological Niches. In Principal Component Analysis—Multidisciplinary Applications; Sanguansat, P., Ed.; InTech: Rijeka, Croatia, 2012. [Google Scholar] [CrossRef]
- Grinnell, J. The Niche-Relationships of the California Thrasher. Auk 1917, 34, 427–433. [Google Scholar] [CrossRef]
- Hutchinson, G.E. Concluding Remarks. Cold Spring Harb. Symp. Quant. Biol. 1957, 22, 415–427. [Google Scholar] [CrossRef]
- Sillero, N.; Arenas-Castro, S.; Enriquez-Urzelai, U.; Vale, C.G.; Sousa-Guedes, D.; Martínez-Freiría, F.; Real, R.; Barbosa, A.M. Want to Model a Species Niche? A Step-by-Step Guideline on Correlative Ecological Niche Modelling. Ecol. Model. 2021, 456, 109671. [Google Scholar] [CrossRef]
- Peterson, A.T. Predicting the Geography of Species’ Invasions via Ecological Niche Modeling. Q. Rev. Biol. 2003, 78, 419–433. [Google Scholar] [CrossRef]
- Chen, S.; Xiao, Y.; Xiao, Z.; Ma, D.; Li, J.; Herrera-Ulloa, A. Prediction of Suitable Habitat Shifts and Assessment of Ecological Niche Overlaps for Three Tridentiger Species with Intertidal and Subtidal Characteristics under Future Climate Changes. Mar. Pollut. Bull. 2024, 198, 115827. [Google Scholar] [CrossRef]
- Pulliam, H.R. Sources, Sinks, and Population Regulation. Am. Nat. 1988, 132, 652–661. [Google Scholar] [CrossRef]
- Araújo, M.B.; Peterson, A.T. Uses and Misuses of Bioclimatic Envelope Modeling. Ecology 2012, 93, 1527–1539. [Google Scholar] [CrossRef]
- Hirzel, A.H.; Le Lay, G.; Helfer, V.; Randin, C.; Guisan, A. Evaluating the Ability of Habitat Suitability Models to Predict Species Presences. Ecol. Model. 2006, 199, 142–152. [Google Scholar] [CrossRef]
- Podenas, S.; Seo, H.Y.; Kim, T.; Hur, J.M.; Kim, A.Y.; Klein, T.A.; Kim, H.C.; Kang, T.H.; Aukštikalnienė, R. Dicranomyia Crane Flies (Diptera: Limoniidae) of Korea. Zootaxa 2019, 4595, 1–110. [Google Scholar] [CrossRef] [PubMed]
- Starý, J. Two New European Species of Dicranomyia Stephens, 1829, Related to D. (s. Str.) Chorea. Bull. Zoöl. Mus. Univ. Van Amst. 1993, 13, 175–182. [Google Scholar]
- Starỳ, J.; Stubbs, A.E. Five Species Under Dicranomyia (Dicranomyia) mitis (Meigen, 1830)(Diptera, Limoniidae). Zootaxa 2015, 3964, 321–334. [Google Scholar] [CrossRef]
- Warren, D.L.; Seifert, S.N. Ecological Niche Modeling in Maxent: The Importance of Model Complexity and the Performance of Model Selection Criteria. Ecol. Appl. 2011, 21, 335–342. [Google Scholar] [CrossRef]
- Phillips, S.J.; Anderson, R.P.; Schapire, R.E. Maximum Entropy Modeling of Species Geographic Distributions. Ecol. Model. 2006, 190, 231–259. [Google Scholar] [CrossRef]
- Elith, J.; Graham, C.H.; Anderson, R.P.; Dudík, M.; Ferrier, S.; Guisan, A.; Hijmans, R.J.; Huettmann, F.; Leathwick, J.R.; Lehmann, A.; et al. Novel Methods Improve Prediction of Species’ Distributions from Occurrence Data. Ecography 2006, 29, 129–151. [Google Scholar] [CrossRef]
- Qiao, H.; Soberón, J.; Peterson, A.T. No Silver Bullets in Correlative Ecological Niche Modelling: Insights from Testing among Many Potential Algorithms for Niche Estimation. Methods Ecol. Evol. 2015, 6, 1126–1136. [Google Scholar] [CrossRef]
- Merow, C.; Smith, M.J.; Silander, J.A. A Practical Guide to MaxEnt for Modeling Species’ Distributions: What It Does, and Why Inputs and Settings Matter. Ecography 2013, 36, 1058–1069. [Google Scholar] [CrossRef]
- GBIF Secretariat. GBIF Home Page. Global Biodiversity Information Facility. Available online: https://www.gbif.org (accessed on 1 December 2024).
- GBIF Occurrence Download. Available online: https://doi.org/10.15468/dl.medpd6 (accessed on 7 December 2024).
- GBIF Occurrence Download. Available online: https://doi.org/10.15468/dl.k8g2z8 (accessed on 10 December 2024).
- GBIF Occurrence Download. Available online: https://doi.org/10.15468/dl.sm7f3u (accessed on 10 December 2024).
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2024; Available online: https://www.R-project.org/ (accessed on 1 November 2024).
- Chamberlain, S.; Oldoni, D.; Waller, J. Rgbif: Interface to the Global Biodiversity Information Facility API, 2012, 3.8.3. [CrossRef]
- Wieczorek, J.; Guo, Q.; Hijmans, R. The Point-Radius Method for Georeferencing Locality Descriptions and Calculating Associated Uncertainty. Int. J. Geogr. Inf. Sci. 2004, 18, 745–767. [Google Scholar] [CrossRef]
- Hijmans, R.J.; Cameron, S.E.; Parra, J.L.; Jones, P.G.; Jarvis, A. Very High Resolution Interpolated Climate Surfaces for Global Land Areas. Int. J. Climatol. 2005, 25, 1965–1978. [Google Scholar] [CrossRef]
- Poggio, L.; De Sousa, L.; Batjes, N.; Heuvelink, G.; Kempen, B.; Ribeiro, E.; Rossiter, D. SoilGrids 2.0: Producing Soil Information for the Globe with Quantified Spatial Uncertainty. Soil 2021, 7, 217–240. [Google Scholar] [CrossRef]
- QGIS Development Team. QGIS Geographic Information System, Open Source Geospatial Foundation, 2009.
- Elith, J.; Phillips, S.J.; Hastie, T.; Dudík, M.; Chee, Y.E.; Yates, C.J. A Statistical Explanation of MaxEnt for Ecologists. Divers. Distrib. 2011, 17, 43–57. [Google Scholar] [CrossRef]
- Dormann, C.F.; Elith, J.; Bacher, S.; Buchmann, C.; Carl, G.; Carré, G.; Marquéz, J.R.G.; Gruber, B.; Lafourcade, B.; Leitão, P.J.; et al. Collinearity: A Review of Methods to Deal with It and a Simulation Study Evaluating Their Performance. Ecography 2013, 36, 27–46. [Google Scholar] [CrossRef]
- Warren, D.L.; Glor, R.E.; Turelli, M. ENMTools: A Toolbox for Comparative Studies of Environmental Niche Models. Ecography 2010, 33, 607–611. [Google Scholar] [CrossRef]
- Phillips, S.J.; Dudík, M. Modeling of Species Distributions with Maxent: New Extensions and a Comprehensive Evaluation. Ecography 2008, 31, 161–175. [Google Scholar] [CrossRef]
- Muscarella, R.; Galante, P.J.; Soley—Guardia, M.; Boria, R.A.; Kass, J.M.; Uriarte, M.; Anderson, R.P. ENMeval: An R Package for Conducting Spatially Independent Evaluations and Estimating Optimal Model Complexity for Maxent Ecological Niche Models. Methods Ecol. Evol. 2014, 5, 1198–1205. [Google Scholar] [CrossRef]
- RStudio Team. RStudio: Integrated Development for R; RStudio, Inc.: Boston, MA, USA, 2019; Available online: http://www.rstudio.com/ (accessed on 20 February 2025).
- Kramer-Schadt, S.; Niedballa, J.; Pilgrim, J.D.; Schröder, B.; Lindenborn, J.; Reinfelder, V.; Stillfried, M.; Heckmann, I.; Scharf, A.K.; Augeri, D.M.; et al. The Importance of Correcting for Sampling Bias in MaxEnt Species Distribution Models. Divers. Distrib. 2013, 19, 1366–1379. [Google Scholar] [CrossRef]
- GBIF Occurence Download. Available online: https://doi.org/10.15468/dl.huhe3y (accessed on 24 December 2024).
- Beale, C.M.; Lennon, J.J. Incorporating Uncertainty in Predictive Species Distribution Modelling. Phil. Trans. R. Soc. B 2012, 367, 247–258. [Google Scholar] [CrossRef] [PubMed]
- Butchart, S.H.M.; Walpole, M.; Collen, B.; Van Strien, A.; Scharlemann, J.P.W.; Almond, R.E.A.; Baillie, J.E.M.; Bomhard, B.; Brown, C.; Bruno, J.; et al. Global Biodiversity: Indicators of Recent Declines. Science 2010, 328, 1164–1168. [Google Scholar] [CrossRef]
- Heikkinen, R.K.; Luoto, M.; Araújo, M.B.; Virkkala, R.; Thuiller, W.; Sykes, M.T. Methods and Uncertainties in Bioclimatic Envelope Modelling under Climate Change. Prog. Phys. Geogr. 2006, 30, 751–777. [Google Scholar] [CrossRef]
- Huntley, B.; Collingham, Y.C.; Willis, S.G.; Green, R.E. Potential Impacts of Climatic Change on European Breeding Birds. PLoS ONE 2008, 3, e1439. [Google Scholar] [CrossRef]
- Murray, J.; Low Choy, S.; McAlpine, C.; Possingham, H.; Goldizen, A. Evaluating Model Transferability for a Threatened Species to Adjacent Areas: Implications for Rock-Wallaby Conservation. Austral. Ecol. 2011, 36, 76–89. [Google Scholar] [CrossRef]
- Peterson, A.T.; Papeş, M.; Eaton, M. Transferability and Model Evaluation in Ecological Niche Modeling: A Comparison of GARP and Maxent. Ecography 2007, 30, 550–560. [Google Scholar] [CrossRef]
- Qiao, H.; Feng, X.; Escobar, L.E.; Peterson, A.T.; Soberón, J.; Zhu, G.; Papeş, M. An Evaluation of Transferability of Ecological Niche Models. Ecography 2019, 42, 521–534. [Google Scholar] [CrossRef]
- Inman, R.; Franklin, J.; Esque, T.; Nussear, K. Comparing Sample Bias Correction Methods for Species Distribution Modeling Using Virtual Species. Ecosphere 2021, 12, e03422. [Google Scholar] [CrossRef]
- Dennis, R.L.; Sparks, T.H.; Hardy, P.B. Bias in Butterfly Distribution Maps: The Effects of Sampling Effort. J. Insect Conserv. 1999, 3, 33–42. [Google Scholar] [CrossRef]
- Prendergast, J.R.; Wood, S.N.; Lawton, J.H.; Eversham, B.C. Correcting for Variation in Recording Effort in Analyses of Diversity Hotspots. Biodivers. Lett. 1993, 1, 39–53. [Google Scholar] [CrossRef]
- Vaughan, I.P.; Ormerod, S.J. The Continuing Challenges of Testing Species Distribution Models. J. Appl. Ecol. 2005, 42, 720–730. [Google Scholar] [CrossRef]
- Veloz, S.D. Spatially Autocorrelated Sampling Falsely Inflates Measures of Accuracy for Presence-Only Niche Models. J. Biogeogr. 2009, 36, 2290–2299. [Google Scholar] [CrossRef]
- Peterson, A.T.; Papeş, M.; Soberón, J. Rethinking Receiver Operating Characteristic Analysis Applications in Ecological Niche Modeling. Ecol. Model. 2008, 213, 63–72. [Google Scholar] [CrossRef]
- Lobo, J.M.; Jiménez-Valverde, A.; Real, R. AUC: A Misleading Measure of the Performance of Predictive Distribution Models. Glob. Ecol. Biogeogr. 2007, 17, 145–151. [Google Scholar] [CrossRef]
- Jiménez, L.; Soberón, J. Leaving the Area under the Receiving Operating Characteristic Curve behind: An Evaluation Method for Species Distribution Modelling Applications Based on Presence—Only Data. Methods Ecol. Evol. 2020, 11, 1571–1586. [Google Scholar] [CrossRef]
- Konowalik, K.; Nosol, A. Evaluation Metrics and Validation of Presence-Only Species Distribution Models Based on Distributional Maps with Varying Coverage. Sci. Rep. 2021, 11, 1482. [Google Scholar] [CrossRef]
- Whitford, A.M.; Shipley, B.R.; McGuire, J.L. The Influence of the Number and Distribution of Background Points in Presence-Background Species Distribution Models. Ecol. Model. 2024, 488, 110604. [Google Scholar] [CrossRef]
- Zhang, L.; Liu, S.; Sun, P.; Wang, T.; Wang, G.; Zhang, X.; Wang, L. Consensus Forecasting of Species Distributions: The Effects of Niche Model Performance and Niche Properties. PLoS ONE 2015, 10, e0120056. [Google Scholar] [CrossRef]
- Araujo, M.; New, M. Ensemble Forecasting of Species Distributions. Trends Ecol. Evol. 2007, 22, 42–47. [Google Scholar] [CrossRef]
- Rhodes, C.G.; Loaiza, J.R.; Romero, L.M.; Gutiérrez Alvarado, J.M.; Delgado, G.; Rojas Salas, O.; Ramírez Rojas, M.; Aguilar-Avendaño, C.; Maynes, E.; Valerín Cordero, J.A.; et al. Anopheles albimanus (Diptera: Culicidae) Ensemble Distribution Modeling: Applications for Malaria Elimination. Insects 2022, 13, 221. [Google Scholar] [CrossRef] [PubMed]
- Grenouillet, G.; Buisson, L.; Casajus, N.; Lek, S. Ensemble Modelling of Species Distribution: The Effects of Geographical and Environmental Ranges. Ecography 2011, 34, 9–17. [Google Scholar] [CrossRef]
- Montoya-Jimenez, J.C.; Valdez-Lazalde, J.R.; Ángeles-Perez, G.; De Los Santos-Posadas, H.M.; Cruz-Cárdenas, G. Predictive Capacity of Nine Algorithms and an Ensemble Model to Determine the Geographic Distribution of Tree Species. IForest-Biogeosci. For. 2022, 15, 363. [Google Scholar] [CrossRef]
- Zhu, G.; Fan, J.; Peterson, A.T. Cautions in Weighting Individual Ecological Niche Models in Ensemble Forecasting. Ecol. Model. 2021, 448, 109502. [Google Scholar] [CrossRef]
- Rundle, H.D.; Nosil, P. Ecological Speciation. Ecol. Lett. 2005, 8, 336–352. [Google Scholar] [CrossRef]
- Nosil, P.; Crespi, B.J.; Sandoval, C.P. Host-Plant Adaptation Drives the Parallel Evolution of Reproductive Isolation. Nature 2002, 417, 440–443. [Google Scholar] [CrossRef] [PubMed]
- Devyatkov, V.I. Data on the crane fly fauna of the family Limoniidae (Diptera) of East Kazakhstan. Subfamily Limoniinae. Euroasian Entomol. J. 2020, 19, 47–55. [Google Scholar] [CrossRef]


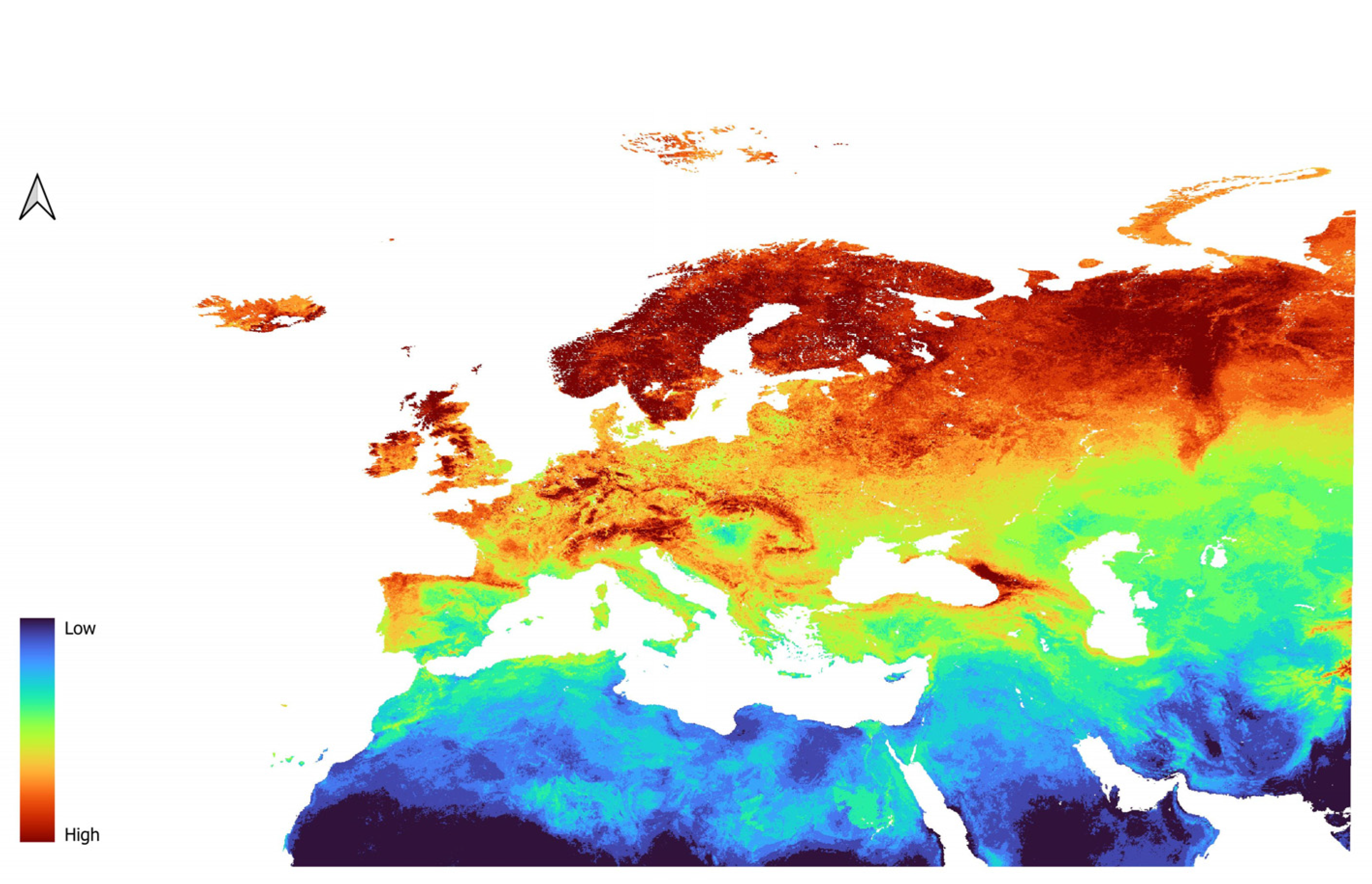
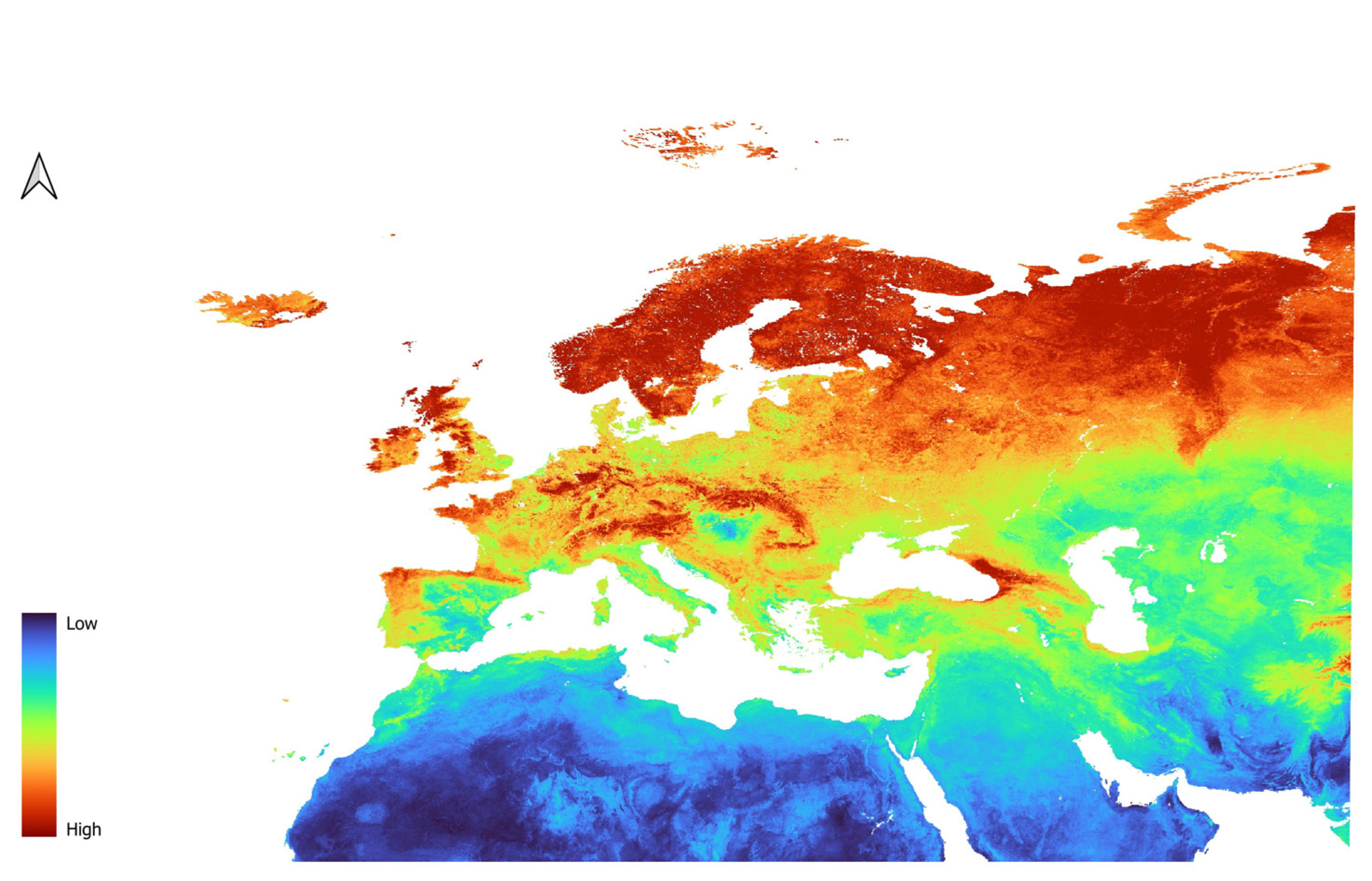

| Environmental Variables | Description | Source |
|---|---|---|
| Bio6 | Min temperature of Coldest Month | WorldClim |
| Bio7 | Temperature Annual Range | WorldClim |
| Bio8 | Mean Temperature of Wettest Quarter | WorldClim |
| Bio13 | Precipitation of Wettest Month | WorldClim |
| Bio15 | Precipitation Seasonality (Coefficient of variation) | WorldClim |
| Clay | Proportion of clay particles (<0.002 mm) in the fine earth fraction (g/kg) at depth 15/30 cm | Soilgrids |
| Nitrogen | Total nitrogen (cg/kg) at depth 15/30 cm | Soilgrids |
| phh2o | Soil pH | Soilgrids |
| Sand | Proportion of sand particles (>0.05/0.063 mm) in the fine earth fraction (g/kg) at depth 15/30 cm | Soilgrids |
| D. mitis | D. chorea | D. affinis | |||||||
|---|---|---|---|---|---|---|---|---|---|
| λ | AiCc | Test AUC | λ | AICc | Test AUC | λ | AICc | Test AUC | Model |
| 7 | 244.6519 | 0.808 | 9 | 262.678500 | 0.910 | 7 | 100.092800 | 0.823 | L1_10k |
| 5 | 237.540500 | 0.824 | 8 | 264.809600 | 0.910 | 5 | 93.570330 | 0.836 | L2_10k |
| 4 | 236.915000 | 0.823 | 8 | 264.809600 | 0.910 | 5 | 93.758980 | 0.839 | L3_10k |
| 4 | 236.975900 | 0.820 | 8 | 265.871800 | 0.909 | 3 | 88.329820 | 0.832 | L4_10k |
| 4 | 237.043300 | 0.818 | 7 | 264.082200 | 0.907 | 3 | 88.466770 | 0.808 | L5_10K |
| 7 | −114.563200 | 0.809 | 9 | −54.147510 | 0.915 | 7 | 4.459933 | 0.823 | L1_500k |
| 6 | −117.191100 | 0.825 | 8 | −56.724220 | 0.916 | 5 | −63.171220 | 0.835 | L2_500k |
| 4 | −122.165000 | 0.823 | 8 | −56.731830 | 0.915 | 5 | 0.489773 | 0.841 | L3_500k |
| 4 | −122.153900 | 0.821 | 8 | −56.760000 | 0.914 | 4 | −2.496222 | 0.837 | L4_500k |
| 4 | −122.143400 | 0.819 | 7 | −59.322370 | 0.912 | 3 | −5.193853 | 0.819 | L5_500k |
| 7 | 244.947100 | 0.791 | 9 | 266.614400 | 0.928 | 8 | 102.453500 | 0.849 | Q1_10k |
| 6 | 241.887700 | 0.810 | 8 | 265.435400 | 0.929 | 7 | 99.925430 | 0.838 | Q2_10k |
| 4 | 236.780600 | 0.819 | 8 | 266.387800 | 0.928 | 6 | 96.902220 | 0.831 | Q3_10k |
| 4 | 236.825000 | 0.816 | 8 | 267.009200 | 0.928 | 5 | 93.853360 | 0.848 | Q4_10k |
| 3 | 234.522700 | 0.814 | 8 | 267.450400 | 0.924 | 4 | 91.024330 | 0.852 | Q5_10k |
| 7 | −114.513700 | 0.790 | 9 | −54.279050 | 0.932 | 8 | 10.786890 | 0.853 | Q1_500k |
| 6 | −117.167500 | 0.814 | 8 | −56.874890 | 0.933 | 7 | 7.012010 | 0.841 | Q2_500k |
| 4 | −122.152500 | 0.819 | 8 | −56.950410 | 0.933 | 7 | 6.908883 | 0.835 | Q3_500k |
| 4 | −122.146500 | 0.816 | 8 | −57.035180 | 0.932 | 5 | 0.328777 | 0.847 | Q4_500k |
| 3 | 124.490600 | 0.814 | 8 | −57.104580 | 0.929 | 4 | −2.555656 | 0.854 | Q5_500k |
| 8 | 247.125600 | 0.820 | 15 | 290.498300 | 0.937 | 10 | 112.573800 | 0.884 | LP1_10k |
| 5 | 239.367900 | 0.818 | 10 | 271.645600 | 0.937 | 10 | 112.664000 | 0.873 | LP2_10k |
| 3 | 234.730600 | 0.812 | 7 | 264.929200 | 0.931 | 6 | 97.043540 | 0.877 | LP3_10k |
| 3 | 234.764400 | 0.811 | 5 | 260.878300 | 0.927 | 4 | 91.126160 | 0.869 | LP4_10k |
| 3 | 234.794500 | 0.811 | 5 | 261.476000 | 0.926 | 3 | 88.510480 | 0.863 | LP5_10k |
| 8 | −111.790300 | 0.824 | 14 | −36.334200 | 0.941 | 10 | 32.729570 | 0.888 | LP1_500k |
| 5 | −119.728800 | 0.821 | 10 | −48.845840 | 0.941 | 11 | 25.108980 | 0.879 | LP2_500k |
| 3 | −124.497500 | 0.813 | 7 | −59.258310 | 0.936 | 7 | 6.969385 | 0.883 | LP3_500k |
| 3 | −124.492800 | 0.813 | 5 | −64.248550 | 0.932 | 4 | −2.524094 | 0.871 | LP4_500k |
| 3 | −124.489200 | 0.813 | 5 | −64.281020 | 0.931 | 3 | −5.200798 | 0.866 | LP5_500k |
| 10 | 253.519000 | 0.797 | 10 | 268.262700 | 0.934 | 9 | 106.090700 | 0.847 | LQ1_10k |
| 8 | 247.210700 | 0.820 | 10 | 269.207500 | 0.930 | 7 | 99.732720 | 0.843 | LQ2_10k |
| 4 | 236.744800 | 0.822 | 10 | 270.577300 | 0.927 | 7 | 99.845550 | 0.864 | LQ3_10k |
| 4 | 236.792300 | 0.820 | 8 | 266.133800 | 0.926 | 6 | 96.583660 | 0.886 | LQ4_10k |
| 4 | 236.847900 | 0.817 | 8 | 266.648700 | 0.924 | 5 | 93.686940 | 0.883 | LQ5_10k |
| 9 | −108.846600 | 0.796 | 12 | −46.288390 | 0.939 | 9 | 14.700140 | 0.849 | LQ1_500k |
| 8 | −111.733600 | 0.823 | 10 | −51.678820 | 0.935 | 7 | 7.154227 | 0.844 | LQ2_500k |
| 4 | −122.160200 | 0.822 | 9 | −54.226530 | 0.932 | 7 | 6.962915 | 0.866 | LQ3_500k |
| 4 | −122.154800 | 0.820 | 8 | −56.937470 | 0.931 | 6 | 3.472355 | 0.889 | LQ4_500k |
| 4 | −122.151500 | 0.817 | 8 | −57.040290 | 0.929 | 5 | 0.324897 | 0.889 | LQ5_500k |
| 7 | 244.295700 | 0.819 | 13 | 275.294900 | 0.945 | 10 | 111.188700 | 0.882 | QP1_10k |
| 5 | 239.164300 | 0.818 | 11 | 273.452600 | 0.940 | 8 | 103.588400 | 0.881 | QP2_10k |
| 3 | 234.495800 | 0.818 | 8 | 267.044100 | 0.934 | 8 | 104.149200 | 0.887 | QP3_10k |
| 3 | 234.557300 | 0.819 | 6 | 262.877900 | 0.932 | 5 | 94.050960 | 0.849 | QP4_10k |
| 3 | 234.620900 | 0.819 | 5 | 261.166300 | 0.931 | 3 | 88.561700 | 0.833 | QP5_10k |
| 8 | −111.762000 | 0.822 | 14 | −38.408350 | 0.950 | 10 | 20.116190 | 0.885 | QP1_500k |
| 5 | −119.711500 | 0.823 | 11 | −48.739510 | 0.945 | 8 | 10.760050 | 0.885 | QP2_500k |
| 3 | −124.481100 | 0.820 | 8 | −57.059590 | 0.939 | 8 | 10.693920 | 0.894 | QP3_500k |
| 3 | −124.479000 | 0.821 | 6 | −62.042200 | 0.937 | 6 | 3.490096 | 0.863 | QP4_500k |
| 3 | −124.477100 | 0.821 | 5 | −64.411760 | 0.936 | 3 | −5.232400 | 0.838 | QP5_500k |
| D. affinis | D. chorea | D. mitis |
|---|---|---|
| Bio7; Bio8 | Bio7; Bio8 | ph |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ciliberti, P.; Starkevich, P.; Podenas, S. Exploring Potential Distribution and Environmental Preferences of Three Species of Dicranomyia (Diptera: Limoniidae: Limoniinae) Across the Western Palaearctic Realm Using Maxent. Insects 2025, 16, 1022. https://doi.org/10.3390/insects16101022
Ciliberti P, Starkevich P, Podenas S. Exploring Potential Distribution and Environmental Preferences of Three Species of Dicranomyia (Diptera: Limoniidae: Limoniinae) Across the Western Palaearctic Realm Using Maxent. Insects. 2025; 16(10):1022. https://doi.org/10.3390/insects16101022
Chicago/Turabian StyleCiliberti, Pasquale, Pavel Starkevich, and Sigitas Podenas. 2025. "Exploring Potential Distribution and Environmental Preferences of Three Species of Dicranomyia (Diptera: Limoniidae: Limoniinae) Across the Western Palaearctic Realm Using Maxent" Insects 16, no. 10: 1022. https://doi.org/10.3390/insects16101022
APA StyleCiliberti, P., Starkevich, P., & Podenas, S. (2025). Exploring Potential Distribution and Environmental Preferences of Three Species of Dicranomyia (Diptera: Limoniidae: Limoniinae) Across the Western Palaearctic Realm Using Maxent. Insects, 16(10), 1022. https://doi.org/10.3390/insects16101022





