Full-Length Transcriptome Profiling of the Complete Mitochondrial Genome of Sericothrips houjii (Thysanoptera: Thripidae: Sericothripinae) Featuring Extensive Gene Rearrangement and Duplicated Control Regions
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Sample Collection
2.2. Acquisition and Annotation of Mitogenome
2.3. Acquisition of Mitochondrial Transcriptome
2.4. Mitochondrial Transcript Identification
2.5. Phylogenetic Analysis
3. Results
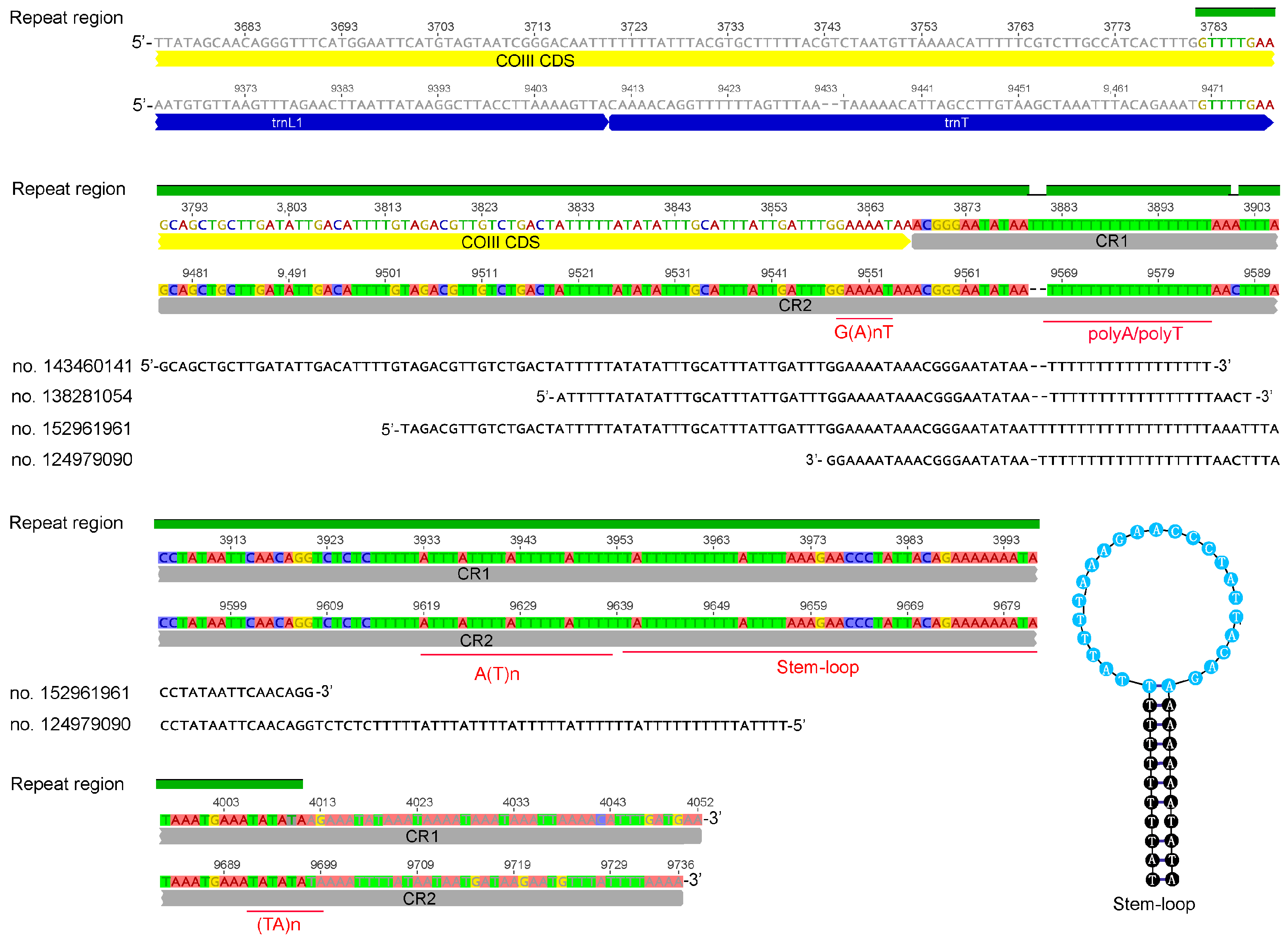
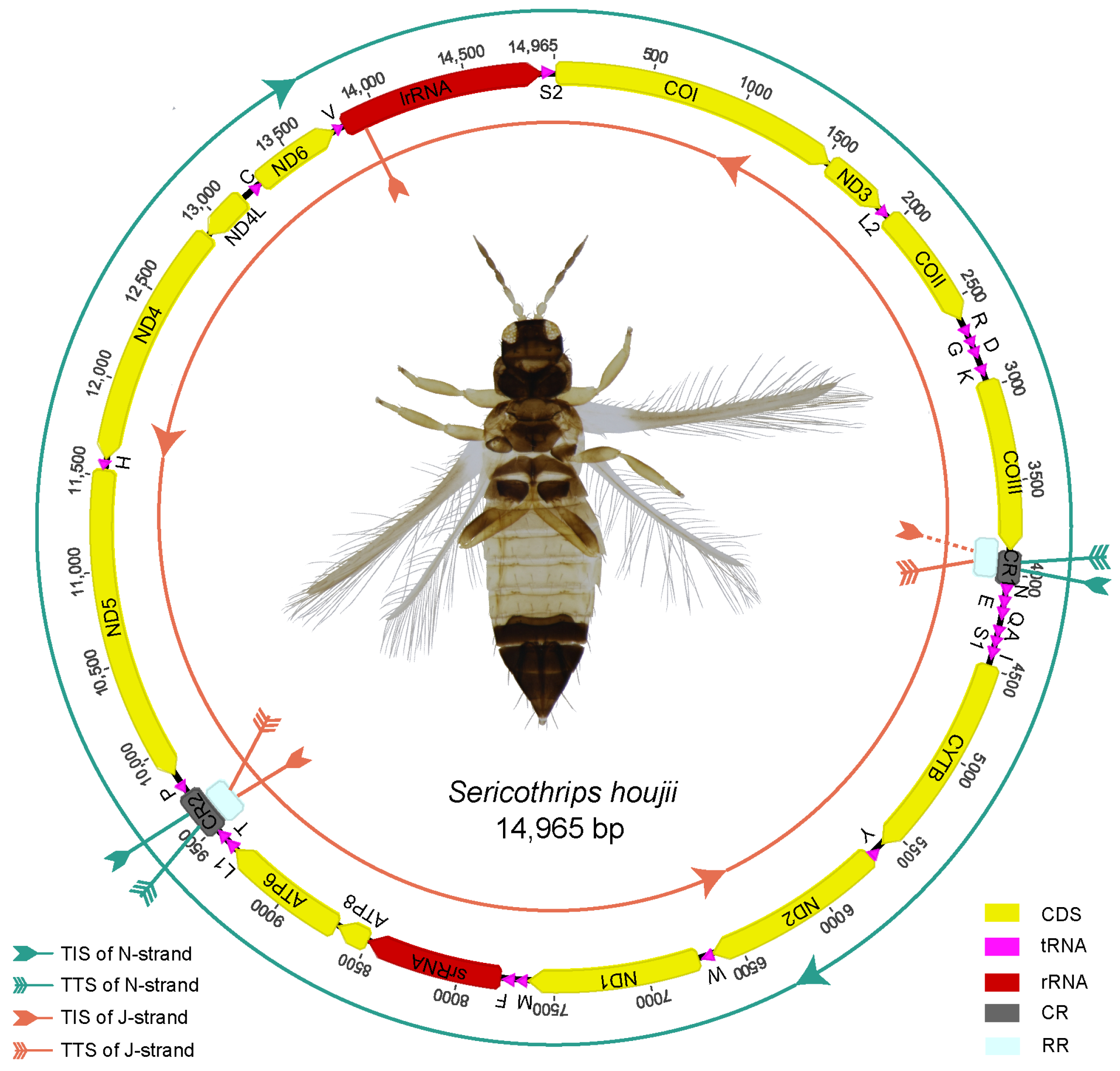
3.1. The Mitogenome Feature of Sericothrips houjii
3.2. The Quantitative Transcription Map of Sericothrips houjii Mitogenome
3.3. Polycistronic RNA and Precursor RNA Cleavage
3.4. Isoform RNA
3.5. The Proposed Model of Mitochondrial Transcription
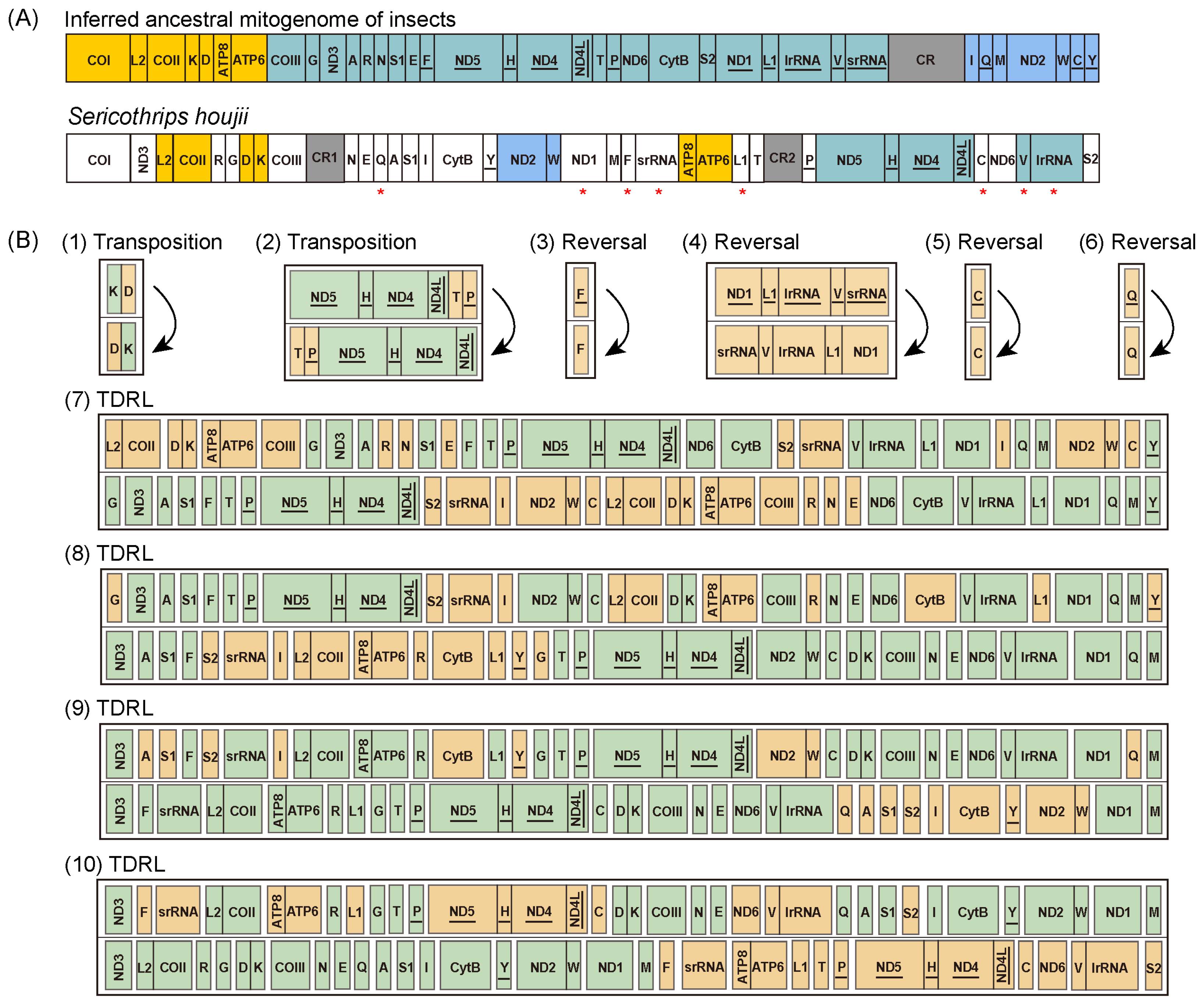
3.6. Phylogenetic Analysis and Mitochondrial Gene Rearrangement of Sericothripinae
4. Discussion
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
Appendix A
| Family | Subfamily | Species | GenBank Accession Number |
|---|---|---|---|
| Aeolothripidae | Aeolothrips fasciatus | ON210959 | |
| Aeolothrips indicus | MW899051 | ||
| Aeolothrips melaleucus | ON210970 | ||
| Aeolothrips sp. | ON210960 | ||
| Aeolothrips xinjiangensis | NC_063848 | ||
| Desmothrips sp. | KY751031 | ||
| Franklinothrips megalops | ON210963 | ||
| Franklinothrips stasseni | ON210964 | ||
| Franklinothrips vespiformis | MN072395 | ||
| Thripidae | Panchaetothripinae | Opimothrips tubulatus | NC_069977 |
| Phibalothrips peringueyi | MW603839 | ||
| Rhipiphorothrips cruentatus | MN072396 | ||
| Selenothrips rubrocinctus | MT872374 | ||
| Dendrothripinae | Dendrothrips minowai | NC_037839 | |
| Pseudodendrothrips mori | NC_050743 | ||
| Sericothripinae | Neohydatothrips samayunkur | MF991901 | |
| Sericothrips houjii | PP697967 | ||
| Thripinae | Anaphothrips obscurus | NC_035510 | |
| Anaphothrips sudanensis | ON210961 | ||
| Aptinothrips stylifer | OQ559124 | ||
| Arorathrips mexicanus | OP913452 | ||
| Bregmatothrips sinensis | OP913450 | ||
| Ctenothrips transeolineae | OP913440 | ||
| Echinothrips americanus | ON210962 | ||
| Ernothrips longitudinalis | OP913449 | ||
| Frankliniella intonsa | JQ917403 | ||
| Frankliniella occidentalis | NC_018370 | ||
| Frankliniella panamensi | NC_081011 | ||
| Frankliniella schultzei | MT872372 | ||
| Lefroyothrips lefroyi | NC_083274 | ||
| Megalurothrips distalis | OP913453 | ||
| Mycterothrips gongshanensis | NC_067744 | ||
| Mycterothrips nilgiriensis | MT872373 | ||
| Odontothrips loti | ON210965 | ||
| Odontothrips pentatrichopus | OP913451 | ||
| Odontothrips phaseoli | NC_084197 | ||
| Scirtothrips dorsalis | NC_025241 | ||
| Scirtothrips hansoni | OR044712 | ||
| Stenchaetothrips bicolor | OP913448 | ||
| Stenchaetothrips biformis | ON653412 | ||
| Stenchaetothrips minutus | OP913447 | ||
| Taeniothrips eucharii | OP913454 | ||
| Taeniothrips sp. | ON210966 | ||
| Taeniothrips tigris | MW751816 | ||
| Thrips palmi | NC_039437 | ||
| Thrips imaginis | AF335993 |
References
- Boore, J.L. Animal mitochondrial genomes. Nucleic Acids Res. 1999, 27, 1767–1780. [Google Scholar] [CrossRef] [PubMed]
- Barshad, G.; Marom, S.; Cohen, T.; Mishmar, D. Mitochondrial DNA transcription and its regulation: an evolutionary perspective. Trends Genet. 2018, 34, 682–692. [Google Scholar] [CrossRef] [PubMed]
- Misof, B.; Liu, S.L.; Meusemann, K.; Peters, R.S.; Donath, A.; Mayer, C.; Frandsen, P.B.; Ware, J.; Flouri, T.; Beutel, R.G.; et al. Phylogenomics resolves the timing and pattern of insect evolution. Science 2014, 346, 763–767. [Google Scholar] [CrossRef] [PubMed]
- Cameron, S.L. Insect mitochondrial genomics: implications for evolution and phylogeny. Annu. Rev. Entomol. 2014, 59, 95–117. [Google Scholar] [CrossRef]
- Sun, G.; Ma, Y.; He, K.; Yu, P.; Yu, D.; Cheng, X.; Zhang, J. The complete mitochondrial genomes of three bristletails (Insecta: Archaeognatha): The paraphyly of Machilidae and insights into Archaeognathan phylogeny. PLoS ONE 2015, 10, e0117669. [Google Scholar]
- Shao, R.F.; Dowton, M.; Murrell, A.; Barker, S.C. Rates of gene rearrangement and nucleotide substitution are correlated in the mitochondrial genomes of insects. Mol. Biol. Evol. 2003, 20, 1612–1619. [Google Scholar] [CrossRef]
- Mao, M.; Austin, A.D.; Johnson, N.F.; Dowton, M. Coexistence of minicircular and a highly rearranged mtDNA molecule suggests that recombination shapes mitochondrial genome organization. Mol. Biol. Evol. 2014, 31, 636–644. [Google Scholar] [CrossRef]
- Dowton, M.; Castro, L.R.; Campbell, S.L.; Bargon, S.D.; Austin, A.D. Frequent mitochondrial gene rearrangements at the hymenopteran nad3-nad5 junction. J. Mol. Evol. 2003, 56, 517–526. [Google Scholar] [CrossRef]
- Song, F.; Li, H.; Liu, G.H.; Wang, W.; James, P.; Colwell, D.D.; Tran, A.; Gong, S.Y.; Cai, W.Z.; Shao, R.F. Mitochondrial genome fragmentation unites the parasitic lice of eutherian mammals. Syst. Biol. 2019, 68, 430–440. [Google Scholar] [CrossRef]
- Dowton, M.; Cameron, S.L.; Dowavic, J.I.; Austin, A.D.; Whiting, M.F. Characterization of 67 mitochondrial tRNA gene rearrangements in the Hymenoptera suggests that mitochondrial tRNA gene position is selectively neutral. Mol. Biol. Evol. 2009, 26, 1607–1617. [Google Scholar] [CrossRef]
- Berthier, F.; Renaud, M.; Alziari, S.; Durand, R. RNA mapping on Drosophila mitochondrial DNA precursors and template strands. Nucleic Acids Res. 1986, 14, 4519–4533. [Google Scholar] [CrossRef] [PubMed]
- Stewart, J.B.; Beckenbach, A.T. Characterization of mature mitochondrial transcripts in Drosophila, and the implications for the tRNA punctuation model in arthropods. Gene 2009, 445, 49–57. [Google Scholar] [CrossRef] [PubMed]
- Roberti, M.; Bruni, F.; Polosa, P.L.; Gadaleta, M.N.; Cantatore, P. The Drosophila termination factor DmTTF regulates in vivo mitochondrial transcription. Nucleic Acids Res. 2006, 34, 2109–2116. [Google Scholar] [CrossRef] [PubMed]
- Ojala, D.; Montoya, J.; Attardi, G. tRNA punctuation model of RNA processing in human mitochondria. Nature 1981, 290, 470–474. [Google Scholar] [CrossRef]
- Gao, S.; Ren, Y.; Sun, Y.; Wu, Z.; Ruan, J.; He, B.; Zhang, T.; Yu, X.; Tian, X.; Bu, W. PacBio full-length transcriptome profiling of insect mitochondrial gene expression. RNA Biol. 2016, 13, 820–825. [Google Scholar] [CrossRef]
- Ji, H.S.; Xu, X.F.; Jin, X.F.; Yin, H.; Luo, J.X.; Liu, G.Y.; Zhao, Q.; Chen, Z.; Bu, W.J.; Gao, S. Using high-resolution annotation of insect mitochondrial DNA to decipher tandem repeats in the control region. RNA Biol. 2019, 16, 830–837. [Google Scholar] [CrossRef]
- Xu, S.W.; Duan, Y.E.; Ma, L.; Song, F.; Tian, L.; Cai, W.Z.; Li, H. Full-length transcriptome profiling of Coridius chinensis mitochondrial genome reveals the transcription of genes with ancestral arrangement in insects. Genes 2023, 14, 225. [Google Scholar] [CrossRef]
- Zhao, X.J.; Xu, S.W.; Li, J.R.; Yang, H.L.; Tian, L.; Song, F.; Cai, W.Z.; Lin, Z.L.; Li, H. Full-length transcriptome profiling of Aphidius gifuensis mitochondrial genome with gene rearrangement and control region duplication. Heliyon 2023, 9, e17070. [Google Scholar] [CrossRef]
- Liu, Q.Q.; Cai, Y.D.; Ma, L.; Liu, H.R.; Linghu, T.; Guo, S.K.; Wei, S.J.; Song, F.; Tian, L.; Cai, W.Z.; et al. Relaxed purifying selection pressure drives accelerated and dynamic gene rearrangements in thrips (Insecta: Thysanoptera) mitochondrial genomes. Int. J. Biol. Macromol. 2023, 253, 126742. [Google Scholar] [CrossRef]
- Mound, L.A.; Morris, D.C. The insect order Thysanoptera: classification versus systematics. Zootaxa 2007, 1668, 395–411. [Google Scholar] [CrossRef]
- Zhang, S.M.; Mound, L.; Feng, J.N. Morphological phylogeny of Thripidae (Thysanoptera: Terebrantia). Invertebr. Syst. 2019, 33, 671–696. [Google Scholar] [CrossRef]
- Buckman, R.S.; Mound, L.A.; Whiting, M.F. Phylogeny of thrips (Insecta: Thysanoptera) based on five molecular loci. Syst. Entomol. 2013, 38, 123–133. [Google Scholar] [CrossRef]
- Pakrashi, A.; Kumar, V.; Stanford-Beale, D.A.C.; Cameron, S.L.; Tyagi, K. Gene arrangement, phylogeny and divergence time estimation of mitogenomes in thrips. Mol. Biol. Rep. 2022, 49, 6269–6283. [Google Scholar] [CrossRef] [PubMed]
- Tyagi, K.; Chakraborty, R.; Cameron, S.L.; Sweet, A.D.; Chandra, K.; Kumar, V. Rearrangement and evolution of mitochondrial genomes in Thysanoptera (Insecta). Sci. Rep. 2020, 10, 695. [Google Scholar] [CrossRef] [PubMed]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef]
- Schmieder, R.; Edwards, R. Quality control and preprocessing of metagenomic datasets. Bioinformatics 2011, 27, 863–864. [Google Scholar] [CrossRef]
- Peng, Y.; Leung, H.C.M.; Yiu, S.M.; Chin, F.Y.L. IDBA-UD: a de novo assembler for single-cell and metagenomic sequencing data with highly uneven depth. Bioinformatics 2012, 28, 1420–1428. [Google Scholar] [CrossRef]
- Chen, Y.; Ye, W.; Zhang, Y.; Xu, Y. High speed BLASTN: an accelerated MegaBLAST search tool. Nucleic Acids Res. 2015, 43, 7762–7768. [Google Scholar] [CrossRef]
- Meng, G.L.; Li, Y.Y.; Yang, C.T.; Liu, S.L. MitoZ: a toolkit for animal mitochondrial genome assembly, annotation and visualization. Nucleic Acids Res. 2019, 47, e63. [Google Scholar] [CrossRef]
- Katoh, K.; Kuma, K.; Toh, H.; Miyata, T. MAFFT version 5: improvement in accuracy of multiple sequence alignment. Nucleic Acids Res. 2005, 33, 511–518. [Google Scholar] [CrossRef]
- Capella-Gutierrez, S.; Silla-Martinez, J.M.; Gabaldon, T. trimAl: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics 2009, 25, 1972–1973. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef] [PubMed]
- Lartillot, N.; Rodrigue, N.; Stubbs, D.; Richer, J. PhyloBayes MPI: Phylogenetic reconstruction with infinite mixtures of profiles in a parallel environment. Syst. Biol. 2013, 62, 611–615. [Google Scholar] [CrossRef]
- Miller, M.A.; Pfeiffer, W.; Schwartz, T. Creating the CIPRES science gateway for inference of large phylogenetic trees. In 2010 Gateway Computing Environments Workshop (GCE); IEEE: Piscataway, NJ, USA, 2010; pp. 1–8. [Google Scholar]
- Taft, R.J.; Glazov, E.A.; Cloonan, N.; Simons, C.; Stephen, S.; Faulkner, G.J.; Lassmann, T.; Forrest, A.R.R.; Grimmond, S.M.; Schroder, K.; et al. Tiny RNAs associated with transcription start sites in animals. Nat. Genet. 2009, 41, 572–578. [Google Scholar] [CrossRef] [PubMed]
- Liang, G.; Mi, D.; Chang, J.; On Yau, T.; Xu, G.; Ruan, J.; Bu, W.; Gao, S. Precise annotation of Drosophila mitochondrial genomes leads to insights into AT-rich regions. Mitochondrion 2022, 65, 145–149. [Google Scholar] [CrossRef] [PubMed]
- Bernt, M.; Merkle, D.; Ramsch, K.; Fritzsch, G.; Perseke, M.; Bernhard, D.; Schlegel, M.; Stadler, P.F.; Middendorf, M. CREx: inferring genomic rearrangements based on common intervals. Bioinformatics 2007, 23, 2957–2958. [Google Scholar] [CrossRef]
- Boore, J.L. The duplication/random loss model for gene rearrangement exemplified by mitochondrial genomes of deuterostome animals. In Comparative Genomics: Empirical and Analytical Approaches to Gene Order Dynamics, Map Alignment and the Evolution of Gene Families; Springer: Dordrecht, The Netherlands, 2000; pp. 133–134. [Google Scholar]
- Lavrov, D.V.; Boore, J.L.; Brown, W.M. Complete mtDNA sequences of two millipedes suggest a new model for mitochondrial gene rearrangements: duplication and nonrandom loss. Mol. Biol. Evol. 2002, 19, 163–169. [Google Scholar] [CrossRef]
- Sun, Z.; Liu, Y.; Wilson, J.J.; Chen, Z.; Song, F.; Cai, W.; Li, H. Mitochondrial genome of Phalantus geniculatus (Hemiptera: Reduviidae): trnT duplication and phylogenetic implications. Int. J. Biol. Macromol. 2019, 129, 110–115. [Google Scholar] [CrossRef]
- Shao, R.; Barker, S.C. The highly rearranged mitochondrial genome of the plague thrips, Thrips imaginis (Insecta: Thysanoptera): convergence of two novel gene boundaries and an extraordinary arrangement of rRNA genes. Mol. Biol. Evol. 2003, 20, 362–370. [Google Scholar] [CrossRef]
- Macey, J.R.; Larson, A.; Ananjeva, N.B.; Fang, Z.L.; Papenfuss, T.J. Two novel gene orders and the role of light-strand replication in rearrangement of the vertebrate mitochondrial genome. Mol. Biol. Evol. 1997, 14, 91–104. [Google Scholar] [CrossRef]
- Geisberg, J.V.; Moqtaderi, Z.; Fan, X.C.; Ozsolak, F.; Struhl, K. Global analysis of mRNA isoform half-lives reveals stabilizing and destabilizing elements in yeast. Cell 2014, 156, 812–824. [Google Scholar] [CrossRef] [PubMed]
- Geisberg, J.V.; Moqtaderi, Z.; Struhl, K. Condition-specific 3′ mRNA isoform half-lives and stability elements in yeast. Proc. Natl. Acad. Sci. USA 2023, 120, e2301117120. [Google Scholar] [CrossRef] [PubMed]
- Ojala, D.; Merkel, C.; Gelfand, R.; Attardi, G. The tRNA genes punctuate the reading of genetic information in human mitochondrial DNA. Cell 1980, 22, 393–403. [Google Scholar] [CrossRef]
- Wolstenholme, D.R. Animal mitochondrial DNA: structure and evolution. Int. Rev. Cytol. 1992, 141, 173–216. [Google Scholar] [PubMed]
- Margam, V.M.; Coates, B.S.; Hellmich, R.L.; Agunbiade, T.; Seufferheld, M.J.; Sun, W.L.; Ba, M.N.; Sanon, A.; Binso-Dabire, C.L.; Baoua, I.; et al. Mitochondrial genome sequence and expression profiling for the legume pod borer Maruca vitrata (Lepidoptera: Crambidae). PLoS ONE 2011, 6, e16444. [Google Scholar] [CrossRef]
- Wang, H.L.; Yang, J.; Boykin, L.M.; Zhao, Q.Y.; Li, Q.; Wang, X.W.; Liu, S.S. The characteristics and expression profiles of the mitochondrial genome for the Mediterranean species of the complex. BMC Genomics 2013, 14, 401. [Google Scholar] [CrossRef]
- Ng, Y.F.; Mound, L.A. Genera of the Scirtothrips genus-group (Thysanoptera, Thripidae) with a new species of Siamothrips from Malaysia. Zootaxa 2015, 4021, 387–394. [Google Scholar] [CrossRef][Green Version]
- Minaei, K.; Mound, L. Scirtothrips genus-group in Iran with an unusual new species of Scirtothrips (Thysanoptera: Thripidae). Zootaxa 2018, 4394, 288–294. [Google Scholar] [CrossRef]
- Priesner, H. Zur vergleichenden Morphologie des Endothorax der Thysanopteren. Zool. Anz. 1957, 7–8, 156–157. [Google Scholar]
- Masumoto, M.; Okajima, S. The genus Scirtothrips Shull (Insecta, Thysanoptera, Thripidae) and three related genera in Japan. Zootaxa 2007, 1552, 1–33. [Google Scholar] [CrossRef]



| Gene | Coding Strand | Annotation Using DNA Sequencing | Annotations According to Transcripts | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Position | Length (bp) | Start Codon | Stop Codon | Position | Length (bp) | Strat Codon | Stop Codon | ||
| COI | J | 1–1539 | 1539 | AUA | UAA | 1–1539 | 1539 | AUA | UAA |
| ND3 | J | 1552–1892 | 341 | AUU | UA | 1552–1891 | 340 | AUU | U |
| trnL2 | J | 1892–1955 | 64 | 1892–1955 | 64 | ||||
| COII | J | 1956–2618 | 663 | AUU | UAA | 1956–2618 | 663 | AUU | UAA |
| trnR | J | 2665–2718 | 54 | 2664–2718 | 55 | ||||
| trnG | J | 2719–2779 | 61 | 2719–2779 | 61 | ||||
| trnD | J | 2798–2858 | 61 | 2798–2858 | 61 | ||||
| trnK | J | 2897–2957 | 61 | 2897–2957 | 61 | ||||
| COIII | J | 3070–3867 | 798 | AUA | UAA | 2959–3867 | 909 | AUA | UAA |
| CR1 | 3868–4052 | 185 | 3868–4052 | 185 | |||||
| trnN | J | 4053–4117 | 65 | 4053–4117 | 65 | ||||
| trnE | J | 4115–4179 | 65 | 4115–4179 | 65 | ||||
| trnQ | J | 4183–4251 | 69 | 4183–4251 | 69 | ||||
| trnA | J | 4291–4353 | 63 | 4291–4353 | 63 | ||||
| trnS1 | J | 4354–4407 | 54 | 4354–4407 | 54 | ||||
| trnI | J | 4409–4477 | 69 | 4409–4477 | 69 | ||||
| CYTB | J | 4482–5588 | 1107 | AUA | UAA | 4479–5588 | 1110 | AUA | UAA |
| trnY | N | 5650–5586 | 65 | 5650–5586 | 65 | ||||
| ND2 | J | 5678–6656 | 979 | AUA | U | 5678–6659 | 982 | AUA | U |
| trnW | J | 6660–6722 | 63 | 6660–6722 | 63 | ||||
| ND1 | J | 6729–7641 | 913 | AUU | U | 6723–7641 | 919 | AUA | U |
| trnM | J | 7642–7702 | 61 | 7642–7702 | 61 | ||||
| trnF | J | 7712–7777 | 66 | 7712–7777 | 66 | ||||
| srRNA | J | 7771–8504 | 734 | 7778–8500 | 723 | ||||
| ATP8 | N | 8501–8669 | 169 | AUU | U | 8501–8669 | 169 | AUU | U |
| ATP6 | J | 8636–9346 | 711 | AUA | UAA | 8696–9346 | 651 | AUU | UAA |
| trnL2 | J | 9347–9410 | 64 | 9347–9410 | 64 | ||||
| trnT | J | 9418–9483 | 66 | 9411–9477 | 66 | ||||
| CR2 | 9484–9736 | 253 | 9478–9736 | 259 | |||||
| trnP | N | 9801–9737 | 65 | 9801–9737 | 65 | ||||
| ND5 | N | 11,532–9856 | 1677 | AUA | UAG | 11,532–9856 | 1677 | AUA | UAG |
| trnH | N | 11,592–11,533 | 60 | 11,592–11,533 | 60 | ||||
| ND4 | N | 12,896–11,593 | 1304 | AUU | UAA | 12,896–11,593 | 1304 | AUU | UAA |
| ND4L | N | 13,168–12,890 | 279 | UAG | UAG | 13,168–12,890 | 279 | UAG | UAG |
| trnC | J | 13,207–13,267 | 61 | 13,207–13,267 | 61 | ||||
| ND6 | J | 13,298–13,762 | 465 | AUU | UAA | 13,283–137,62 | 480 | AUA | UAA |
| trnV | J | 13,764–13,819 | 56 | 13,764–13,819 | 56 | ||||
| lrRNA | J | 13,819–14,907 | 1087 | 13,820–14,902 | 1083 | ||||
| trnS1 | J | 14,903–14,965 | 63 | 14,903–14,965 | 63 | ||||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, Q.; Xu, S.; He, J.; Cai, W.; Wang, X.; Song, F. Full-Length Transcriptome Profiling of the Complete Mitochondrial Genome of Sericothrips houjii (Thysanoptera: Thripidae: Sericothripinae) Featuring Extensive Gene Rearrangement and Duplicated Control Regions. Insects 2024, 15, 700. https://doi.org/10.3390/insects15090700
Liu Q, Xu S, He J, Cai W, Wang X, Song F. Full-Length Transcriptome Profiling of the Complete Mitochondrial Genome of Sericothrips houjii (Thysanoptera: Thripidae: Sericothripinae) Featuring Extensive Gene Rearrangement and Duplicated Control Regions. Insects. 2024; 15(9):700. https://doi.org/10.3390/insects15090700
Chicago/Turabian StyleLiu, Qiaoqiao, Shiwen Xu, Jia He, Wanzhi Cai, Xingmin Wang, and Fan Song. 2024. "Full-Length Transcriptome Profiling of the Complete Mitochondrial Genome of Sericothrips houjii (Thysanoptera: Thripidae: Sericothripinae) Featuring Extensive Gene Rearrangement and Duplicated Control Regions" Insects 15, no. 9: 700. https://doi.org/10.3390/insects15090700
APA StyleLiu, Q., Xu, S., He, J., Cai, W., Wang, X., & Song, F. (2024). Full-Length Transcriptome Profiling of the Complete Mitochondrial Genome of Sericothrips houjii (Thysanoptera: Thripidae: Sericothripinae) Featuring Extensive Gene Rearrangement and Duplicated Control Regions. Insects, 15(9), 700. https://doi.org/10.3390/insects15090700







