Mapping and Characterization of Target-Site Resistance to Cyclic Ketoenol Insecticides in Cabbage Whiteflies, Aleyrodes proletella (Hemiptera: Aleyrodidae)
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Insects
2.2. Chemicals
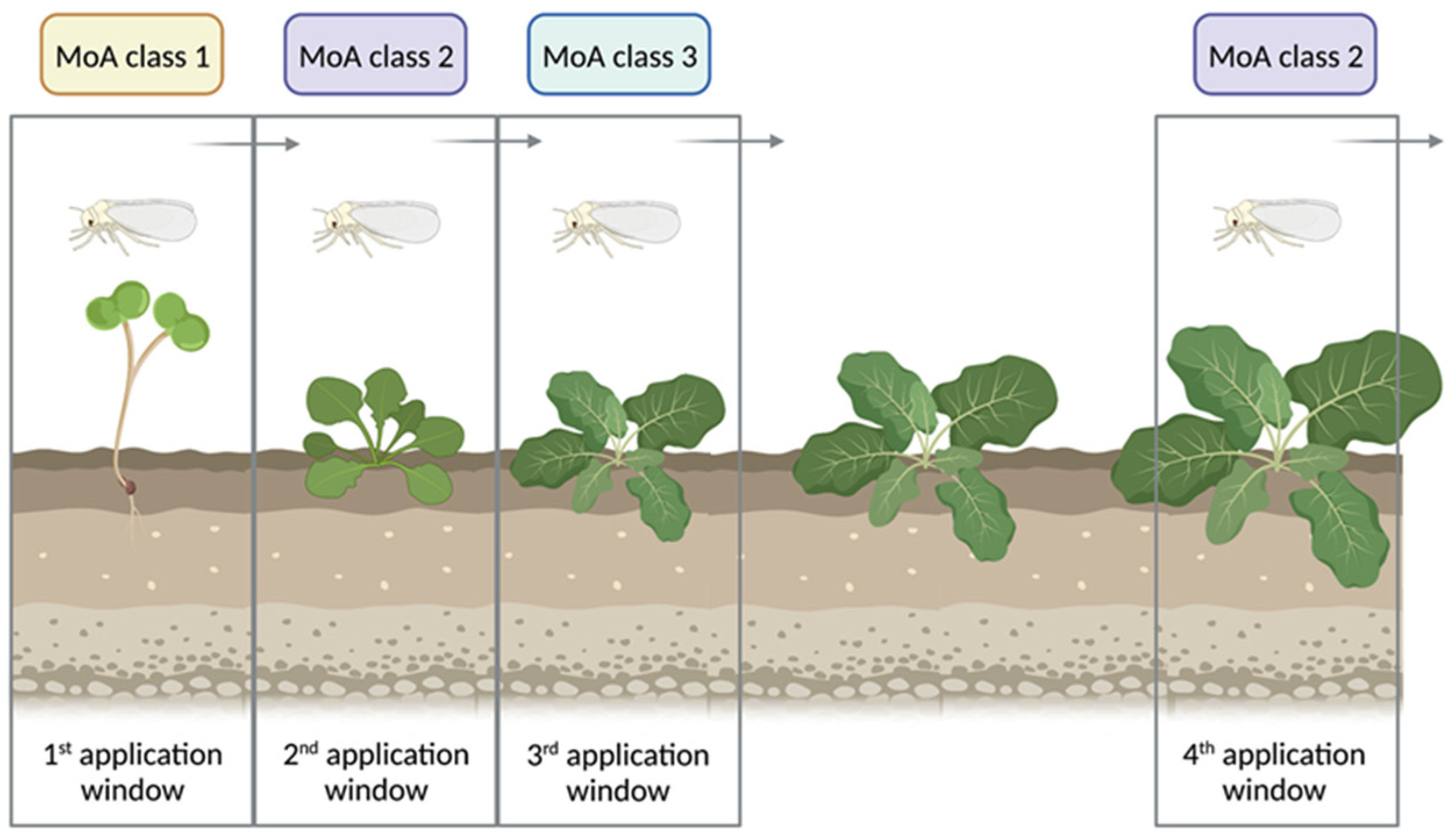
2.3. Nymph Bioassays
2.4. Adult Bioassays
2.5. Reciprocal Crossing Experiments
2.6. RNA Sequencing and Acetyl-CoA Carboxylase Assembly
2.7. RT-qPCR
2.8. Genotyping by Pyrosequencing
2.9. Computational Modeling and Docking
2.10. Statistical Analysis
3. Results
3.1. Bioassays
3.2. Reciprocal Crossing Experiments
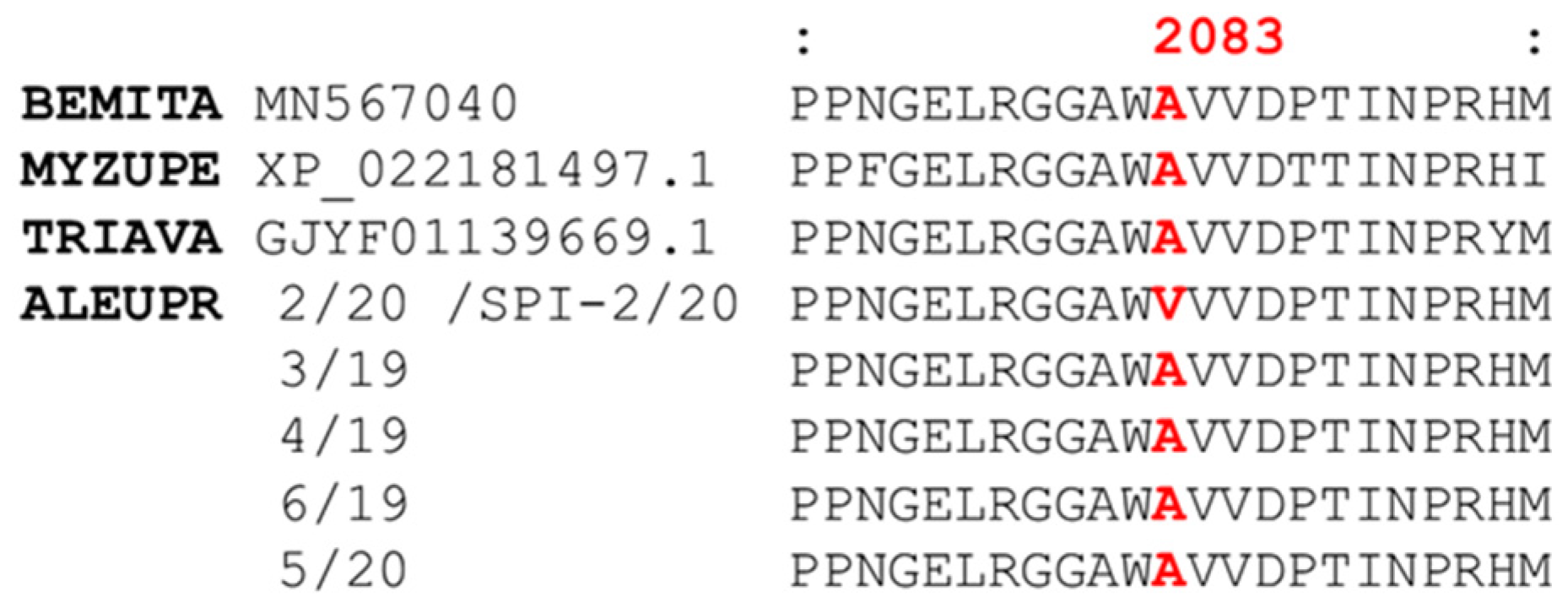
3.3. RNAseq and acetyl-CoA Carboxylase (ACC) Assembly
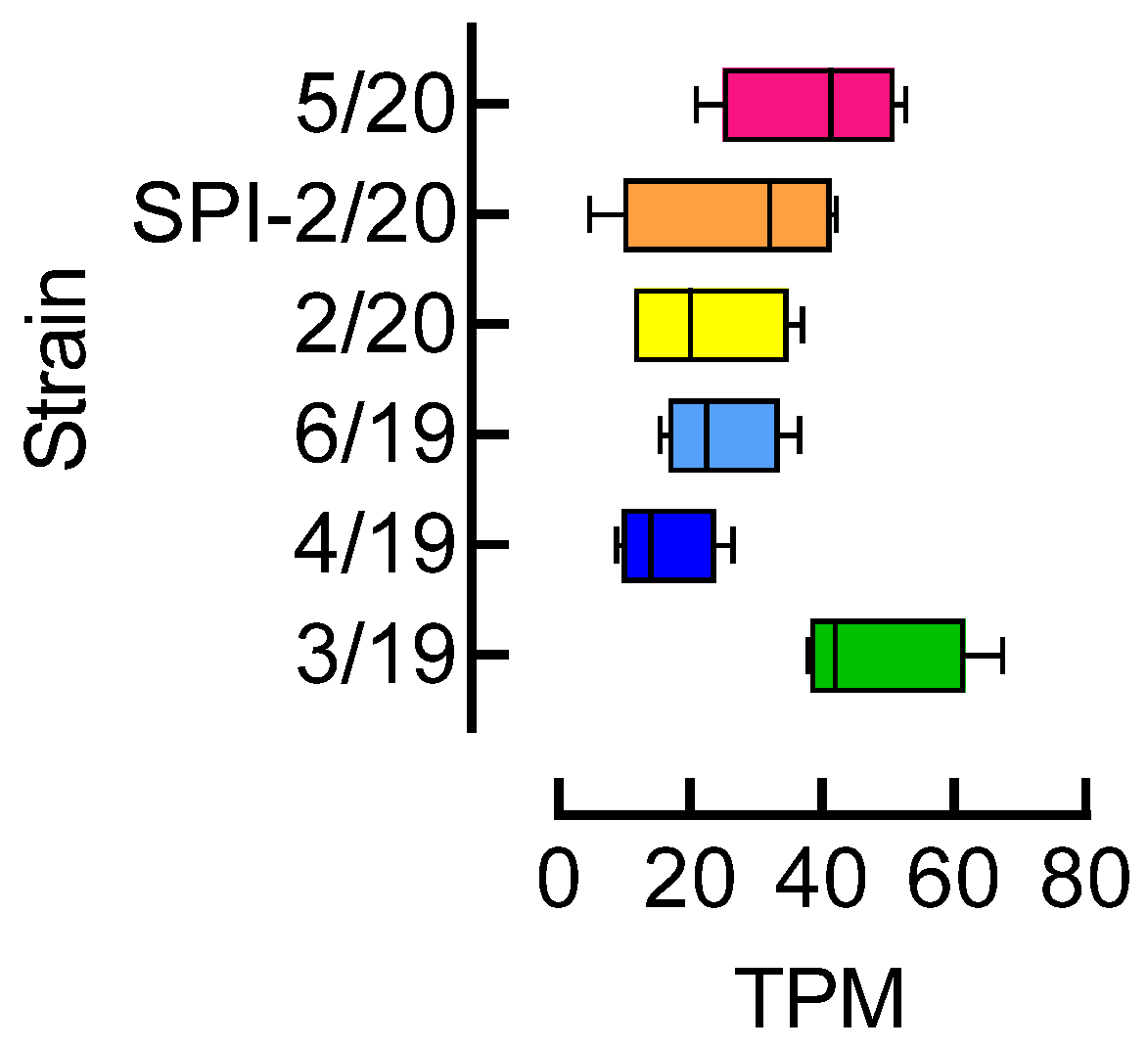
3.4. Acetyl-CoA Carboxylase (ACC) Expression Level
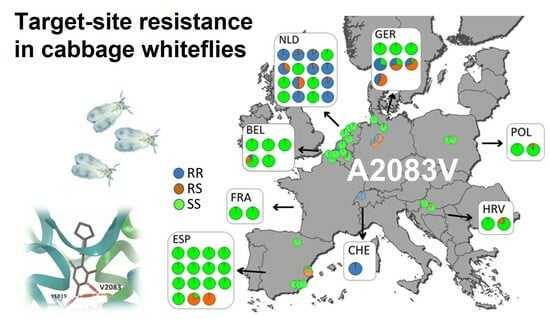
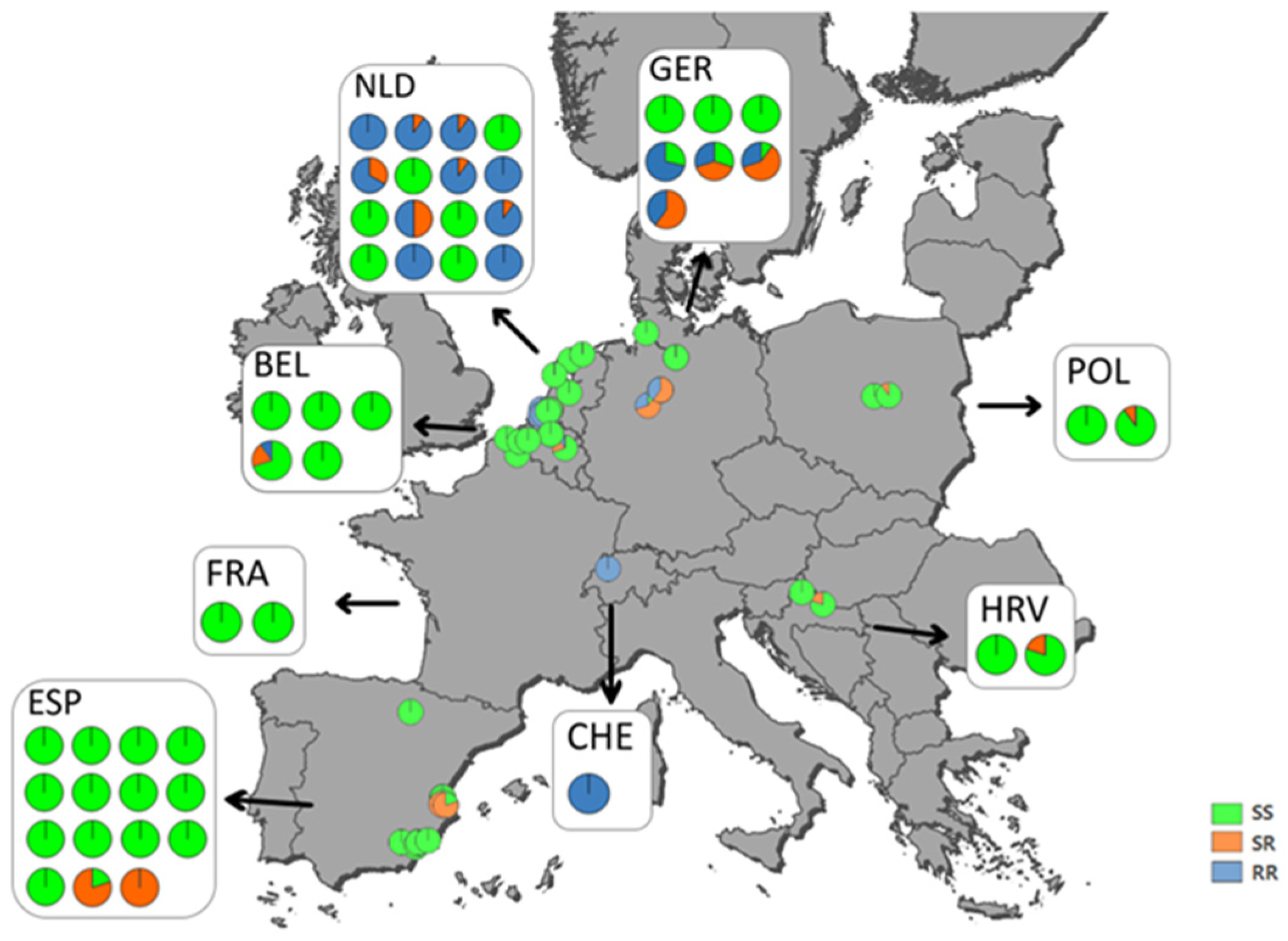
3.5. Mapping of Ketoenol Resistance by Analyzing Alcohol Persevered A. proletella Field Samples
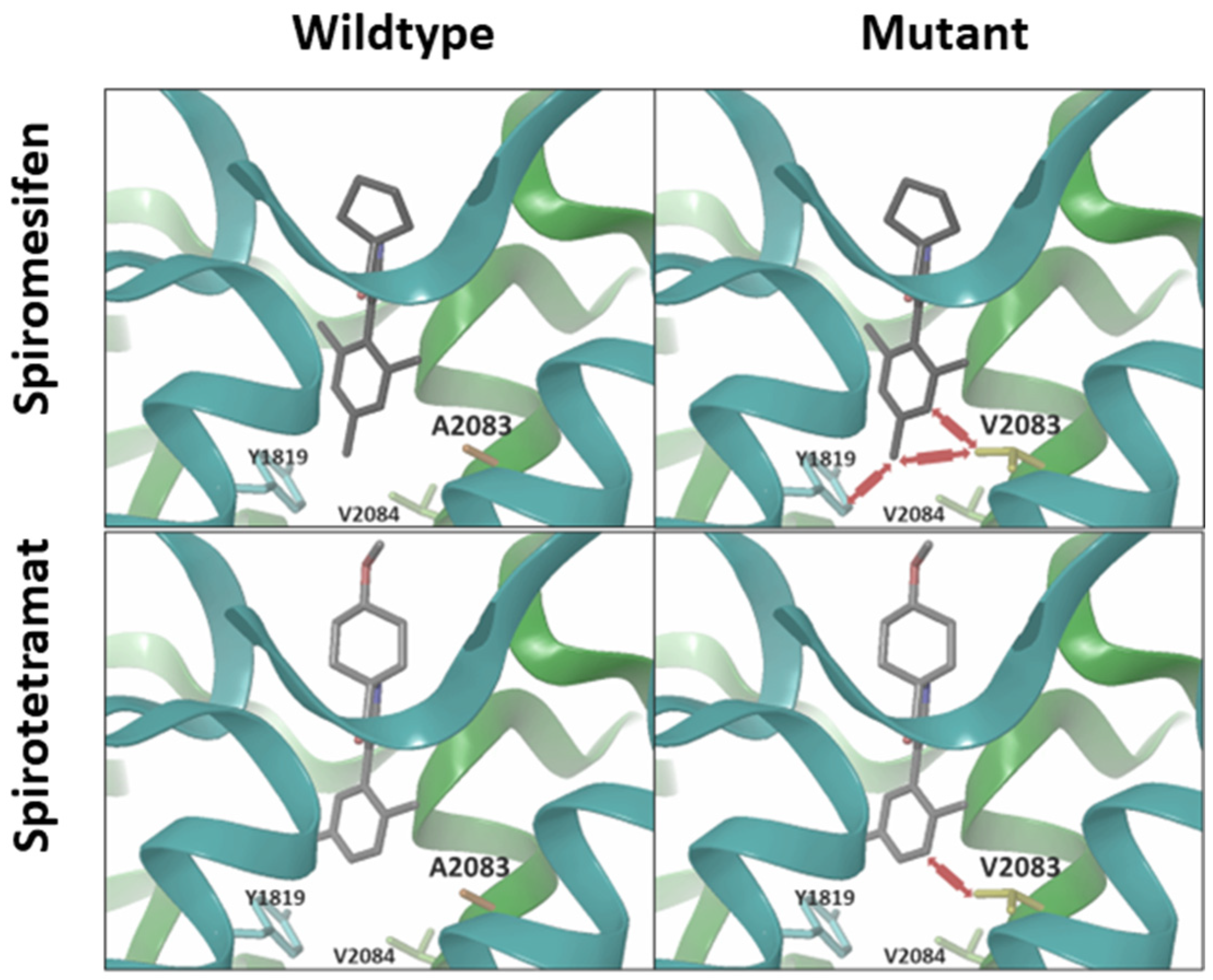
3.6. Computational Modelling and Ketoenol Docking Analysis
4. Discussion
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Martin, J.H.; Mifsud, D.; Rapisarda, C. The Whiteflies (Hemiptera: Aleyrodidae) of Europe and the Mediterranean Basin. Bull. Entomol. Res. 2000, 90, 407–448. [Google Scholar] [CrossRef]
- Martin, J.H.; Mound, L.A. An Annotated Check List of the World’s Whiteflies (Insecta: Hemiptera: Aleyrodidae). Zootaxa 2007, 1492, 1–84. [Google Scholar] [CrossRef]
- Chen, M.-M.; Guo, R.; Zhang, J.-L.; Wan, G.-H.; Yang, J.; Zhang, G.-F. Rapid Identification of Aleyrodes proletella (Hemiptera: Aleyrodidae), a New Invasive Whitefly Species in Mainland China, Based on SS-COI Marker. Acta Entomol. Sin. 2015, 58, 579–586. [Google Scholar]
- Zhang, G.F.; Xian, X.Q.; Zhang, J.L.; Li, X.F.; Ma, D.Y.; Wan, F.H. Cabbage Whitefly, Aleyrodes proletella (L.) (Hemiptera: Aleyrodidae), Invaded Mainland China. Entomol. J. East China 2014, 23, 66–70. [Google Scholar]
- Alexander Jesudasan, R.W.; David, B.V. Taxonomic Studies on Indian Aleyrodidae (Insecta: Homoptera). Orient. Insects 1991, 25, 231–434. [Google Scholar] [CrossRef]
- De Barro, P.J.; Carver, M. Cabbage Whitefly, Aleyrodes proletella (L.) (Hemiptera: Aleyrodidae), Newly Discovered in Australia. Aust. J. Entomol. 1997, 36, 255–256. [Google Scholar] [CrossRef]
- Trdan, S.; Papler, U. Susceptibility of Four Different Vegetable Brassicas to Cabbage Whitefly (Aleyrodes proletella L., Aleyrodidae) Attack. Meded. Rijksuniv. Te Gent Fak. Van Landbouwkd. En Toegepaste Biol. Wet. 2002, 67, 531–535. [Google Scholar]
- Richter, E.; Hirthe, G. Hibernation and Migration of Aleyrodes proletella in Germany. IOBC/WPRS Bull. 2014, 107, 143–149. [Google Scholar]
- OEPP/EPPO PP 1/311 (1) Aleyrodes proletella on Brassica Crops. EPPO Bull. 2019, 49, 37–39. [CrossRef]
- Saucke, H.; Schultz, B.; Wedemeyer, R.; Liebig, N.; Zimmermann, O.; Katz, P. Biotechnische Regulierung der Kohlmottenschildlaus in Kohlgemüse—Sachstand und Perspektiven. Gesunde Pflanz. 2011, 63, 183–189. [Google Scholar] [CrossRef]
- Carden, P.W. New or Uncommon Plant Diseases and Pests. Plant Pathol. 1972, 21, 145–146. [Google Scholar] [CrossRef]
- Alonso, D.; Gómez, A.A.; Nombela, G.; Muñiz, M. Temperature-Dependent Development of Aleyrodes proletella (Homoptera: Aleyrodidae) on Two Cultivars of Broccoli under Constant Temperatures. Environ. Entomol. 2009, 38, 11–17. [Google Scholar] [CrossRef]
- Butler, C.G. On the Ecology of Aleurodes brassicae Walk. (Hemiptera). Trans. R. Entomol. Soc. Lond. 1938, 87, 291–311. [Google Scholar] [CrossRef]
- Pajović, I. Seasonal Dynamics of Most Detrimental Pest Insects Species on Cabbage Plants in Montenegro. Agric. For. 2011, 51, 25–42. [Google Scholar]
- Kovaříková, K.; Holý, K.; Skuhrovec, J.; Saska, P. The Efficacy of Insecticides against Eggs and Nymphs of Aleyrodes Proletella (Hemiptera: Aleyrodidae) under Laboratory Conditions. Crop Prot. 2017, 98, 40–45. [Google Scholar] [CrossRef]
- Laurenz, S.; Schmidt, S.; Balkenhol, B.; Meyhofer, R. Natural Enemies Associated with the Cabbage Whitefly Aleyrodes proletella in Germany. J. Plant Dis. Prot. 2019, 126, 47–54. [Google Scholar] [CrossRef]
- Koca, A.S.; Kütük, H. Distribution, Host Plants and Natural Enemies of Aleyrodes proletella L. (Hemiptera: Aleyrodidae) on Collard (Brassica oleracea L. Var. Acephala) in Düzce Province of Turkey. Plant Prot. Bull. 2020, 60, 17–24. [Google Scholar] [CrossRef]
- Laurenz, S.; Meyhöfer, R. Banker Plants Promote Functional Biodiversity and Decrease Populations of the Cabbage Whitefly Aleyrodes proletella. J. Appl. Entomol. 2021, 145, 36–45. [Google Scholar] [CrossRef]
- Broekgaarden, C.; Riviere, P.; Steenhuis, G.; del sol Cuenca, M.; Kos, M.; Vosman, B. Phloem-Specific Resistance in Brassica Oleracea against the Whitefly Aleyrodes proletella. Entomol. Exp. Appl. 2012, 142, 153–164. [Google Scholar] [CrossRef]
- Hondelmann, P.; Paul, C.; Schreiner, M.; Meyhöfer, R. Importance of Antixenosis and Antibiosis Resistance to the Cabbage Whitefly (Aleyrodes proletella) in Brussels Sprout Cultivars. Insects 2020, 11, 56. [Google Scholar] [CrossRef]
- Richter, E.; Hirthe, G. First Results on Population Dynamics and Chemical Control of Aleyrodes proletella in Germany. IOBC-WPRS Bull. 2014, 107, 63–70. [Google Scholar]
- Bretschneider, T.; Benet-Buchholz, J.; Fischer, R.; Nauen, R. Spirodiclofen and Spiromesifen—Novel Acaricidal and Insecticidal Tetronic Acid Derivatives with a New Mode of Action. Chimia 2003, 57, 697. [Google Scholar] [CrossRef]
- Marcic, D.; Peric, P.; Petronijevic, S.; Prijovic, M.; Drobnjakovic, T. Cyclic Ketoenols: Acaricides and Insecticides with a Novel Mode of Action. Pestic. Fitomed. 2011, 26, 185–195. [Google Scholar] [CrossRef]
- Brück, E.; Elbert, A.; Fischer, R.; Krueger, S.; Kühnhold, J.; Klueken, A.M.; Nauen, R.; Niebes, J.-F.; Reckmann, U.; Schnorbach, H.-J.; et al. Movento®, an Innovative Ambimobile Insecticide for Sucking Insect Pest Control in Agriculture: Biological Profile and Field Performance. Crop Prot. 2009, 28, 838–844. [Google Scholar] [CrossRef]
- Nauen, R.; Konanz, S. Spiromesifen as a New Chemical Option for Resistance Management in Whiteflies and Spider Mites. Bayer Crop. J. 2005, 58, 485–502. [Google Scholar]
- Nauen, R.; Reckmann, U.; Thonzik, J.; Thielert, W. Biological Profile of Spirotetramat (Movento)—A New Two-Way Systemic (Ambimobile) Insecticide against Sucking Pest Species. Bayer Crop. J. 2008, 61, 245–277. [Google Scholar]
- Muehlebach, M.; Buchholz, A.; Zambach, W.; Schaetzer, J.; Daniels, M.; Hueter, O.; Kloer, D.P.; Lind, R.; Maienfisch, P.; Pierce, A.; et al. Spiro N-Methoxy Piperidine Ring Containing Aryldiones for the Control of Sucking Insects and Mites: Discovery of Spiropidion. Pest Manag. Sci. 2020, 76, 3440–3450. [Google Scholar] [CrossRef]
- Parvy, J.-P.; Napal, L.; Rubin, T.; Poidevin, M.; Perrin, L.; Wicker-Thomas, C.; Montagne, J. Drosophila Melanogaster Acetyl-CoA-Carboxylase Sustains a Fatty Acid-Dependent Remote Signal to Waterproof the Respiratory System. PLoS Genet. 2012, 8, e1002925. [Google Scholar] [CrossRef]
- Lümmen, P.; Khajehali, J.; Luther, K.; Van Leeuwen, T. The Cyclic Keto-Enol Insecticide Spirotetramat Inhibits Insect and Spider Mite Acetyl-CoA Carboxylases by Interfering with the Carboxyltransferase Partial Reaction. Insect Biochem. Mol. Biol. 2014, 55, 1–8. [Google Scholar] [CrossRef]
- Karatolos, N.; Williamson, M.S.; Denholm, I.; Gorman, K.; Ffrench-Constant, R.; Nauen, R. Resistance to Spiromesifen in Trialeurodes vaporariorum Is Associated with a Single Amino Acid Replacement in Its Target Enzyme Acetyl-Coenzyme A Carboxylase. Insect Mol. Biol. 2012, 21, 327–334. [Google Scholar] [CrossRef] [PubMed]
- Bielza, P.; Moreno, I.; Belando, A.; Grávalos, C.; Izquierdo, J.; Nauen, R. Spiromesifen and Spirotetramat Resistance in Field Populations of Bemisia tabaci Gennadius in Spain. Pest Manag. Sci. 2019, 75, 45–52. [Google Scholar] [CrossRef] [PubMed]
- Lueke, B.; Douris, V.; Hopkinson, J.E.; Maiwald, F.; Hertlein, G.; Papapostolou, K.-M.; Bielza, P.; Tsagkarakou, A.; Van Leeuwen, T.; Bass, C.; et al. Identification and Functional Characterization of a Novel Acetyl-CoA Carboxylase Mutation Associated with Ketoenol Resistance in Bemisia tabaci. Pestic. Biochem. Physiol. 2020, 166, 104583. [Google Scholar] [CrossRef] [PubMed]
- Guest, M.; Kriek, N.; Flemming, A.J. Studies of an Insecticidal Inhibitor of Acetyl-CoA Carboxylase in the Nematode C. Elegans. Pestic. Biochem. Physiol. 2020, 169, 104604. [Google Scholar] [CrossRef] [PubMed]
- Hopkinson, J.; Balzer, J.; Fang, C.; Walsh, T. Insecticide Resistance Management of Bemisia tabaci (Hemiptera: Aleyrodidae) in Australian Cotton—Pyriproxyfen, Spirotetramat and Buprofezin. Pest Manag. Sci. 2023, 79, 1829–1839. [Google Scholar] [CrossRef] [PubMed]
- Mavridis, K.; Papapostolou, K.M.; Ilias, A.; Michaelidou, K.; Stavrakaki, M.; Roditakis, E.; Tsagkarakou, A.; Bass, C.; Vontas, J. Next-Generation Molecular Diagnostics (TaqMan qPCR and ddPCR) for Monitoring Insecticide Resistance in Bemisia tabaci. Pest Manag. Sci. 2022, 78, 4994–5001. [Google Scholar] [CrossRef]
- Kapantaidaki, D.E.; Sadikoglou, E.; Tsakireli, D.; Kampanis, V.; Stavrakaki, M.; Schorn, C.; Ilias, A.; Riga, M.; Tsiamis, G.; Nauen, R.; et al. Insecticide Resistance in Trialeurodes vaporariorum Populations and Novel Diagnostics for Kdr Mutations. Pest Manag. Sci. 2018, 74, 59–69. [Google Scholar] [CrossRef]
- Horowitz, A.R.; Ghanim, M.; Roditakis, E.; Nauen, R.; Ishaaya, I. Insecticide Resistance and Its Management in Bemisia tabaci Species. J. Pest Sci. 2020, 93, 893–910. [Google Scholar] [CrossRef]
- De Rouck, S.; İnak, E.; Dermauw, W.; Van Leeuwen, T. A Review of the Molecular Mechanisms of Acaricide Resistance in Mites and Ticks. Insect Biochem. Mol. Biol. 2023, 159, 103981. [Google Scholar] [CrossRef]
- İnak, E.; Demirci, B.; Vandenhole, M.; Söylemezoğlu, G.; Van Leeuwen, T.; Toprak, U. Molecular Mechanisms of Resistance to Spirodiclofen and Spiromesifen in Tetranychus urticae. Crop Prot. 2023, 172, 106343. [Google Scholar] [CrossRef]
- Springate, S.; Colvin, J. Pyrethroid Insecticide Resistance in British Populations of the Cabbage Whitefly, Aleyrodes proletella. Pest Manag. Sci. 2012, 68, 260–267. [Google Scholar] [CrossRef]
- Horowitz, A.R.; Gorman, K.; Ross, G.; Denholm, I. Inheritance of Pyriproxyfen Resistance in the Whitefly, Bemisia tabaci (Q Biotype). Arch. Insect Biochem. Physiol. 2003, 54, 177–186. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Tabashnik, B.E. Inheritance of Resistance to the Bacillus Thuringiensis Toxin Cry1C in the Diamondback Moth. Appl. Environ. Microbiol. 1997, 63, 2218–2223. [Google Scholar] [CrossRef] [PubMed]
- Grabherr, M.G.; Haas, B.J.; Yassour, M.; Levin, J.Z.; Thompson, D.A.; Amit, I.; Adiconis, X.; Fan, L.; Raychowdhury, R.; Zeng, Q.; et al. Full-Length Transcriptome Assembly from RNA-Seq Data without a Reference Genome. Nat. Biotechnol. 2011, 29, 644–652. [Google Scholar] [CrossRef] [PubMed]
- Wu, T.D.; Watanabe, C.K. GMAP: A Genomic Mapping and Alignment Program for mRNA and EST Sequences. Bioinformatics 2005, 21, 1859–1875. [Google Scholar] [CrossRef] [PubMed]
- Sievers, F.; Wilm, A.; Dineen, D.; Gibson, T.J.; Karplus, K.; Li, W.; Lopez, R.; McWilliam, H.; Remmert, M.; Söding, J.; et al. Fast, Scalable Generation of High-Quality Protein Multiple Sequence Alignments Using Clustal Omega. Mol. Syst. Biol. 2011, 7, 539. [Google Scholar] [CrossRef]
- Yu, L.P.C.; Kim, Y.S.; Tong, L. Mechanism for the Inhibition of the Carboxyltransferase Domain of Acetyl-Coenzyme A Carboxylase by Pinoxaden. Proc. Natl. Acad. Sci. USA 2010, 107, 22072–22077. [Google Scholar] [CrossRef]
- Schneider, N.; Lange, G.; Hindle, S.; Klein, R.; Rarey, M. A Consistent Description of HYdrogen Bond and DEhydration Energies in Protein-Ligand Complexes: Methods behind the HYDE Scoring Function. J. Comput. Aided Mol. Des. 2013, 27, 15–29. [Google Scholar] [CrossRef]
- Abbott, W.S. A Method of Computing the Effectiveness of an Insecticide. J. Econ. Entomol. 1925, 18, 265–267. [Google Scholar] [CrossRef]
- Kaundun, S.S. Resistance to Acetyl-CoA Carboxylase-Inhibiting Herbicides. Pest Manag. Sci. 2014, 70, 1405–1417. [Google Scholar] [CrossRef]
- Singh, K.S.; Cordeiro, E.M.G.; Troczka, B.J.; Pym, A.; Mackisack, J.; Mathers, T.C.; Duarte, A.; Legeai, F.; Robin, S.; Bielza, P.; et al. Global Patterns in Genomic Diversity Underpinning the Evolution of Insecticide Resistance in the Aphid Crop Pest Myzus persicae. Commun. Biol. 2021, 4, 847. [Google Scholar] [CrossRef] [PubMed]
- Umina, P.A.; Bass, C.; van Rooyen, A.; Chirgwin, E.; Arthur, A.L.; Pym, A.; Mackisack, J.; Mathews, A.; Kirkland, L. Spirotetramat Resistance in Myzus persicae (Sulzer) (Hemiptera: Aphididae) and Its Association with the Presence of the A2666V Mutation. Pest Manag. Sci. 2022, 78, 4822–4831. [Google Scholar] [CrossRef]
- Pan, Y.; Zhu, E.; Gao, X.; Nauen, R.; Xi, J.; Peng, T.; Wei, X.; Zheng, C.; Shang, Q. Novel Mutations and Expression Changes of Acetyl-Coenzyme A Carboxylase Are Associated with Spirotetramat Resistance in Aphis gossypii Glover: Overexpression of Mutated ACC Account for Spirotetramat Resistance. Insect Mol. Biol. 2017, 26, 383–391. [Google Scholar] [CrossRef]
- Bajda, S.A.; De Clercq, P.; Van Leeuwen, T. Selectivity and Molecular Stress Responses to Classical and Botanical Acaricides in the Predatory Mite Phytoseiulus Persimilis Athias-Henriot (Acari: Phytoseiidae). Pest Manag. Sci. 2022, 78, 881–895. [Google Scholar] [CrossRef] [PubMed]
- Buitenhuis, R.; Murphy, G.; Shipp, L.; Scott-Dupree, C. Amblyseius swirskii in Greenhouse Production Systems: A Floricultural Perspective. Exp. Appl. Acarol. 2015, 65, 451–464. [Google Scholar] [CrossRef] [PubMed]
- Helps, J.C.; Paveley, N.D.; van den Bosch, F. Identifying Circumstances under Which High Insecticide Dose Increases or Decreases Resistance Selection. J. Theor. Biol. 2017, 428, 153–167. [Google Scholar] [CrossRef] [PubMed]
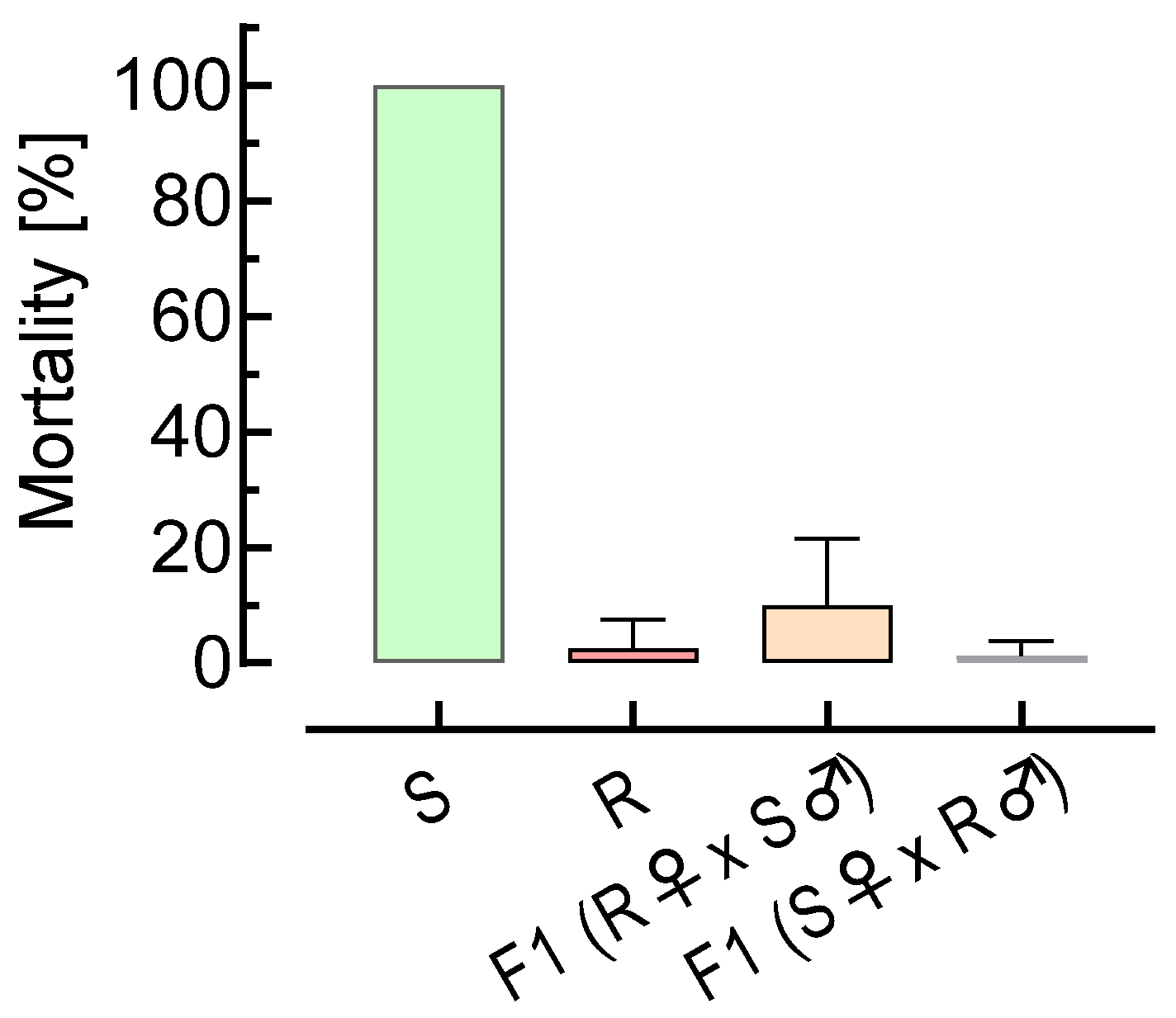
| STRAIN | YEAR | COUNTRY | VENUE | HOST PLANT |
|---|---|---|---|---|
| 1/19 | 2019 | France | Richebourg | Cauliflower |
| 3/19 | 2019 | Croatia | Varazdin | Green cabbage |
| 4/19 | 2019 | France | Warrem | White cabbage |
| 5/19 | 2019 | Belgium | Borgworm | White cabbage |
| SPI-5/19 | Selected 5/19 | |||
| 6/19 | 2019 | Belgium | Lier | White cabbage |
| 1/20 | 2020 | Germany | Helse | White cabbage |
| 2/20 | 2020 | Germany | Blomberg | Green cabbage |
| SPI-2/20 | Selected 2/20 | |||
| 4/20 | 2020 | Germany | Bardowick | White cabbage |
| 5/20 | 2020 | Germany | Bardowick | White cabbage |
| 6/20 | 2020 | Germany | Hannover | Green cabbage |
| SPI-6/20 | Selected 6/20 |
| Insecticide | Strain | LC50 [mg/L] | 95% CI a | Slope ± SE | RR |
|---|---|---|---|---|---|
| Spiromesifen | 1/19 | 3.96 | 3.39–4.63 | 3.42 ± 0.33 | 1 |
| 3/19 * | 2.76 | 1.71–4.49 | 1.87 ± 0.15 | 1 | |
| 4/19 | 4.33 | 1.74–11.6 | 2.29 ± 0.19 | 2 | |
| 5/19 | 154 | 45.4–1235 | 0.9 ± 0.07 | 56 | |
| SPI-5/19 | >200 | - | - | >72 | |
| 6/19 | 5.7 | 3.42–9.33 | 2.1 ± 0.17 | 2 | |
| 1/20 | 13.8 | 8.88–21.3 | 2.24 ± 0.2 | 5 | |
| 2/20 | >200 | - | - | >72 | |
| SPI-2/20 | >200 | - | - | >72 | |
| 4/20 | 9.85 | 5.23–20.8 | 2.82 ± 0.26 | 4 | |
| 5/20 | 4.13 | 2.34–7.42 | 1.78 ± 0.14 | 1 | |
| 6/20 | >200 | - | - | >72 | |
| SPI-6/20 | >200 | - | - | >72 | |
| Spirotetramat | 1/19 | 11.3 | 6.36–20.9 | 2.42 ± 0.21 | 3 |
| 3/19 | 6.95 | 5.92–8.17 | 3.18 ± 0.34 | 2 | |
| 4/19 | 6.15 | 4.53–8.41 | 2.66 ± 0.25 | 2 | |
| 5/19 | 57.8 | 37.2–91 | 1.57 ± 0.12 | 16 | |
| SPI-5/19 | >200 | - | - | >56 | |
| 6/19 | 4.24 | 1.98–9.43 | 2.48 ± 0.22 | 1 | |
| 1/20 * | 3.58 | 3.08–4.16 | 3.67 ± 0.34 | 1 | |
| 2/20 | >200 | - | - | >56 | |
| SPI-2/20 | >200 | - | - | >56 | |
| 4/20 | 3.7 | 1.69–7.79 | 3.26 ± 0.3 | 1 | |
| 5/20 | 3.63 | 1.7–8.17 | 2.55 ± 0.22 | 1 | |
| 6/20 | 123 | 103–146 | 2.76 ± 0.26 | 34 | |
| SPI-6/20 | >200 | - | - | >56 |
| Insecticide | Strain | LC50 [mg L−1] | 95% CI a | Slope ± SE | RR |
|---|---|---|---|---|---|
| Acetamiprid | 3/19 | 40.5 | 7.81–255 | 1.21 ± 0.08 | 1 |
| 4/19 * | 37.8 | 8.03–244 | 1.1 ± 0.07 | 1 | |
| 5/19 | 75.1 | 61.3–92.2 | 1.93 ± 0.15 | 2 | |
| SPI-5/19 | 42.1 | 35.8–49.5 | 3.09 ± 0.3 | 1 | |
| 6/19 | 99.6 | 47.8–220 | 1.72 ± 0.13 | 3 | |
| 6/20 | 60.7 | 31.6–123 | 1.86 ± 0.14 | 2 | |
| SPI-6/20 | 61.7 | 29.9–135 | 1.90 ± 0.15 | 2 | |
| λ-cyhalothrin | 3/19 | 52.7 | 12.6–299 | 1.29 ± 0.08 | 2 |
| 4/19 * | 27.9 | 3.59–387 | 1.02 ± 0.06 | 1 | |
| 5/19 | 137 | 95.1–195 | 2.1 ± 0.18 | 5 | |
| SPI-5/19 | 223 | 100–460 | 2.24 ± 0.18 | 8 | |
| 6/19 | 174 | 142–212 | 1.98 ± 0.16 | 6 | |
| 6/20 | 94.8 | 40.1–245 | 1.2 ± 0.08 | 3 | |
| SPI-6/20 | 74.7 | 52.2–108 | 1.73 ± 0.13 | 3 |
| ACC Genotype (%) | |||
|---|---|---|---|
| Strain | A/A | A/V | V/V |
| 1/19 | 100 | 0 | 0 |
| 3/19 | 100 | 0 | 0 |
| 4/19 | 100 | 0 | 0 |
| 5/19 | 70 | 20 | 10 |
| SPI-5/19 | 0 | 20 | 80 |
| 6/19 | 100 | 0 | 0 |
| 1/20 | 100 | 0 | 0 |
| 2/20 | 10 | 60 | 30 |
| SPI-2/20 | 0 | 10 | 90 |
| 4/20 | 100 | 0 | 0 |
| 5/20 | 100 | 0 | 0 |
| 6/20 | 30 | 0 | 70 |
| SPI-6/20 | 0 | 0 | 100 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Müller, V.; Maiwald, F.; Lange, G.; Nauen, R. Mapping and Characterization of Target-Site Resistance to Cyclic Ketoenol Insecticides in Cabbage Whiteflies, Aleyrodes proletella (Hemiptera: Aleyrodidae). Insects 2024, 15, 178. https://doi.org/10.3390/insects15030178
Müller V, Maiwald F, Lange G, Nauen R. Mapping and Characterization of Target-Site Resistance to Cyclic Ketoenol Insecticides in Cabbage Whiteflies, Aleyrodes proletella (Hemiptera: Aleyrodidae). Insects. 2024; 15(3):178. https://doi.org/10.3390/insects15030178
Chicago/Turabian StyleMüller, Viola, Frank Maiwald, Gudrun Lange, and Ralf Nauen. 2024. "Mapping and Characterization of Target-Site Resistance to Cyclic Ketoenol Insecticides in Cabbage Whiteflies, Aleyrodes proletella (Hemiptera: Aleyrodidae)" Insects 15, no. 3: 178. https://doi.org/10.3390/insects15030178
APA StyleMüller, V., Maiwald, F., Lange, G., & Nauen, R. (2024). Mapping and Characterization of Target-Site Resistance to Cyclic Ketoenol Insecticides in Cabbage Whiteflies, Aleyrodes proletella (Hemiptera: Aleyrodidae). Insects, 15(3), 178. https://doi.org/10.3390/insects15030178