A Novel Polymerase Chain Reaction (PCR)-Based Method for the Rapid Identification of Chrysodeixis includens and Rachiplusia nu
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Insect Populations
2.2. Genomic DNA Extraction, Sequence Amplification, and Sequencing
2.3. Species-Specific Primer Development
3. Results
3.1. mtDNA COI Partial Gene Sequencing
3.2. Species-Specific Primer
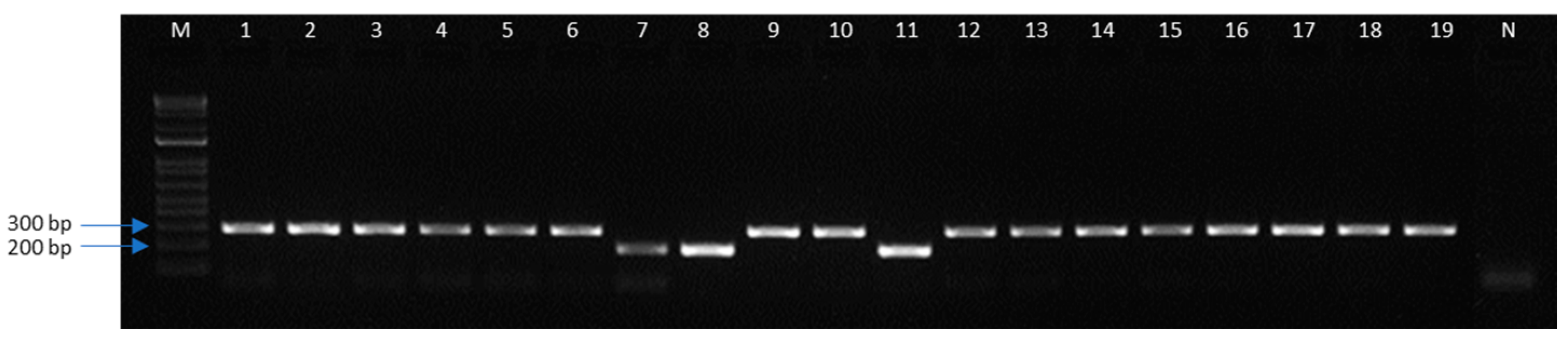
3.3. Field Collection Validation
4. Discussion
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Amer, A.M.; El Torkey, A.M. Revision of Subfamilies ‘Acronictinae, Heliothinae, Metoponiinae, Noctuinae, Oncocnemidinae and Plusiinae’ of Egypt (Lepidoptera, Noctuidae). Egypt. Acad. J. Biolog. Sci. A. Entomol. 2019, 12, 29–67. [Google Scholar] [CrossRef]
- Eichlin, T.D.; Cunningham, H.B. The Plusiinae (Lepidoptera: Noctuidae) of America North of Mexico, Emphasizing Genitalic and Larval Morphology. U.S. Dep. Agric. Tech. Bull. 1978, 1567, 1–128. [Google Scholar]
- Pastrana, J.A. Los Lepidópteros Argentinos: Sus Plantas Hospedadoras y Otros Sustratos Alimenticios, 1st ed.; Sociedad Entomologica Argentina: Buenos Aires, Argentina, 2004; pp. 1–334. [Google Scholar]
- Alford, R.A.; Hammond, A.M., Jr. Plusiinae (Lepidoptera: Noctuidae) populations in Louisiana soybean ecosystems as determined with looplure-baited traps. J. Econ. Entomol. 1982, 75, 647–650. [Google Scholar] [CrossRef]
- Baldin, E.L.L.; Lourenção, A.L.; Schlick-Souza, E.C. Outbreaks of Chrysodeixis includens (Walker) (Lepidoptera: Noctuidae) in common bean and castor bean in São Paulo State. Bragantia 2014, 73, 458–461. [Google Scholar] [CrossRef]
- Specht, A.; Paula-Moraes, S.V.; Sosa-Gómez, D.R. Host plants of Chrysodeixis includens (Walker) (Lepidoptera, Noctuidae, Plusiinae). Rev. Bras. Entomol. 2015, 59, 343–345. [Google Scholar] [CrossRef]
- Hoffmann-Campo, C.B.; Moscardi, F.; Corso, I.C. Artrópodes que atacam as folhas da soja. In Soja: Manejo Integrado de Insetos e Outros Artrópodes-Praga, 1st ed.; Panizzi, A.R., Corrêa-Ferreira, B.S., Eds.; Embrapa: Brasília, Brazil, 2012; pp. 153–179. [Google Scholar]
- Moonga, M.N.; Davis, J.A. Partial life history of Chrysodeixis includens (Lepidoptera: Noctuidae) on summer hosts. J. Econ. Entomol. 2016, 109, 1713–1719. [Google Scholar] [CrossRef] [PubMed]
- Palma, J.; Maebe, K.; Guedes, J.V.C.; Smagghe, G. Molecular variability and genetic structure of Chrysodeixis includens (Lepidoptera: Noctuidae), an important soybean defoliator in Brazil. PLoS ONE 2015, 10, e0121260. [Google Scholar] [CrossRef]
- Horikoshi, R.J.; Dourado, P.M.; Berger, G.U.; de S. Fernandes, D.; Omoto, C.; Willse, A.; Martinelli, S.; Head, G.P.; Corrêa, A.S. Large-scale assessment of lepidopteran soybean pests and efficacy of Cry1Ac soybean in Brazil. Sci. Rep. 2021, 11, 15956. [Google Scholar] [CrossRef]
- Silva, C.S.; Cordeiro, E.M.; de Paiva, J.B.; Dourado, P.M.; Carvalho, R.A.; Head, G.; Martinelli, S.; Correa, A.S. Population expansion and genomic adaptation to agricultural environments of the soybean looper, Chrysodeixis includens. Evol. Appl. 2020, 13, 2071–2085. [Google Scholar] [CrossRef]
- Pelizza, S.A.; Schalamuk, S.; Simón, M.R.; Stenglein, S.A.; Pacheco-Marino, S.G.; Scorsetti, A.C. Compatibility of chemical insecticides and entomopathogenic fungi for control of soybean defoliating pest, Rachiplusia nu. Rev. Argent Microbiol. 2018, 50, 189–201. [Google Scholar] [CrossRef]
- Specht, A.; Sosa-Gómez, D.R.; Roque-Specht, V.F.; Valduga, E.; Gonzatti, F.; Schuh, S.M.; Carneiro, E. Biotic Potential and Life Tables of Chrysodeixis includens (Lepidoptera: Noctuidae), Rachiplusia nu, and Trichoplusia ni on Soybean and Forage Turnip. J. Insect Sci. 2019, 19, 8. [Google Scholar] [CrossRef] [PubMed]
- Santos, S.R.; Specht, A.; Carneiro, E.; Casagrande, M.M. The influence of agricultural occupation and climate on the spatial distribution of Plusiinae (Lepidoptera: Noctuidae) on a latitudinal gradient in Brazil. Rev. Bras. Entomol. 2021, 65, e20200103. [Google Scholar] [CrossRef]
- Guedes, J.V.C.; Perini, C.R.; Stacke, R.F.; Curioletti, L.E.; Arnemann, J.A.; Alende, V.P. Lagartas da soja: Das lições do passado ao manejo do futuro. Plantio Direto 2014, 178, 20–24. [Google Scholar]
- Braga, L.E.; Warpechowski, L.F.; Diniz, L.H.M.; Dallanora, A.; Reis, A.C.; Farias, J.R.; Bernardi, O. Characterizing the differential susceptibility and resistance to insecticides in populations of Chrysodeixis includens and Rachiplusia nu (Lepidoptera: Noctuidae) in Brazil. Pest Manag. Sci. 2024, 80, 4853–4862. [Google Scholar] [CrossRef]
- Specht, A.; Vogt, T.G.; Corseuil, E. Biological aspects of Autoplusia egena (Guenée) (Lepidoptera: Noctuidae: Plusiinae). Neotrop. Entomol. 2007, 36, 1–4. [Google Scholar] [CrossRef]
- Rolim, A.A.S.G.; Sosa-Gómez, D.R.; Yano, S.A.C.; Specht, A.; Andrade, C.G.T.J. Morphological and Molecular Characterization of the Eggs of Some Noctuid Species Associated with Soybean in Brazil. Ann. Entomol. Soc. Am. 2013, 106, 643–651. [Google Scholar] [CrossRef]
- Frézal, L.; Leblois, R. Four years of DNA barcoding: Current advances and prospects. Infect. Genet. Evol. 2008, 8, 727–736. [Google Scholar] [CrossRef]
- Portillo, P.D.; Thomas, C.M.; Martinez, E.; Marañón, C.; Valladares, B.; Patarroyo, E.M.; López, C.M. Multiprimer PCR system for differential identification of mycobacteria in clinical samples. J. Clin. Microbiol. 1996, 34, 324–328. [Google Scholar] [CrossRef]
- Roehrdanz, R.L. Multiplex polymerase chain reaction method for differentiating western and northern corn rootworm larvae (Coleoptera: Chrysomelidae). J. Econ. Entom. 2003, 96, 669–672. [Google Scholar] [CrossRef]
- Hashiyama, A. Clarification of Factors Contributing to the Low Effectiveness of the Mating Disruption for Asiatic Common Looper, Autographa nigrisigna, and Application of Molecular Techniques to Identify Plusiine Species. Ph.D. Thesis, Chiba University, Chiba, Japan, 2013. [Google Scholar]
- Kaur, R.; Singh, D. Molecular markers a valuable tool for species identification of insects: A review. Ann. Entomol. 2020, 38, 1–20. [Google Scholar]
- Kekkonen, M.; Hebert, P.D.N. DNA barcode-based delineation of putative species: Efficient start for taxonomic workflows. Mol. Ecol. Resour. 2014, 14, 706–715. [Google Scholar] [CrossRef] [PubMed]
- Perera, O.P.; Allen, K.C.; Jain, D.; Purcell, M.; Little, N.S.; Luttrell, R.G. Rapid Identification of Helicoverpa armigera and Helicoverpa zea (Lepidoptera: Noctuidae) Using Ribosomal RNA Internal Transcribed Spacer 1. J. Insect Sci. 2015, 15, 155. [Google Scholar] [CrossRef] [PubMed]
- Kar, O.; Mukherjee, A.; Ghosh, D.; Mukherjee, K.; Pramanik, D.; Naskar, A.; Banerjee, D. DNA Barcoding of Economically Important Fruit Flies (Diptera: Tephritidae) from the Lower Gangetic Plains of Eastern India. J. Adv. Biol. Biotechnol. 2024, 27, 587–604. [Google Scholar] [CrossRef]
- Herzog, D.C. Sampling Soybean Looper on Soybean. In Sampling Methods in Soybean Entomology, 1st ed.; Kogan, M., Herzog, D.C., Eds.; Springer: Berlin/Heidelberg, Germany, 1980; pp. 141–168. [Google Scholar] [CrossRef]
- Folmer, O.; Black, M.; Hoeh, W.; Lutz, R.; Vrijenhoek, R. DNA primers for amplification of mitochondrial cytochrome c oxidase subunit I from diverse metazoan invertebrates. Mol. Mar. Biol. Biotechnol. 1994, 3, 294–299. [Google Scholar] [PubMed]
- Sosa-Gómez, D.R.; Côrrea-Ferreira, B.S.; Hoffmann-Campo, C.B.; Corso, I.C.; Oliveira, L.J.; Moscardi, F.; Panizzi, A.R.; Bueno, A.F.; Hirose, E.; Roggia, S. Manual de identificação de insetos e outros invertebrados da cultura da soja. Embrapa Soja 2014, 3, 1–100. Available online: https://www.embrapa.br/busca-de-publicacoes/-/publicacao/991685/manual-de-identificacao-de-insetos-e-outros-invertebrados-da-cultura-da-soja (accessed on 3 September 2024).
- Tembrock, L.R.; Roxanne, E.F.; Ledezma, L.; Barr, N.B.; Gilligan, T.M. A Real-Time PCR Assay for the Separation of Autographa gamma (Noctuidae: Plusiinae) From Morphologically Similar Species in North America. J. Econ. Entom. 2017, 110, 2609–2617. [Google Scholar] [CrossRef]
- Smith, M.A.; Rodriguez, J.J.; Whitfield, J.B.; Deans, A.R.; Janzen, D.H.; Hallwachs, W.; Hebert, P.D.N. Extreme diversity of tropical parasitoid wasps exposed by iterative integration of natural history, DNA barcoding, morphology, and collections. Proc. Natl. Acad. Sci. USA 2008, 105, 12359–12364. [Google Scholar] [CrossRef]
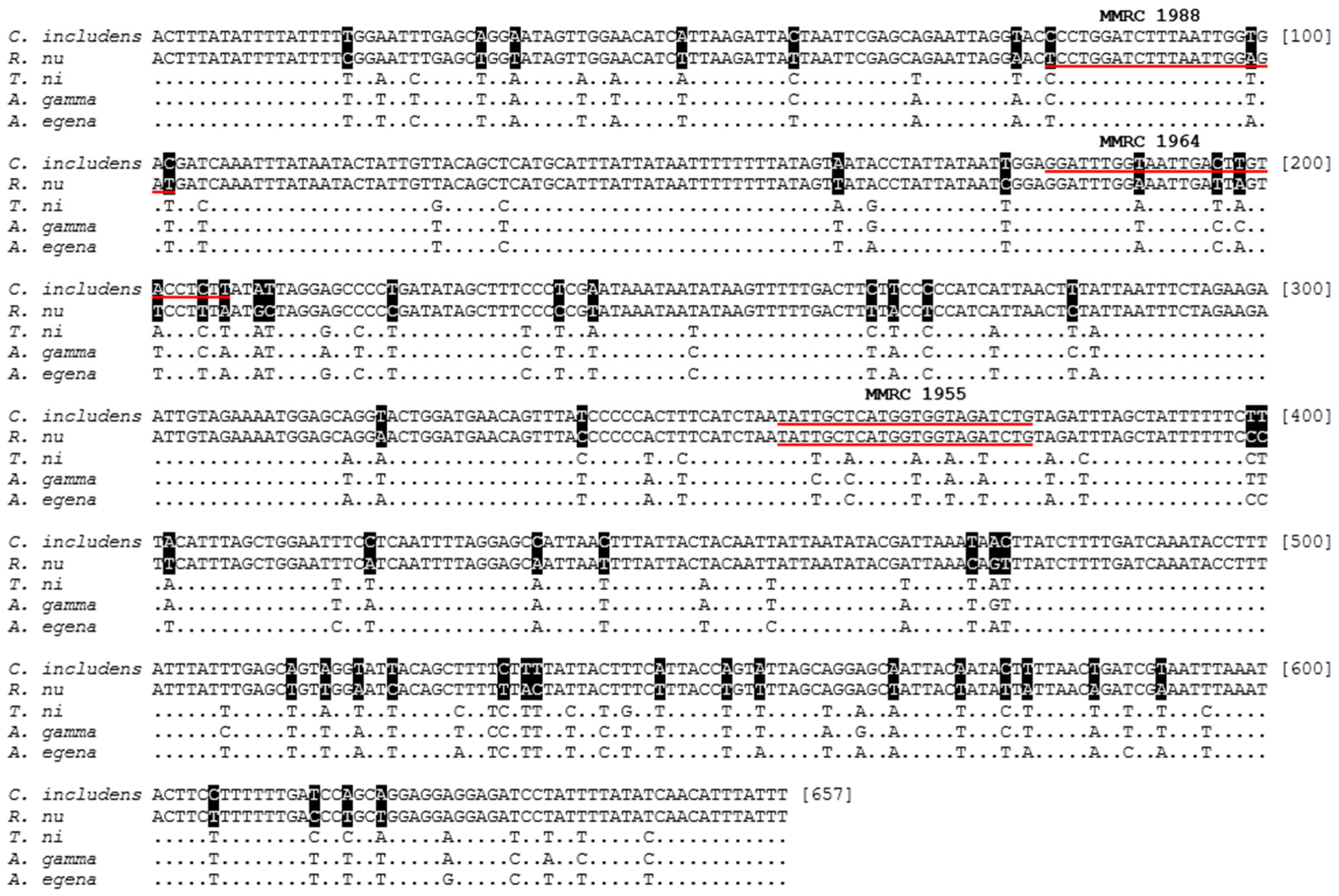


| Primer Name | Primer Sequence (5′ > 3′) | Purpose |
|---|---|---|
| LCO 1490 | GGTCAACAAATCATAAAGATATTGG | mtDNA COI amplification and sequencing |
| HCO 2198 | TAAACTTCAGGGTGACCAAAAAATCA | mtDNA COI amplification and sequencing |
| MMRC_1955 | CAGATCTACCACCATGAGCAATA | Species Identification—Common reverse |
| MMRC_1964 | GGATTTGGTAATTGACTTGTACCTCTT | Species Identification—C. includens-specific forward |
| MMRC_1988 | TCCTGGATCTTTAATTGGAGAT | Species Identification—R. nu-specific forward |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gotardi, G.A.; Batista, N.R.F.; Ishizuka, T.K.; Marques, L.H.; Dal Pogetto, M.H.; Sethi, A.; Dahmer, M.L.; Nowatzki, T. A Novel Polymerase Chain Reaction (PCR)-Based Method for the Rapid Identification of Chrysodeixis includens and Rachiplusia nu. Insects 2024, 15, 969. https://doi.org/10.3390/insects15120969
Gotardi GA, Batista NRF, Ishizuka TK, Marques LH, Dal Pogetto MH, Sethi A, Dahmer ML, Nowatzki T. A Novel Polymerase Chain Reaction (PCR)-Based Method for the Rapid Identification of Chrysodeixis includens and Rachiplusia nu. Insects. 2024; 15(12):969. https://doi.org/10.3390/insects15120969
Chicago/Turabian StyleGotardi, Guilherme A., Natália R. F. Batista, Tamylin Kaori Ishizuka, Luiz H. Marques, Mário H. Dal Pogetto, Amit Sethi, Mark L. Dahmer, and Timothy Nowatzki. 2024. "A Novel Polymerase Chain Reaction (PCR)-Based Method for the Rapid Identification of Chrysodeixis includens and Rachiplusia nu" Insects 15, no. 12: 969. https://doi.org/10.3390/insects15120969
APA StyleGotardi, G. A., Batista, N. R. F., Ishizuka, T. K., Marques, L. H., Dal Pogetto, M. H., Sethi, A., Dahmer, M. L., & Nowatzki, T. (2024). A Novel Polymerase Chain Reaction (PCR)-Based Method for the Rapid Identification of Chrysodeixis includens and Rachiplusia nu. Insects, 15(12), 969. https://doi.org/10.3390/insects15120969






