Transposable Elements Contribute to the Regulation of Long Noncoding RNAs in Drosophila melanogaster
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
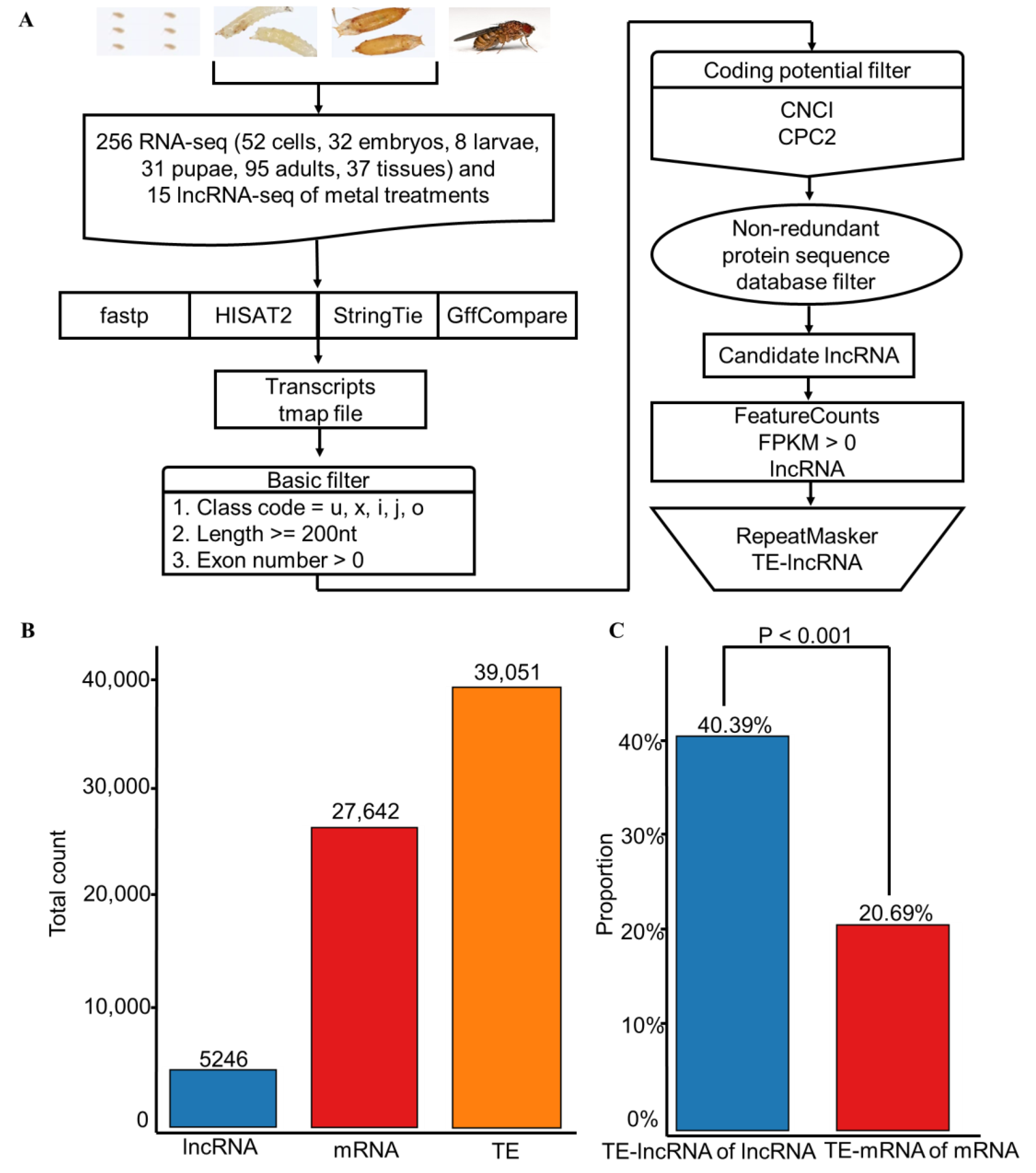
2.1. Data
2.2. LncRNA Identification Pipeline
2.3. TE Annotation
2.4. General Bioinformatical Analysis
2.5. ATAC-Seq Analysis
2.6. MeRIP-Seq Analysis
2.7. Ribo-Seq Analysis
2.8. Proteomic Analysis
3. Results
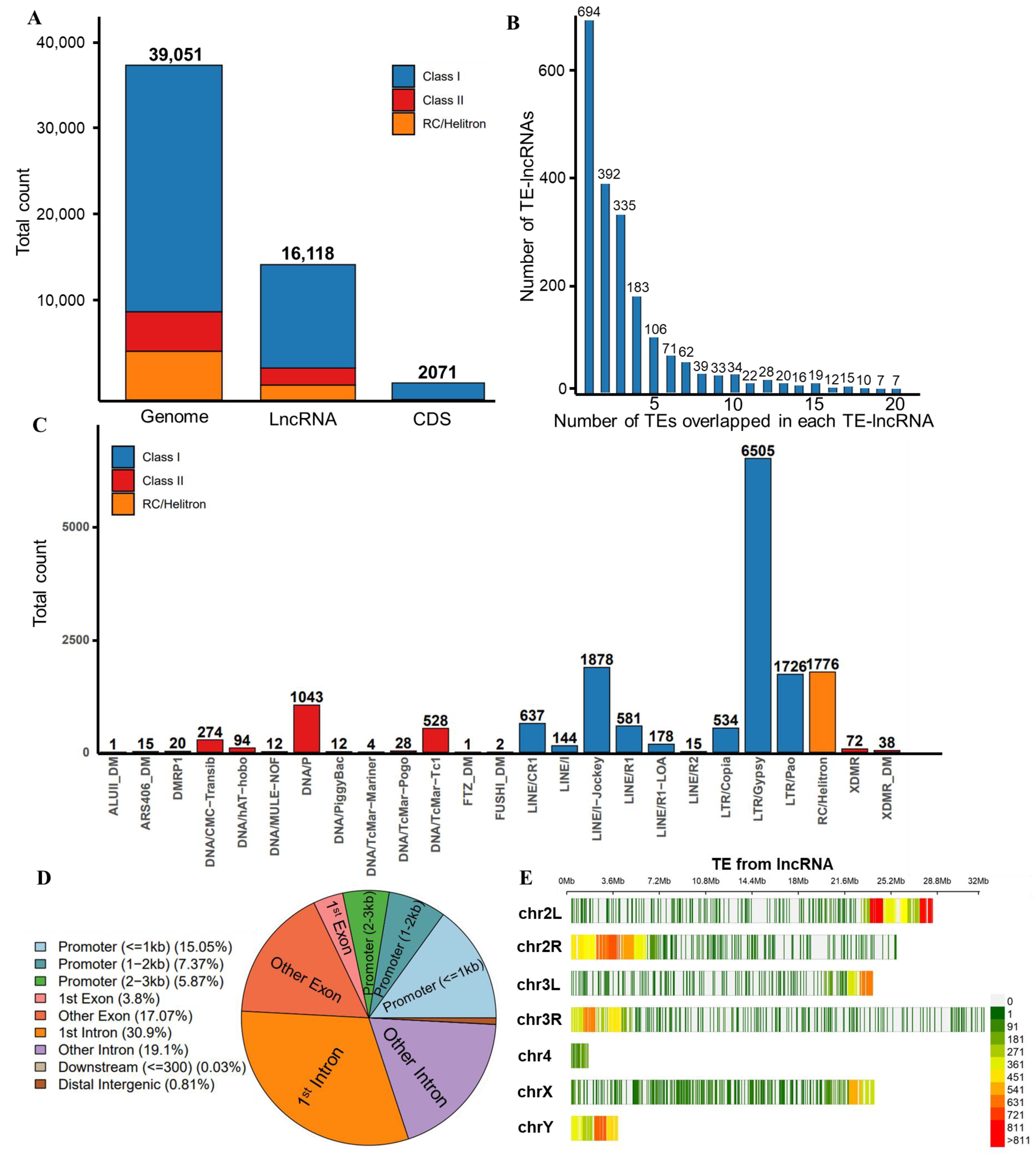
3.1. Identification of lncRNAs and TEs in D. melanogaster
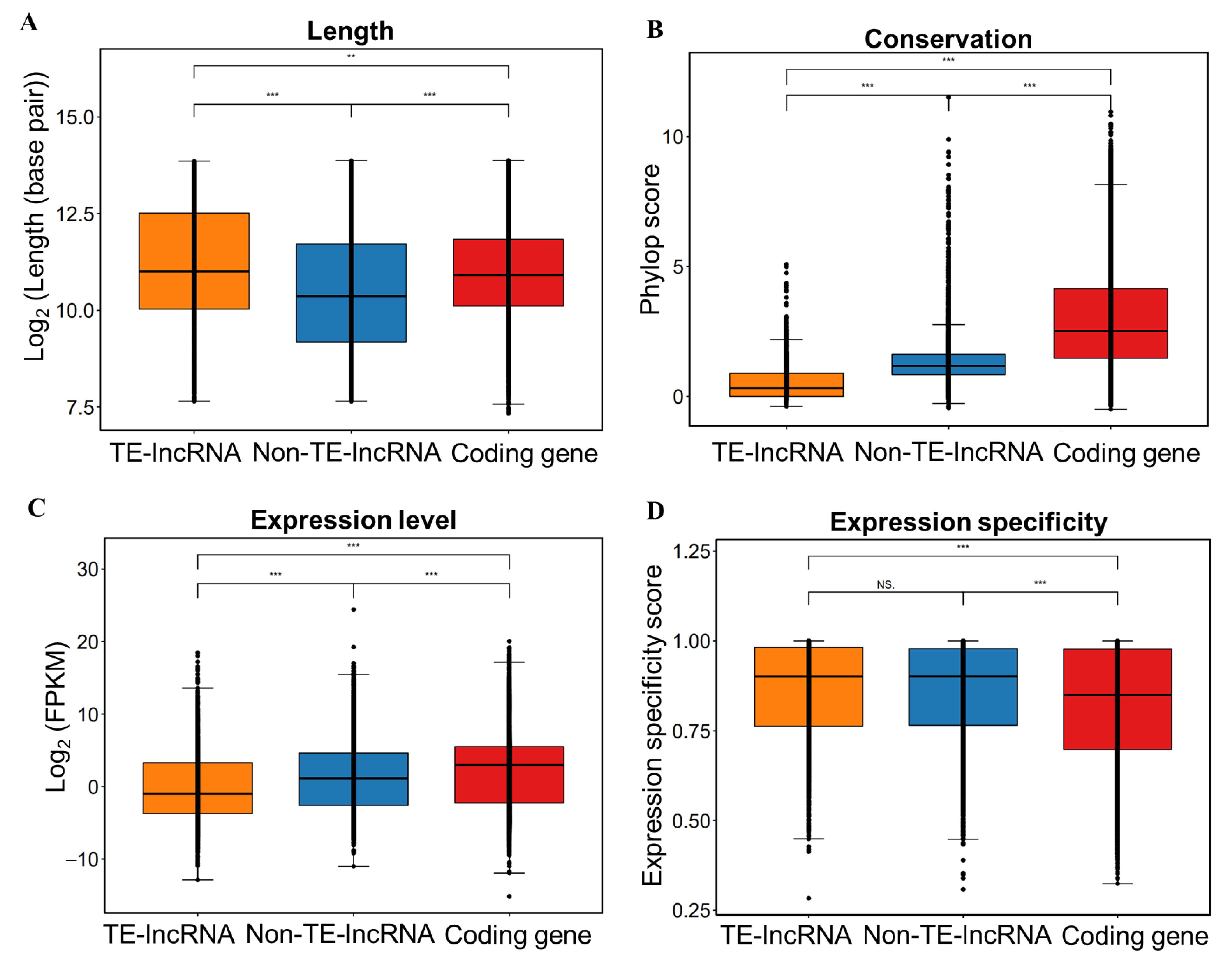
3.2. Characterization of TE-lncRNAs
3.3. Characteristics of TE Insertions in lncRNAs
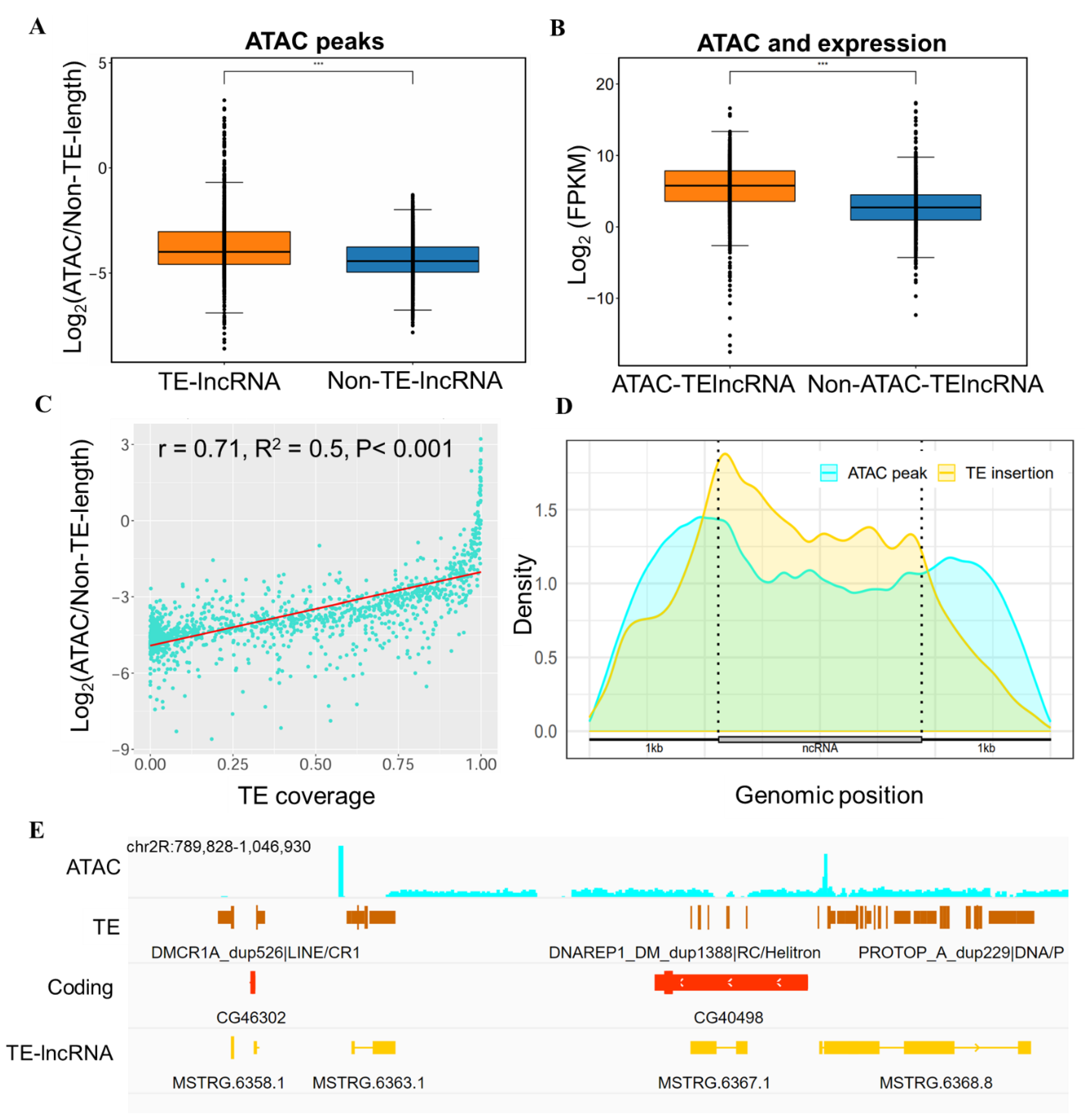
3.4. Epigenetic Regulation Involving TE-lncRNAs
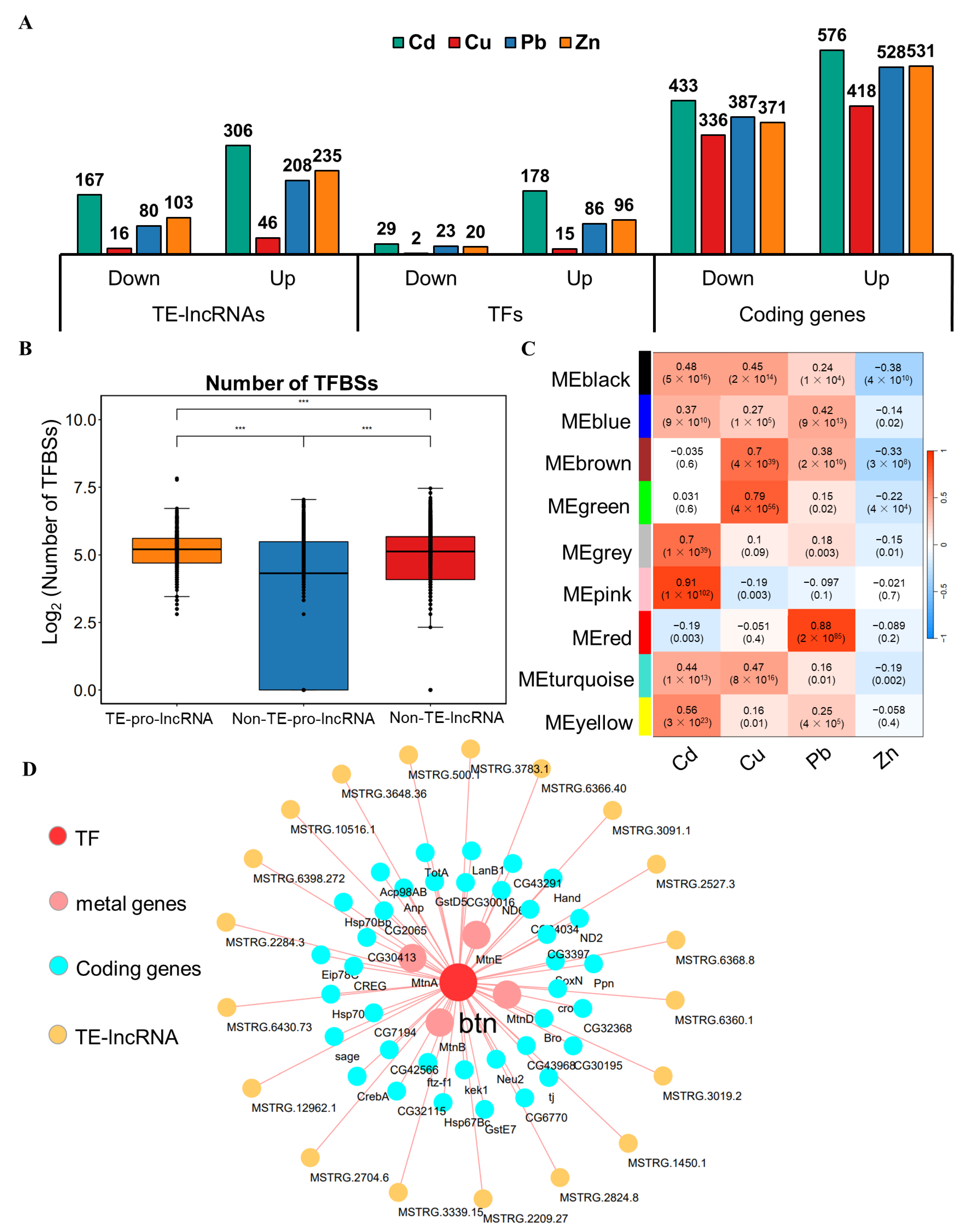
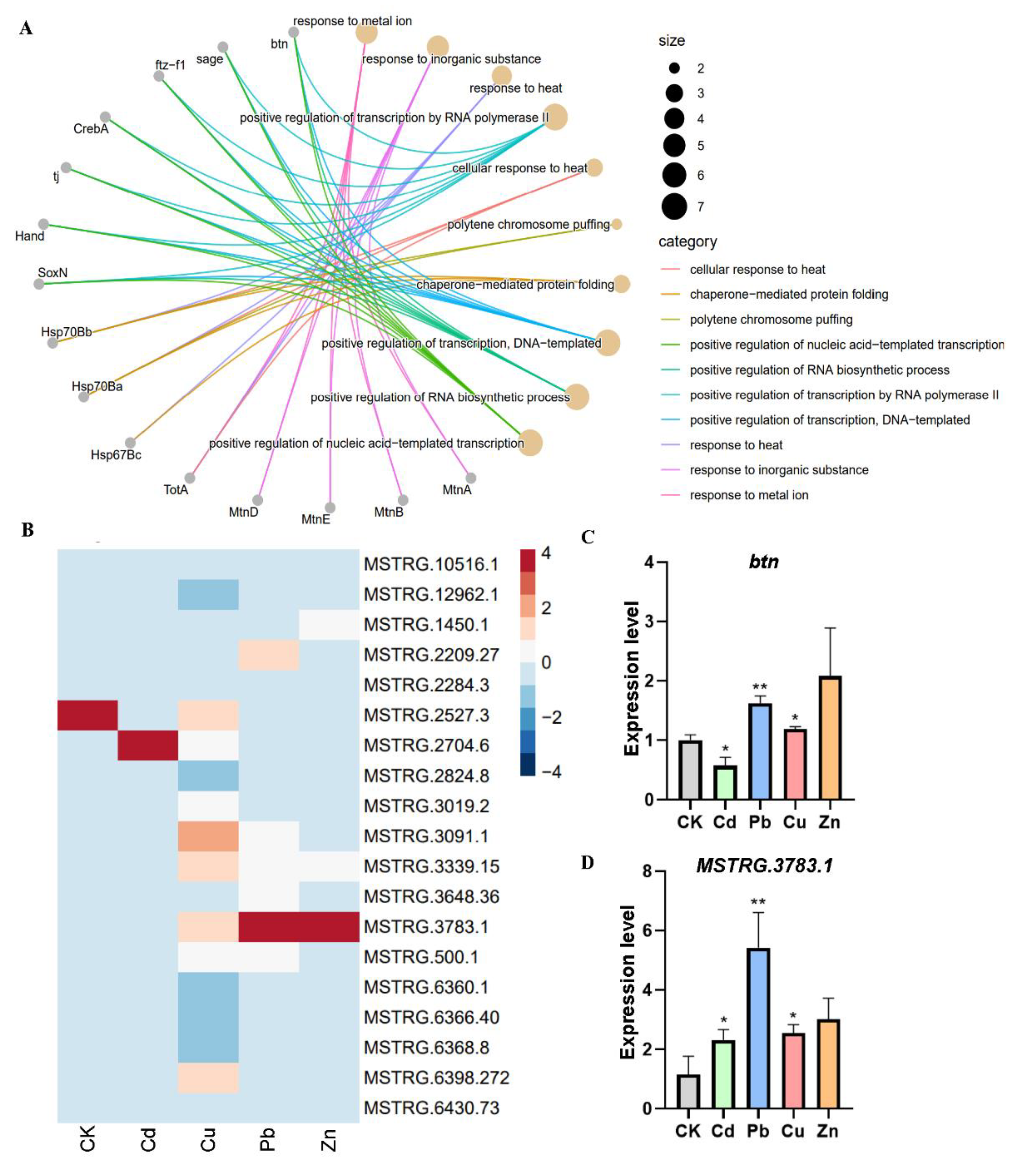
3.5. Transcriptional Regulation and Co-Expression Analysis for TE-lncRNAs
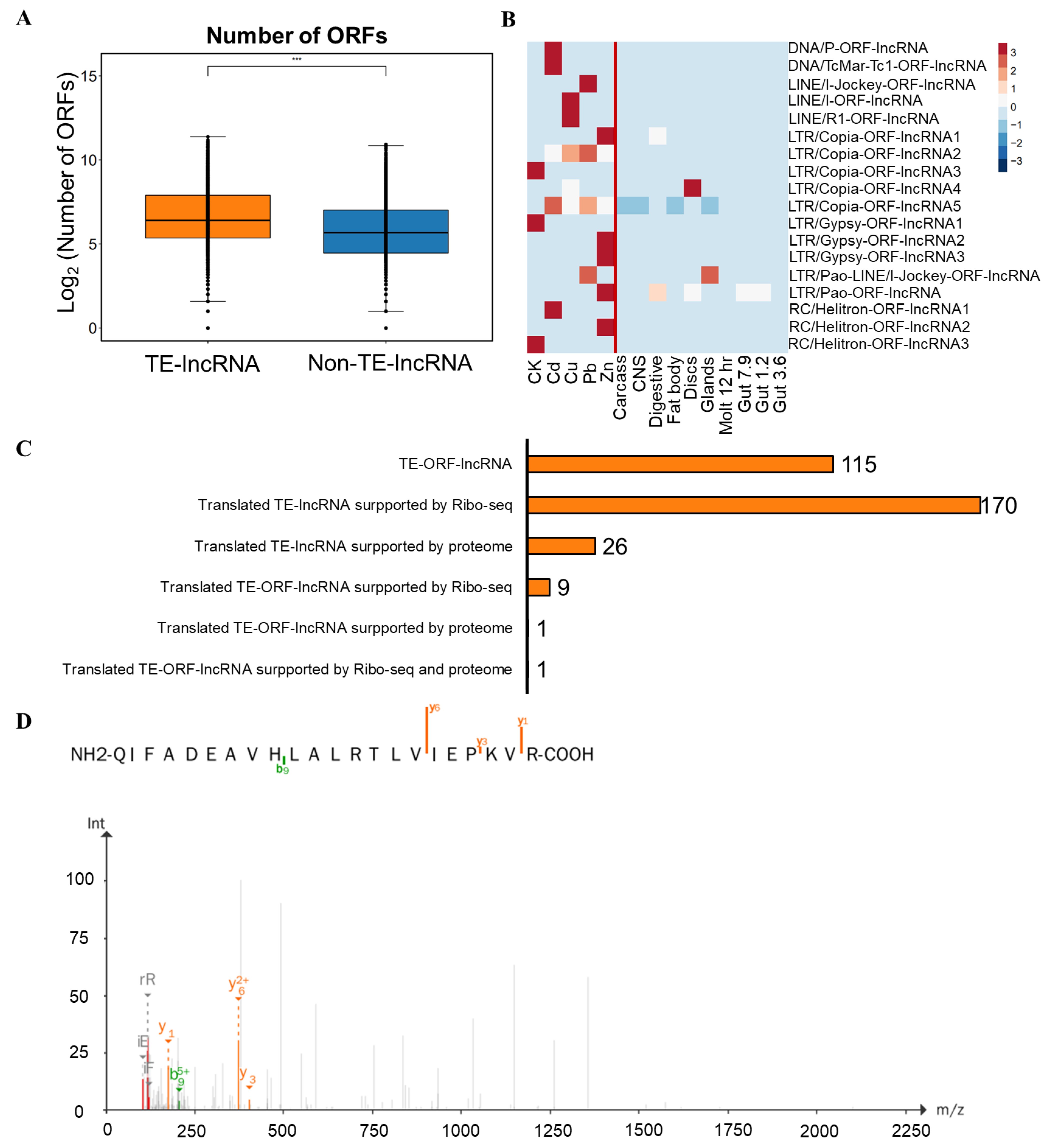
3.6. Post-Transcriptional Regulation of TE-lncRNAs
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Birney, E.; Stamatoyannopoulos, J.A.; Dutta, A.; Guigo, R.; Gingeras, T.R.; Margulies, E.H.; Weng, Z.; Snyder, M.; Dermitzakis, E.T.; Stamatoyannopoulos, J.A.; et al. Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature 2007, 447, 799–816. [Google Scholar] [CrossRef] [PubMed]
- Scheuermann, J.C.; Boyer, L.A. Getting to the heart of the matter: Long non-coding RNAs in cardiac development and disease. EMBO J. 2013, 32, 1805–1816. [Google Scholar] [CrossRef] [PubMed]
- Wilusz, J.E.; Sunwoo, H.; Spector, D.L. Long noncoding RNAs: Functional surprises from the RNA world. Genes Dev. 2009, 23, 1494–1504. [Google Scholar] [CrossRef] [PubMed]
- Zuo, L.; Su, H.; Zhang, Q.; Wu, W.-Y.; Zeng, Y.; Li, X.-M.; Xiong, J.; Chen, L.-F.; Zhou, Y. Comprehensive analysis of lncRNAs N6-methyladenosine modification in colorectal cancer. Aging 2021, 13, 4182–4198. [Google Scholar] [CrossRef]
- Liu, Y.; Shi, M.; He, X.; Cao, Y.; Liu, P.; Li, F.; Zou, S.; Wen, C.; Zhan, Q.; Xu, Z.; et al. LncRNA-PACERR induces pro-tumour macrophages via interacting with miR-671-3p and m6A-reader IGF2BP2 in pancreatic ductal adenocarcinoma. J. Hematol. Oncol. 2022, 15, 52. [Google Scholar] [CrossRef]
- Choi, S.-W.; Kim, H.-W.; Nam, J.-W. The small peptide world in long noncoding RNAs. Brief. Bioinform. 2019, 20, 1853–1864. [Google Scholar] [CrossRef]
- Kong, S.; Tao, M.; Shen, X.; Ju, S. Translatable circRNAs and lncRNAs: Driving mechanisms and functions of their translation products. Cancer Lett. 2020, 483, 59–65. [Google Scholar] [CrossRef]
- Matsumoto, A.; Pasut, A.; Matsumoto, M.; Yamashita, R.; Fung, J.; Monteleone, E.; Saghatelian, A.; Nakayama, K.I.; Clohessy, J.G.; Pandolfi, P.P. mTORC1 and muscle regeneration are regulated by the LINC00961-encoded SPAR polypeptide. Nature 2017, 541, 228–232. [Google Scholar] [CrossRef]
- Zhang, Q.; Vashisht, A.A.; O’Rourke, J.; Corbel, S.Y.; Moran, R.; Romero, A.; Miraglia, L.; Zhang, J.; Durrant, E.; Schmedt, C.; et al. The microprotein Minion controls cell fusion and muscle formation. Nat. Commun. 2017, 8, 15664. [Google Scholar] [CrossRef]
- Huang, J.-Z.; Chen, M.; Chen, D.; Gao, X.-C.; Zhu, S.; Huang, H.; Hu, M.; Zhu, H.; Yan, G.-R. A Peptide Encoded by a Putative lncRNA HOXB-AS3 Suppresses Colon Cancer Growth. Mol. Cell 2017, 68, 171–184. [Google Scholar] [CrossRef]
- D’Lima, N.G.; Ma, J.; Winkler, L.; Chu, Q.; Loh, K.H.; Corpuz, E.O.; Budnik, B.A.; Lykke-Andersen, J.; Saghatelian, A.; Slavoff, S.A. A human microprotein that interacts with the mRNA decapping complex. Nat. Chem. Biol. 2017, 13, 174–180. [Google Scholar] [CrossRef] [PubMed]
- Sun, L.; Wang, W.; Han, C.; Huang, W.; Sun, Y.; Fang, K.; Zeng, Z.; Yang, Q.; Pan, Q.; Chen, T.; et al. The oncomicropeptide APPLE promotes hematopoietic malignancy by enhancing translation initiation. Mol. Cell 2021, 81, 4493–4508. [Google Scholar] [CrossRef] [PubMed]
- Anderson, D.M.; Anderson, K.M.; Chang, C.-L.; Makarewich, C.A.; Nelson, B.R.; McAnally, J.R.; Kasaragod, P.; Shelton, J.M.; Liou, J.; Bassel-Duby, R.; et al. A Micropeptide Encoded by a Putative Long Noncoding RNA Regulates Muscle Performance. Cell 2015, 160, 595–606. [Google Scholar] [CrossRef] [PubMed]
- Nelson, B.R.; Makarewich, C.A.; Anderson, D.M.; Winders, B.R.; Troupes, C.D.; Wu, F.; Reese, A.L.; McAnally, J.R.; Chen, X.; Kavalali, E.T.; et al. A peptide encoded by a transcript annotated as long noncoding RNA enhances SERCA activity in muscle. Science 2016, 351, 271–275. [Google Scholar] [CrossRef] [PubMed]
- Pauli, A.; Norris, M.L.; Valen, E.; Chew, G.-L.; Gagnon, J.A.; Zimmerman, S.; Mitchell, A.; Ma, J.; Dubrulle, J.; Reyon, D.; et al. Toddler: An Embryonic Signal That Promotes Cell Movement via Apelin Receptors. Science 2014, 343, 1248636. [Google Scholar] [CrossRef]
- Kondo, T.; Hashimoto, Y.; Kato, K.; Inagaki, S.; Hayashi, S.; Kageyama, Y. Small peptide regulators of actin-based cell morphogenesis encoded by a polycistronic mRNA. Nat Cell Biol 2007, 9, 660–665. [Google Scholar] [CrossRef]
- Magny, E.G.; Pueyo, J.I.; Pearl, F.M.; Cespedes, M.A.; Niven, J.E.; Bishop, S.A.; Couso, J.P. Conserved regulation of cardiac calcium uptake by peptides encoded in small open reading frames. Science 2013, 341, 1116–1120. [Google Scholar] [CrossRef]
- Hanyu-Nakamura, K.; Sonobe-Nojima, H.; Tanigawa, A.; Lasko, P.; Nakamura, A. Drosophila Pgc protein inhibits P-TEFb recruitment to chromatin in primordial germ cells. Nature 2008, 451, 730–733. [Google Scholar] [CrossRef]
- Shao, Z.; Hu, J.; Jandura, A.; Wilk, R.; Jachimowicz, M.; Ma, L.; Hu, C.; Sundquist, A.; Das, I.; Samuel-Larbi, P.; et al. Spatially revealed roles for lncRNAs in Drosophila spermatogenesis, Y chromosome function and evolution. Nat. Commun. 2024, 15, 3806. [Google Scholar] [CrossRef]
- Lisch, D. How important are transposons for plant evolution? Nat. Rev. Genet. 2013, 14, 49–61. [Google Scholar] [CrossRef]
- Piégu, B.; Bire, S.; Arensburger, P.; Bigot, Y. A survey of transposable element classification systems—A call for a fundamental update to meet the challenge of their diversity and complexity. Mol. Phylogenetics Evol. 2015, 86, 90–109. [Google Scholar] [CrossRef] [PubMed]
- Arensburger, P.; Piégu, B.; Bigot, Y. The future of transposable element annotation and their classification in the light of functional genomics—What we can learn from the fables of Jean de la Fontaine? Mob. Genet. Elem. 2016, 6, e1256852. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Wicker, T.; Sabot, F.; Hua-Van, A.; Bennetzen, J.L.; Capy, P.; Chalhoub, B.; Flavell, A.; Leroy, P.; Morgante, M.; Panaud, O.; et al. A unified classification system for eukaryotic transposable elements. Nat. Rev. Genet. 2007, 8, 973–982. [Google Scholar] [CrossRef] [PubMed]
- Hua-Van, A.; Capy, P. Transposable Elements and Genome Evolution; John Wiley & Sons: Hoboken, NJ, USA, 2024. [Google Scholar]
- Walser, J.-C.; Chen, B.; Feder, M.E. Heat-Shock Promoters: Targets for Evolution by P Transposable Elements in Drosophila. PLoS Genet. 2006, 2, e165. [Google Scholar] [CrossRef] [PubMed]
- Chen, B.; Zhang, B.; Xu, L.; Li, Q.; Jiang, F.; Yang, P.; Xu, Y.; Kang, L. Transposable Element-Mediated Balancing Selection at Hsp90 Underlies Embryo Developmental Variation. Mol. Biol. Evol. 2017, 34, 1127–1139. [Google Scholar] [CrossRef]
- Chen, B.; Walser, J.C.; Rodgers, T.H.; Sobota, R.S.; Burke, M.K.; Rose, M.R.; Feder, M.E. Abundant, diverse, and consequentialPelements segregate in promoters of small heat-shock genes inDrosophilapopulations. J. Evol. Biol. 2007, 20, 2056–2066. [Google Scholar] [CrossRef]
- Chen, B.; Shilova, V.Y.; Zatsepina, O.G.; Evgen’ev, M.B.; Feder, M.E. Location of P element insertions in the proximal promoter region of Hsp70A is consequential for gene expression and correlated with fecundity in Drosophila melanogaster. Cell Stress Chaperones 2008, 13, 11–17. [Google Scholar] [CrossRef]
- Wang, K.; Hua, G.; Li, J.; Yang, Y.; Zhang, C.; Yang, L.; Hu, X.; Scheben, A.; Wu, Y.; Gong, P.; et al. Duck pan-genome reveals two transposon insertions caused bodyweight enlarging and white plumage phenotype formation during evolution. iMeta 2023, 3, e154. [Google Scholar] [CrossRef]
- Xia, B.; Zhang, W.; Zhao, G.; Zhang, X.; Bai, J.; Brosh, R.; Wudzinska, A.; Huang, E.; Ashe, H.; Ellis, G.; et al. On the genetic basis of tail-loss evolution in humans and apes. Nature 2024, 626, 1042–1048. [Google Scholar] [CrossRef]
- Zhang, X.; Zhu, Y.N.; Chen, B.; Kang, L. A Gypsy element contributes to the nuclear retention and transcriptional regulation of the resident lncRNA in locusts. Rna Biol. 2022, 19, 206–220. [Google Scholar] [CrossRef]
- Kelley, D.; Rinn, J. Transposable elements reveal a stem cell-specific class of long noncoding RNAs. Genome Biol. 2012, 13, R107. [Google Scholar] [CrossRef] [PubMed]
- Kapusta, A.; Kronenberg, Z.; Lynch, V.J.; Zhuo, X.Y.; Ramsay, L.; Bourque, G.; Yandell, M.; Feschotte, C. Transposable Elements Are Major Contributors to the Origin, Diversification, and Regulation of Vertebrate Long Noncoding RNAs. PloS Genet. 2013, 9, e1003470. [Google Scholar] [CrossRef] [PubMed]
- Ramsay, L.; Marchetto, M.C.; Caron, M.; Chen, S.H.; Busche, S.; Kwan, T.; Pastinen, T.; Gage, F.H.; Bourque, G. Conserved expression of transposon-derived non-coding transcripts in primate stem cells. BMC Genom. 2017, 18, 214. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Ai, G.; Zhang, C.; Cui, L.; Wang, J.; Li, H.; Zhang, J.; Ye, Z. Expression and diversification analysis reveals transposable elements play important roles in the origin of Lycopersicon-specific lncRNAs in tomato. New Phytol. 2016, 209, 1442–1455. [Google Scholar] [CrossRef]
- Wang, Z.X.; Schwacke, R.; Kunze, R. DNA Damage-Induced Transcription of Transposable Elements and Long Non-coding RNAs in Arabidopsis Is Rare and ATM-Dependent. Mol. Plant 2016, 9, 1142–1155. [Google Scholar] [CrossRef]
- Zhang, X.; Xu, Y.n.; Chen, B.; Kang, L. Long noncoding RNA PAHAL modulates locust behavioural plasticity through the feedback regulation of dopamine biosynthesis. PLoS Genet. 2020, 16, e1008771. [Google Scholar] [CrossRef]
- Melo, E.S.; Wallau, G.L. Mosquito long non-coding RNAs are enriched with Transposable Elements. Genet Mol Biol 2022, 45, e20210215. [Google Scholar] [CrossRef]
- Graveley, B.R.; Brooks, A.N.; Carlson, J.; Duff, M.O.; Landolin, J.M.; Yang, L.; Artieri, C.G.; van Baren, M.J.; Boley, N.; Booth, B.W.; et al. The developmental transcriptome of Drosophila melanogaster. Nature 2011, 471, 473–479. [Google Scholar] [CrossRef]
- Brown, J.B.; Boley, N.; Eisman, R.; May, G.E.; Stoiber, M.H.; Duff, M.O.; Booth, B.W.; Wen, J.; Park, S.; Suzuki, A.M.; et al. Diversity and dynamics of the Drosophila transcriptome. Nature 2014, 512, 393–399. [Google Scholar] [CrossRef]
- Guo, B.; Li, T.; Wang, L.; Liu, F.; Chen, B. Long non-coding RNAs regulate heavy metal-induced apoptosis in embryo-derived cells. Environ. Pollut. 2023, 333, 121956. [Google Scholar] [CrossRef]
- Chen, S.; Zhou, Y.; Chen, Y.; Gu, J. fastp: An ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 2018, 34, 884–890. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.; Paggi, J.M.; Park, C.; Bennett, C.; Salzberg, S.L. Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat. Biotechnol. 2019, 37, 907–915. [Google Scholar] [CrossRef] [PubMed]
- Pertea, M.; Pertea, G.M.; Antonescu, C.M.; Chang, T.-C.; Mendell, J.T.; Salzberg, S.L. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat. Biotechnol. 2015, 33, 290–295. [Google Scholar] [CrossRef] [PubMed]
- Pertea, G.; Pertea, M. GFF Utilities: GffRead and GffCompare. F1000Research 2020, 9, 304. [Google Scholar] [CrossRef]
- Kang, Y.-J.; Yang, D.-C.; Kong, L.; Hou, M.; Meng, Y.-Q.; Wei, L.; Gao, G. CPC2: A fast and accurate coding potential calculator based on sequence intrinsic features. Nucleic Acids Res. 2017, 45, W12–W16. [Google Scholar] [CrossRef]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Liao, Y.; Smyth, G.K.; Shi, W. featureCounts: An efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 2014, 30, 923–930. [Google Scholar] [CrossRef]
- Smit, A.H.R.; Green, P. RepeatMasker Open-4.0. Available online: http://www.repeatmasker.org (accessed on 9 April 2019).
- Tempel, S. Using and understanding RepeatMasker. Methods Mol. Biol. 2012, 859, 29–51. [Google Scholar] [CrossRef]
- Jurka, J.; Kapitonov, V.V.; Pavlicek, A.; Klonowski, P.; Kohany, O.; Walichiewicz, J. Repbase update, a database of eukaryotic repetitive elements. Cytogenet. Genome Res. 2005, 110, 462–467. [Google Scholar] [CrossRef]
- Bao, W.; Kojima, K.K.; Kohany, O. Repbase Update, a database of repetitive elements in eukaryotic genomes. Mob. DNA 2015, 6, 11. [Google Scholar] [CrossRef]
- Yu, G.C.; Wang, L.G.; He, Q.Y. ChIPseeker: An R/Bioconductor package for ChIP peak annotation, comparison and visualization. Bioinformatics 2015, 31, 2382–2383. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An Integrative Toolkit Developed for Interactive Analyses of Big Biological Data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef] [PubMed]
- Palmer, D.; Fabris, F.; Doherty, A.; Freitas, A.A.; de Magalhães, J.P. Ageing transcriptome meta-analysis reveals similarities and differences between key mammalian tissues. Aging 2021, 13, 3313–3341. [Google Scholar] [CrossRef] [PubMed]
- Langfelder, P.; Horvath, S. WGCNA: An R package for weighted correlation network analysis. Bmc Bioinform. 2008, 9, 559. [Google Scholar] [CrossRef]
- Yu, G.C.; Wang, L.G.; Han, Y.Y.; He, Q.Y. clusterProfiler: An R Package for Comparing Biological Themes Among Gene Clusters. Omics-A J. Integr. Biol. 2012, 16, 284–287. [Google Scholar] [CrossRef]
- Ramirez, F.; Duendar, F.; Diehl, S.; Gruening, B.A.; Manke, T. deepTools: A flexible platform for exploring deep-sequencing data. Nucleic Acids Res. 2014, 42, W187–W191. [Google Scholar] [CrossRef]
- Zhang, Y.; Liu, T.; Meyer, C.A.; Eeckhoute, J.; Johnson, D.S.; Bernstein, B.E.; Nussbaum, C.; Myers, R.M.; Brown, M.; Li, W.; et al. Model-based Analysis of ChIP-Seq (MACS). Genome Biol. 2008, 9, R137. [Google Scholar] [CrossRef]
- Quinlan, A.R. BEDTools: The Swiss-Army Tool for Genome Feature Analysis. Curr. Protoc. Bioinform. 2014, 47, 11.12.1–11.12.34. [Google Scholar] [CrossRef]
- Meng, J.; Cui, X.; Rao, M.K.; Chen, Y.; Huang, Y. Exome-based analysis for RNA epigenome sequencing data. Bioinformatics 2013, 29, 1565–1567. [Google Scholar] [CrossRef]
- Liu, Q.; Gregory, R.I. RNAmod: An integrated system for the annotation of mRNA modifications. Nucleic Acids Res. 2019, 47, W548–W555. [Google Scholar] [CrossRef]
- Langmead, B.; Salzberg, S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 2012, 9, 357–359. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Durbin, R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 2009, 25, 1754–1760. [Google Scholar] [CrossRef] [PubMed]
- Zhang, P.; He, D.; Xu, Y.; Hou, J.; Pan, B.-F.; Wang, Y.; Liu, T.; Davis, C.M.; Ehli, E.A.; Tan, L.; et al. Genome-wide identification and differential analysis of translational initiation. Nat. Commun. 2017, 8, 1749. [Google Scholar] [CrossRef] [PubMed]
- Lauria, F.; Tebaldi, T.; Bernabo, P.; Groen, E.J.N.; Gillingwater, T.H.; Viero, G. riboWaltz: Optimization of ribosome P-site positioning in ribosome profiling data. PLoS Comput. Biol. 2018, 14, e1006169. [Google Scholar] [CrossRef] [PubMed]
- Ji, Z. RibORF: Identifying Genome-Wide Translated Open Reading Frames Using Ribosome Profiling. Curr. Protoc. Mol. Biol. 2018, 124, e67. [Google Scholar] [CrossRef]
- Gregory, R.I.; Sliz, P.; Shvarts, T.; Liu, Q. RiboToolkit: An integrated platform for analysis and annotation of ribosome profiling data to decode mRNA translation at codon resolution. Nucleic Acids Res. 2020, 48, W218–W229. [Google Scholar] [CrossRef]
- Tyanova, S.; Temu, T.; Cox, J. The MaxQuant computational platform for mass spectrometry-based shotgun proteomics. Nat. Protoc. 2016, 11, 2301–2319. [Google Scholar] [CrossRef]
- Li, K.; Vaudel, M.; Zhang, B.; Ren, Y.; Wen, B. PDV: An integrative proteomics data viewer. Bioinformatics 2019, 35, 1249–1251. [Google Scholar] [CrossRef]
- Luo, H.; Bu, D.; Sun, L.; Chen, R.; Zhao, Y. De novo approach to classify protein-coding and noncoding transcripts based on sequence composition. Methods Mol. Biol. 2014, 1182, 203–207. [Google Scholar] [CrossRef]
- Anver, S.; Sumit, A.F.; Sun, X.M.; Hatimy, A.; Thalassinos, K.; Marguerat, S.; Alic, N.; Bahler, J. Ageing-associated long non-coding RNA extends lifespan and reduces translation in non-dividing cells. EMBO Rep 2024, 25, 4921–4949. [Google Scholar] [CrossRef]
- Huang, Q.; Li, J.; Qi, Y.; He, X.; Shen, C.; Wang, C.; Wang, X.; Xia, Q.; Zhang, Y.; Pan, Z.; et al. Copper overload exacerbates testicular aging mediated by lncRNA:CR43306 deficiency through ferroptosis in Drosophila. Redox Biol. 2024, 76, 103315. [Google Scholar] [CrossRef] [PubMed]
- Perez-Lluch, S.; Klein, C.C.; Breschi, A.; Ruiz-Romero, M.; Abad, A.; Palumbo, E.; Bekish, L.; Arnan, C.; Guigo, R. bsAS, an antisense long non-coding RNA, essential for correct wing development through regulation of blistered/DSRF isoform usage. PLoS Genet 2020, 16, e1009245. [Google Scholar] [CrossRef] [PubMed]
- Camilleri-Robles, C.; Amador, R.; Tiebe, M.; Teleman, A.A.; Serras, F.; Guigó, R.; Corominas, M. Long non-coding RNAs involved in Drosophila development and regeneration. NAR Genom. Bioinform. 2024, 6, lqae091. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Duan, Y.; Duan, G.; Wang, Q.; Zhang, K.; Deng, X.; Qian, B.; Gu, J.; Ma, Z.; Zhang, S.; et al. Stress Induces Dynamic, Cytotoxicity-Antagonizing TDP-43 Nuclear Bodies via Paraspeckle LncRNA NEAT1-Mediated Liquid-Liquid Phase Separation. Mol. Cell 2020, 79, 443–458.e7. [Google Scholar] [CrossRef]
- Dayal Aggarwal, D.; Mishra, P.; Yadav, G.; Mitra, S.; Patel, Y.; Singh, M.; Sahu, R.K.; Sharma, V. Decoding the connection between lncRNA and obesity: Perspective from humans and Drosophila. Heliyon 2024, 10, e35327. [Google Scholar] [CrossRef]
- Chen, J.; Huang, Y.; Qi, G. LncRNA-IRAR-mediated regulation of insulin receptor transcripts in Drosophila melanogaster during nutritional stress. Insect Mol. Biol. 2022, 31, 261–272. [Google Scholar] [CrossRef]
- Valanne, S.; Salminen, T.S.; Jarvela-Stolting, M.; Vesala, L.; Ramet, M. Immune-inducible non-coding RNA molecule lincRNA-IBIN connects immunity and metabolism in Drosophila melanogaster. PLoS Pathog 2019, 15, e1007504. [Google Scholar] [CrossRef]
- Zhang, L.; Xu, W.; Gao, X.; Li, W.; Qi, S.; Guo, D.; Ajayi, O.E.; Ding, S.W.; Wu, Q. lncRNA Sensing of a Viral Suppressor of RNAi Activates Non-canonical Innate Immune Signaling in Drosophila. Cell Host Microbe 2020, 27, 115–128.e8. [Google Scholar] [CrossRef]
- Kaur, R.; McGarry, A.; Shropshire, J.D.; Leigh, B.A.; Bordenstein, S.R. Prophage proteins alter long noncoding RNA and DNA of developing sperm to induce a paternal-effect lethality. Science 2024, 383, 1111–1117. [Google Scholar] [CrossRef]
- Becker, P.B.; Thomae, A.W.; Villa, R.; Krause, S.; Schauer, T.; Müller, M. Two-step mechanism for selective incorporation of lncRNA into a chromatin modifier. Nucleic Acids Res. 2020, 48, 7483–7501. [Google Scholar] [CrossRef]
- Patraquim, P.; Magny, E.G.; Pueyo, J.I.; Platero, A.I.; Couso, J.P. Translation and natural selection of micropeptides from long non-canonical RNAs. Nat. Commun. 2022, 13, 6515. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.; Qu, Z.P.; Yang, L.; Zhang, Q.Z.; Liu, Z.H.; Do, T.; Adelson, D.L.; Wang, Z.Y.; Searle, I.; Zhu, J.K. Transposable elements (TEs) contribute to stress-related long intergenic noncoding RNAs in plants. Plant J. 2017, 90, 133–146. [Google Scholar] [CrossRef] [PubMed]
- Zhao, T.; Tao, X.; Feng, S.; Wang, L.; Hong, H.; Ma, W.; Shang, G.; Guo, S.; He, Y.; Zhou, B.; et al. LncRNAs in polyploid cotton interspecific hybrids are derived from transposon neofunctionalization. Genome Biol 2018, 19, 195. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Chen, Y.; Wang, Y.; Zhao, J.; Wang, Y. Gypsy retrotransposon-derived maize lncRNA GARR2 modulates gibberellin response. Plant J. 2022, 110, 1433–1446. [Google Scholar] [CrossRef]
- Chen, X.; Wang, C.; He, B.; Wan, Z.; Zhao, Y.; Hu, F.; Lv, Y. Transcriptome Profiling of Transposon-Derived Long Non-coding RNAs Response to Hormone in Strawberry Fruit Development. Front. Plant Sci. 2022, 13, 915569. [Google Scholar] [CrossRef]
- Zhu, Y.; Chen, L.; Hong, X.; Shi, H.; Li, X. Revealing the novel complexity of plant long non-coding RNA by strand-specific and whole transcriptome sequencing for evolutionarily representative plant species. BMC Genom. 2022, 23, 381. [Google Scholar] [CrossRef]
- Cerqueira de Araujo, A.; Huguet, E.; Herniou, E.A.; Drezen, J.M.; Josse, T. Transposable element repression using piRNAs, and its relevance to endogenous viral elements (EVEs) and immunity in insects. Curr. Opin. Insect Sci. 2022, 50, 100876. [Google Scholar] [CrossRef]
- Kwapisz, M.; Morillon, A. Subtelomeric Transcription and its Regulation. J. Mol. Biol. 2020, 432, 4199–4219. [Google Scholar] [CrossRef]
- Günther, V.; Lindert, U.; Schaffner, W. The taste of heavy metals: Gene regulation by MTF-1. Biochim. Et Biophys. Acta (BBA)—Mol. Cell Res. 2012, 1823, 1416–1425. [Google Scholar] [CrossRef]
- Navarro, J.A.; Schneuwly, S. Copper and Zinc Homeostasis: Lessons from Drosophila melanogaster. Front. Genet. 2017, 8, 223. [Google Scholar] [CrossRef]
- Slobodian, M.R.; Petahtegoose, J.D.; Wallis, A.L.; Levesque, D.C.; Merritt, T.J.S. The Effects of Essential and Non-Essential Metal Toxicity in the Drosophila melanogaster Insect Model: A Review. Toxics 2021, 9, 269. [Google Scholar] [CrossRef] [PubMed]
- Kokotovic, T.; Lenartowicz, E.M.; Langeslag, M.; Ciotu, C.I.; Fell, C.W.; Scaramuzza, A.; Fischer, M.J.M.; Kress, M.; Penninger, J.M.; Nagy, V. Transcription factor mesenchyme homeobox protein 2 (MEOX2) modulates nociceptor function. FEBS J. 2022, 289, 3457–3476. [Google Scholar] [CrossRef] [PubMed]
- Johnson, R.; Guigo, R. The RIDL hypothesis: Transposable elements as functional domains of long noncoding RNAs. RNA 2014, 20, 959–976. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gan, Y.; Wang, L.; Liu, G.; Guo, X.; Zhou, Y.; Chang, K.; Zhang, Z.; Yan, F.; Liu, Q.; Chen, B. Transposable Elements Contribute to the Regulation of Long Noncoding RNAs in Drosophila melanogaster. Insects 2024, 15, 950. https://doi.org/10.3390/insects15120950
Gan Y, Wang L, Liu G, Guo X, Zhou Y, Chang K, Zhang Z, Yan F, Liu Q, Chen B. Transposable Elements Contribute to the Regulation of Long Noncoding RNAs in Drosophila melanogaster. Insects. 2024; 15(12):950. https://doi.org/10.3390/insects15120950
Chicago/Turabian StyleGan, Yuli, Lingyan Wang, Guoxian Liu, Xiruo Guo, Yiming Zhou, Kexin Chang, Zhonghui Zhang, Fang Yan, Qi Liu, and Bing Chen. 2024. "Transposable Elements Contribute to the Regulation of Long Noncoding RNAs in Drosophila melanogaster" Insects 15, no. 12: 950. https://doi.org/10.3390/insects15120950
APA StyleGan, Y., Wang, L., Liu, G., Guo, X., Zhou, Y., Chang, K., Zhang, Z., Yan, F., Liu, Q., & Chen, B. (2024). Transposable Elements Contribute to the Regulation of Long Noncoding RNAs in Drosophila melanogaster. Insects, 15(12), 950. https://doi.org/10.3390/insects15120950






