The Selection and Validation of Reference Genes for RT-qPCR Analysis of the Predatory Natural Enemy Orius nagaii (Hemiptera: Anthocoridae)
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Preparation of Insects
2.2. Sample Handling and Retrieval
2.3. Production of cDNA Template
2.4. Primer Design and Verification
2.5. RT-qPCR
2.6. Stability Assessment
2.7. Validation of Optimal Reference Genes in Various Tissues
3. Results
3.1. Dependability of Primers for CRGs
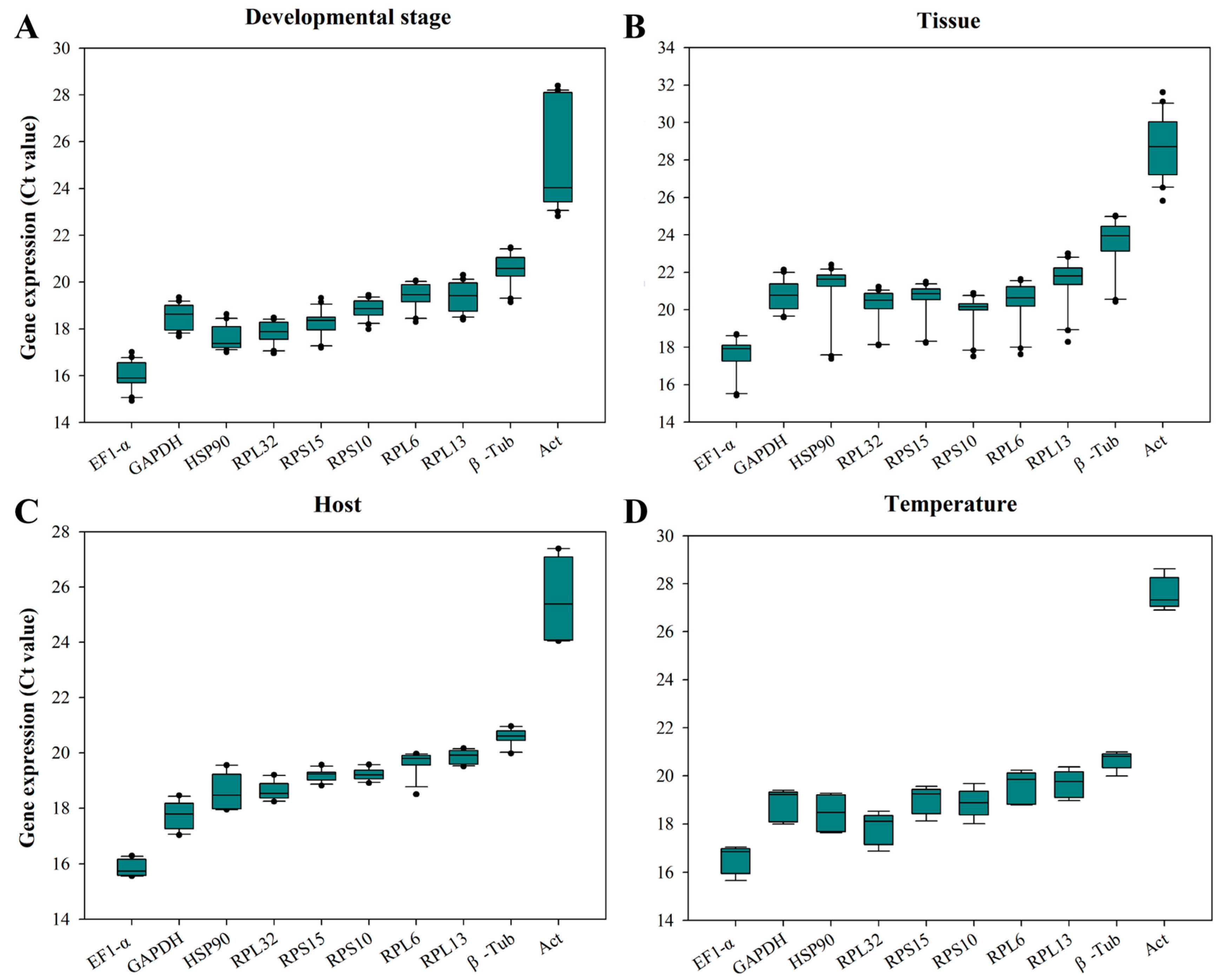
3.2. Expression Profile of CRGs
3.3. Assessment of the Expression Stability of the CRGs
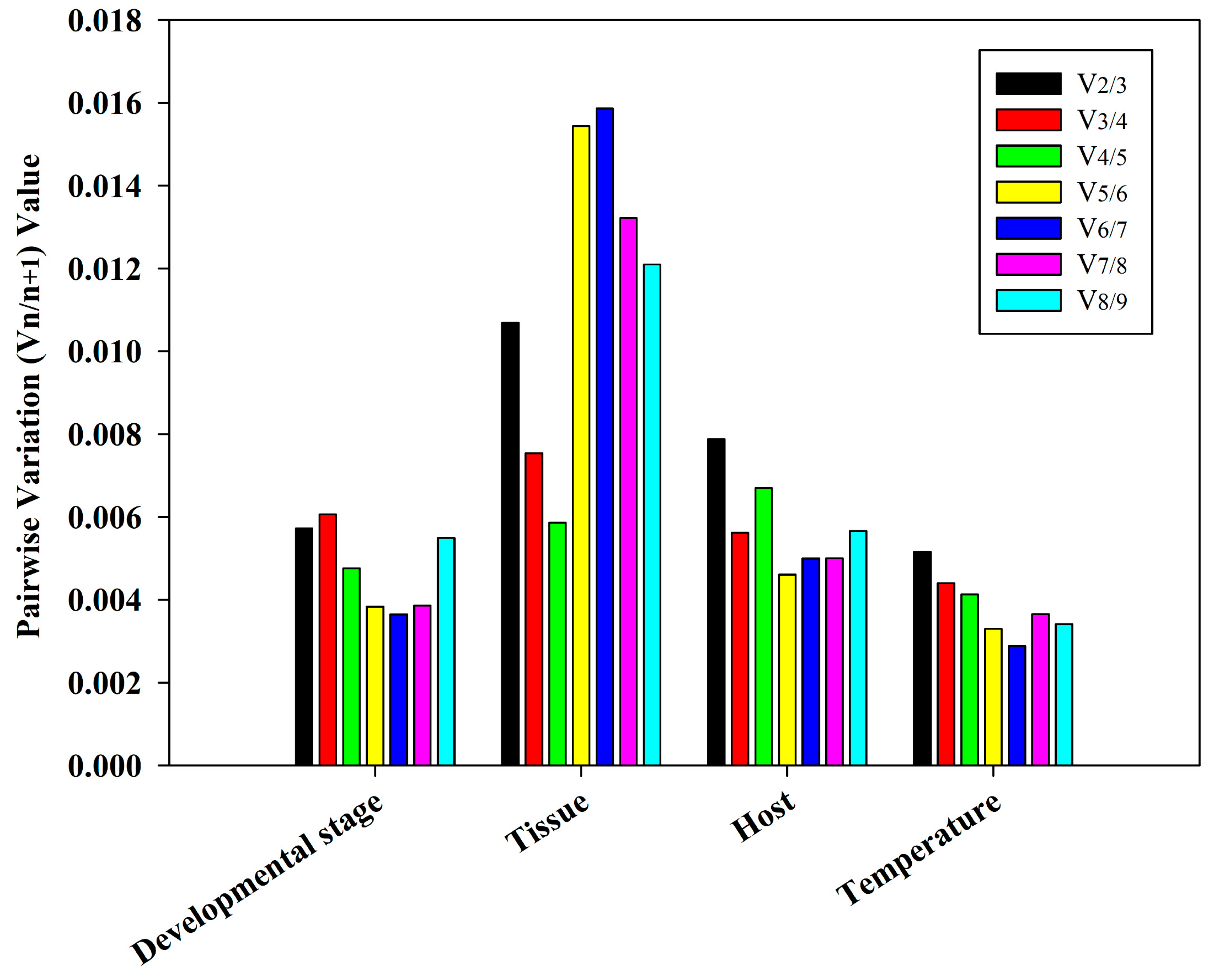
3.4. Ideal Number of Reference Genes Across Various Experimental Conditions
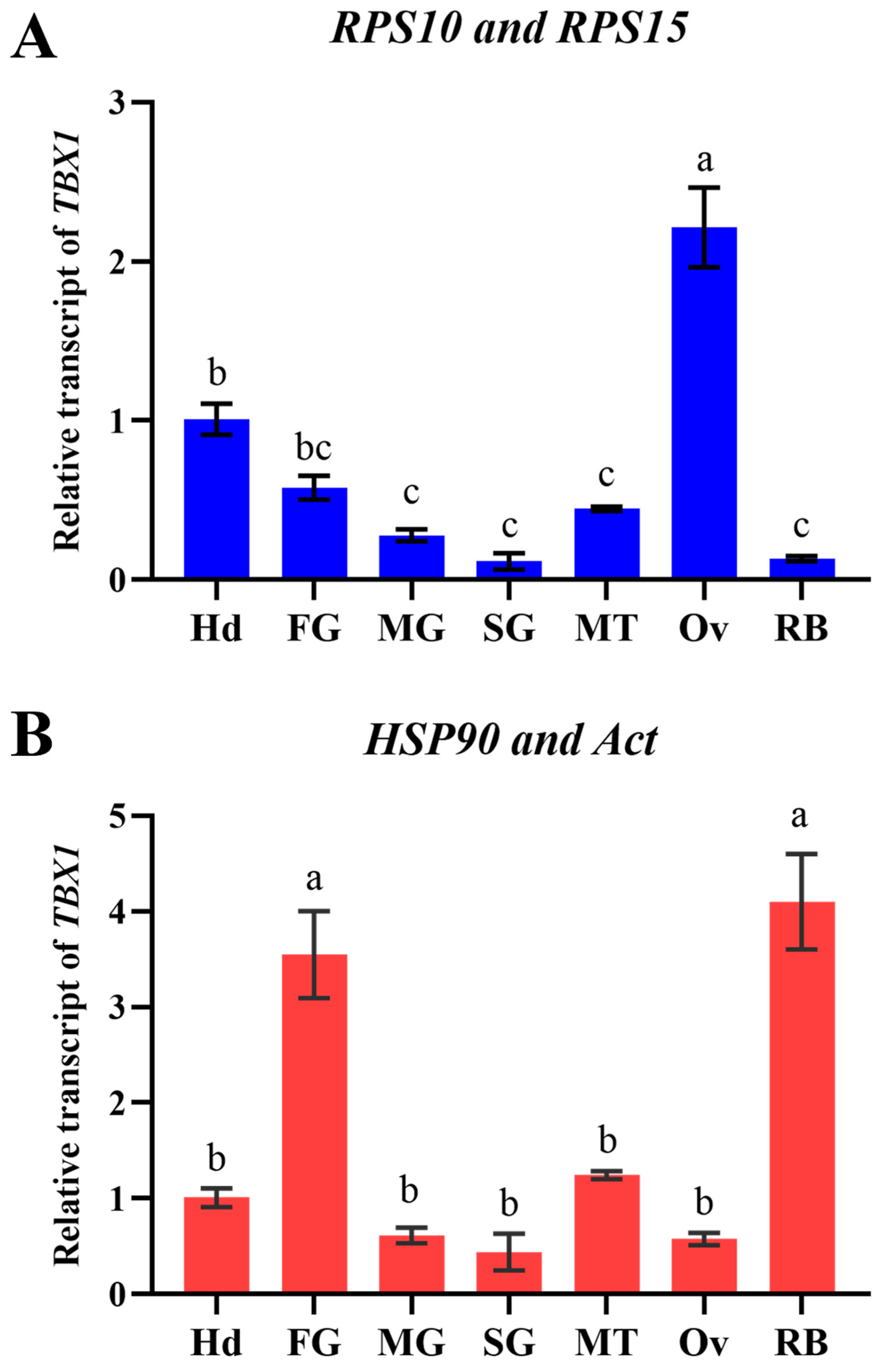
3.5. Confirmation of Optimal Reference Genes
4. Discussion
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Hu, C.; Li, Y.; Chen, G.; Duan, P.; Wu, D.; Liu, Q.; Yin, H.; Xu, T.; Zhang, X. Population dynamics of Frankliniella occidentalis Pergrande and its predator Orius similis Zheng on common crops and surrounding plants. J. Asia-Pac. Entomol. 2021, 24, 555–563. [Google Scholar] [CrossRef]
- Dai, X.; Wang, R.; Liu, Y.; Su, L.; Yin, Z.; Wu, M.; Chen, H.; Zheng, L.; Zhai, Y. Control effect and field application of four predatory Orius species on Megalurothrips usitatus (Thysanoptera: Thripidae). J. Econ. Entomol. 2024, 117, 448–456. [Google Scholar] [CrossRef] [PubMed]
- Fathi, S.A.A.; Nouri-Ganbalani, G. Assessing the potential for biological control of potato field pests in Ardabil, Iran: Functional responses of Orius niger (Wolf.) and O. minutus (L.) (Hemiptera: Anthocoridae). J. Pest Sci. 2010, 83, 47–52. [Google Scholar] [CrossRef]
- Ge, Y.; Camara, I.; Wang, Y.; Liu, P.; Zhang, L.; Xing, Y.; Li, A.; Shi, W. Predation of Aphis craccivora (Hemiptera: Aphididae) by Orius sauteri (Hemiptera: Anthocoridae) under different temperatures. J. Econ. Entomol. 2018, 111, 2599–2604. [Google Scholar] [CrossRef]
- Sharifi, M.; Malkeshi, S.H.; Madahi, K.; Mobasheri, M.T.; Malek Shahkoei, S.; Ghaderi, K.; Rajaei, A.; Khamar, E. Evaluation of predator and prey preference of Orius niger (Wolff) (Hemiptera: Anthocoridae) in the control of important sucking pests of oilseeds. J. Plant Protect. Res. 2021, 8, 107–118. [Google Scholar]
- Silva, L.P.; Souza, I.L.; Marucci, R.C.; Guzman-Martinez, M. Doru luteipes (Dermaptera: Forficulidae) and Orius insidiosus (Hemiptera: Anthocoridae) as nocturnal and diurnal predators of thrips. Neotrop. Entomol. 2023, 52, 263–272. [Google Scholar] [CrossRef]
- Lins Jr, J.; Bueno, V.; Silva, D.; van Lenteren, J.; Calixto, A.; Sidney, L. Tuta absoluta egg predation by Orius insidiosus. IOBC/WPRS Bull. 2011, 68, 101–104. [Google Scholar]
- Salehi, Z.; Yarahmadi, F.; Rasekh, A.; Sohani, N. Functional responses of Orius albidipennis Reuter (Hemiptera, Anthocoridae) to Tuta absoluta Meyrick (Lepidoptera, Gelechiidae) on two tomato cultivars with different leaf morphological characteristics. Entomol. Gen. 2016, 36, 127–136. [Google Scholar] [CrossRef]
- Desneux, N.; Wajnberg, E.; Wyckhuys, K.A.G.; Burgio, G.; Arpaia, S.; Narváez-Vasquez, C.A.; González-Cabrera, J.; Catalán Ruescas, D.; Tabone, E.; Frandon, J.; et al. Biological invasion of European tomato crops by Tuta absoluta: Ecology, geographic expansion and prospects for biological control. J. Pest Sci. 2010, 83, 197–215. [Google Scholar] [CrossRef]
- Yue-Li, J.; Yu-Qing, W.U.; Yun, D.; Xin-Guo, G. Control efficiencies of releasing Orius sauteri (Heteroptera: Anthocoridae) on some pests in greenhouse pepper. Chin. J. Biol. Control 2011, 27, 414–417. [Google Scholar]
- Jian, Y.; Xinguo, G.; Yuqing, W.U.; Yuli, J.; Shuntong, L.; Aiju, D.; Ziqi, Z.; Changying, L. Thrips control on the greenhouse eggplant by releasing Orius sauteri (Heteroptera: Anthocoridae). Chin. J. Biol. Control 2013, 29, 459–462. [Google Scholar]
- Gomez, R.; Sendín, L. Relative expression analysis of target genes by using reverse transcription-quantitative PCR. Methods Mol. Biol. 2020, 2072, 51–63. [Google Scholar]
- Bustin, S.A.; Benes, V.; Nolan, T.; Pfaffl, M.W. Quantitative real-time RT-PCR—A perspective. J. Mol. Endocrinol. 2005, 34, 597–601. [Google Scholar] [CrossRef]
- Suzuki, T.; Higgins, P.J.; Crawford, D.R. Control selection for RNA quantitation. Biotechniques 2000, 29, 332–337. [Google Scholar] [CrossRef] [PubMed]
- Yuan, M.; Lu, Y.; Zhu, X.; Wan, H.; Shakeel, M.; Zhan, S.; Jin, B.; Li, J. Selection and evaluation of potential reference genes for gene expression analysis in the brown planthopper, Nilaparvata lugens (Hemiptera: Delphacidae) using reverse-transcription quantitative PCR. PLoS ONE 2014, 9, e86503. [Google Scholar] [CrossRef]
- Dong, R.; Cao, F.; Yu, J.; Yuan, Y.; Wang, J.; Li, Z.; Zhu, C.; Li, S.; Li, N. Selection and validation of reference genes for quantitative real-time PCR analysis in cockroach parasitoid Tetrastichus hagenowii (Ratzeburg). Insects 2024, 15, 668. [Google Scholar] [CrossRef]
- Sagri, E.; Koskinioti, P.; Gregoriou, M.; Tsoumani, K.T.; Bassiakos, Y.C.; Mathiopoulos, K.D. Housekeeping in Tephritid insects: The best gene choice for expression analyses in the medfly and the olive fly. Sci. Rep. 2017, 7, 45634. [Google Scholar] [CrossRef] [PubMed]
- Lu, Y.; Yuan, M.; Gao, X.; Kang, T.; Zhan, S.; Wan, H.; Li, J. Identification and validation of reference genes for gene expression analysis using quantitative PCR in Spodoptera litura (Lepidoptera: Noctuidae). PLoS ONE 2013, 8, e68059. [Google Scholar] [CrossRef]
- Lü, J.; Chen, S.; Guo, M.; Ye, C.; Qiu, B.; Wu, J.; Yang, C.; Pan, H. Selection and validation of reference genes for RT-qPCR analysis of the Ladybird Beetle Henosepilachna vigintioctomaculata. Front. Physiol. 2018, 9, 1614. [Google Scholar] [CrossRef]
- Shen, C.; Tang, M.; Li, X.; Zhu, L.; Li, W.; Deng, P.; Zhai, Q.; Wu, G.; Yan, X. Evaluation of reference genes for quantitative expression analysis in Mylabris sibirica (Coleoptera, Meloidae). Front. Physiol. 2024, 15, 1345836. [Google Scholar] [CrossRef]
- Lü, J.; Yang, C.; Zhang, Y.; Pan, H. Selection of reference genes for the normalization of RT-qPCR data in gene expression studies in insects: A systematic review. Front. Physiol. 2018, 9, 1560. [Google Scholar] [CrossRef] [PubMed]
- Du, H.; Wang, R.; Dai, X.; Yin, Z.; Liu, Y.; Su, L.; Chen, H.; Zhao, S.; Zheng, L.; Dong, X.; et al. Effect of guanylate cyclase-22-like on ovarian development of Orius nagaii (Hemiptera: Anthocoridae). Insects 2024, 15, 110. [Google Scholar] [CrossRef] [PubMed]
- Shen, X.; Zhang, G.; Zhao, Y.; Zhu, X.; Yu, X.; Yang, M.; Zhang, F. Selection and validation of optimal reference genes for RT-qPCR analyses in Aphidoletes aphidimyza Rondani (Diptera: Cecidomyiidae). Front. Physiol. 2023, 14, 1277942. [Google Scholar] [CrossRef] [PubMed]
- Xie, F.; Wang, J.; Zhang, B. RefFinder: A web-based tool for comprehensively analyzing and identifying reference genes. Funct. Integr. Genom. 2023, 23, 125. [Google Scholar] [CrossRef] [PubMed]
- Vandesompele, J.; De Preter, K.; Pattyn, F.; Poppe, B.; Van Roy, N.; De Paepe, A.; Speleman, F. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002, 3, h31–h34. [Google Scholar] [CrossRef]
- Andersen, C.L.; Jensen, J.L.; Ørntoft, T.F. Normalization of real-time quantitative reverse transcription-PCR data: A model-based variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer data sets. Cancer Res. 2004, 64, 5245–5250. [Google Scholar] [CrossRef] [PubMed]
- Pfaffl, M.W.; Tichopad, A.; Prgomet, C.; Neuvians, T.P. Determination of stable housekeeping genes, differentially regulated target genes and sample integrity: BestKeeper-Excel-based tool using pair-wise correlations. Biotechnol. Lett. 2004, 26, 509–515. [Google Scholar] [CrossRef] [PubMed]
- Silver, N.; Best, S.; Jiang, J.; Thein, S.L. Selection of housekeeping genes for gene expression studies in human reticulocytes using real-time PCR. BMC Mol. Biol. 2006, 7, 33. [Google Scholar] [CrossRef]
- Plageman, T.F., Jr.; Yutzey, K.E. T-box genes and heart development: Putting the “T” in heart. Dev. Dyn. 2005, 232, 11–20. [Google Scholar] [CrossRef]
- Porsch, M.; Hofmeyer, K.; Bausenwein, B.S.; Grimm, S.; Weber, B.H.F.; Miassod, R.; Pflugfelder, G.O. Isolation of a Drosophila T-box gene closely related to human TBX1. Gene 1998, 212, 237–248. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Yang, G.; Yu, X.; Zhang, Y.; Luo, J.; Li, X.; Zhu, L.; Zhang, H.; Jin, L.; Wu, G.; Yan, X.; et al. Screening and validation of stable reference genes for qRT-PCR analysis in Epicauta gorhami (Coleoptera: Meloidae). Preprints 2024. [Google Scholar] [CrossRef]
- Zhou, L.; Meng, J.; Ruan, H.; Yang, C.; Zhang, C. Expression stability of candidate RT-qPCR housekeeping genes in Spodoptera frugiperda (Lepidoptera: Noctuidae). Arch. Insect Biochem. Physiol. 2021, 108, e21831. [Google Scholar] [CrossRef]
- Huggett, J.; Dheda, K.; Bustin, S.; Zumla, A. Real-time RT-PCR normalisation; strategies and considerations. Genes Immun. 2005, 6, 279–284. [Google Scholar] [CrossRef] [PubMed]
- Liang, P.; Guo, Y.; Zhou, X.; Gao, X. Expression profiling in Bemisia tabaci under insecticide treatment: Indicating the necessity for custom reference gene selection. PLoS ONE 2014, 9, e87514. [Google Scholar] [CrossRef] [PubMed]
- Anger, A.M.; Armache, J.; Berninghausen, O.; Habeck, M.; Subklewe, M.; Wilson, D.N.; Beckmann, R. Structures of the human and Drosophila 80S ribosome. Nature 2013, 497, 80–85. [Google Scholar] [CrossRef] [PubMed]
- Liang, X.; Zuo, M.; Zhang, Y.; Li, N.; Ma, C.; Dong, M.; Gao, N. Structural snapshots of human pre-60S ribosomal particles before and after nuclear export. Nat. Commun. 2020, 11, 3542. [Google Scholar] [CrossRef] [PubMed]
- Zhi-Chuang, L.; Liu-Hao, W.; Rui-Lin, D.; Gui-Fen, Z.; Jian-Ying, G.; Fang-Hao, W. Evaluation of endogenous reference genes of Bactrocera (Tetradacus) minax by gene expression profiling under various experimental conditions. Fla. Entomol. 2014, 97, 597–604. [Google Scholar]
- Gong, Z.; Zhang, J.; Chen, Q.; Li, H.; Zhang, Z.; Duan, Y.; Jiang, Y.; Li, T.; Miao, J.; Wu, Y. Comprehensive screening and validation of stable internal reference genes for accurate qRT-PCR analysis in Holotrichia parallela under diverse biological conditions and environmental stresses. Insects 2024, 15, 661. [Google Scholar] [CrossRef]
- Zhang, S.; An, S.; Li, Z.; Wu, F.; Yang, Q.; Liu, Y.; Cao, J.; Zhang, H.; Zhang, Q.; Liu, X. Identification and validation of reference genes for normalization of gene expression analysis using qRT-PCR in Helicoverpa armigera (Lepidoptera: Noctuidae). Gene 2015, 555, 393–402. [Google Scholar] [CrossRef] [PubMed]
- Shakeel, M.; Zhu, X.; Kang, T.; Wan, H.; Li, J. Selection and evaluation of reference genes for quantitative gene expression studies in cotton bollworm, Helicoverpa armigera (Lepidoptera: Noctuidae). J. Asia-Pac. Entomol. 2015, 18, 123–130. [Google Scholar] [CrossRef]
- Majerowicz, D.; Alves-Bezerra, M.; Logullo, R.; Fonseca-de-Souza, A.L.; Meyer-Fernandes, J.R.; Braz, G.R.C.; Gondim, K.C. Looking for reference genes for real-time quantitative PCR experiments in Rhodnius prolixus (Hemiptera: Reduviidae). Insect Mol. Biol. 2011, 20, 713–722. [Google Scholar] [CrossRef] [PubMed]
- Rajarapu, S.P.; Mamidala, P.; Mittapalli, O. Validation of reference genes for gene expression studies in the emerald ash borer (Agrilus planipennis). Insect Sci. 2012, 19, 41–46. [Google Scholar] [CrossRef]
- Yano, A.; Nicol, B.; Guerin, A.; Guiguen, Y. The duplicated rainbow trout (Oncorhynchus mykiss) T-box transcription factors 1, tbx1a and tbx1b, are up-regulated during testicular development. Mol. Reprod. Dev. 2011, 78, 172–180. [Google Scholar] [CrossRef]
- Hu, Q.; Meng, Y.; Wang, D.; Tian, H.; Xiao, H. Characterization and function of the T-box 1 gene in Chinese giant salamander Andrias davidianus. Genomics 2019, 111, 1351–1359. [Google Scholar] [CrossRef] [PubMed]

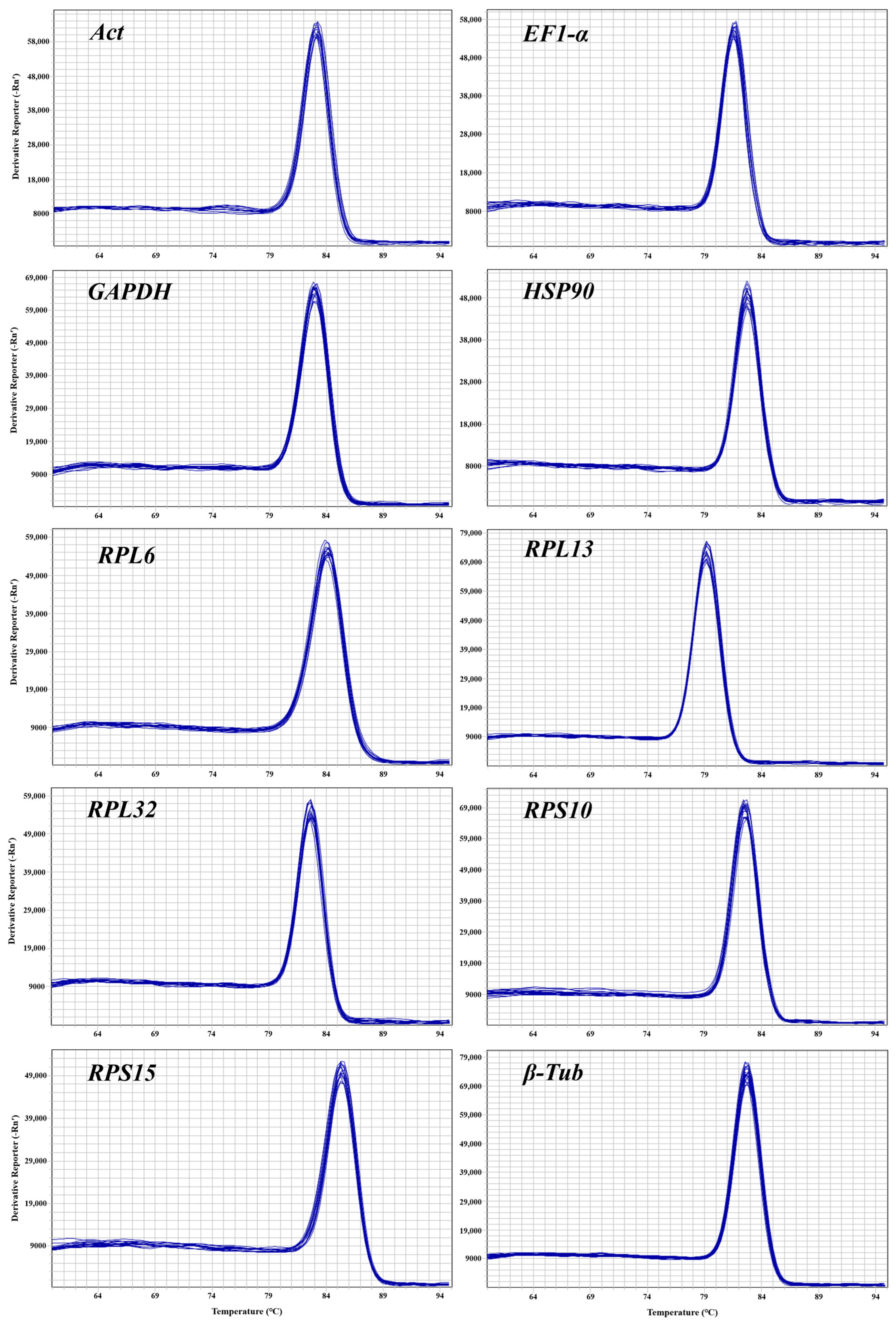

| Gene | Primer Sequences (5′–3′) | Length (bp) | Efficiency (%) | R2 | Linear Regression |
|---|---|---|---|---|---|
| Act | F: GACTGCTGAGCGTGAAATAG | 107 | 106.73 | 0.9956 | y = −3.1705x + 33.694 |
| R: GACCGTCTGGAAGTTCGTAG | |||||
| GAPDH | F: ATTTGTTGTTGAGCGGGATT | 100 | 106.59 | 0.999 | y = −3.1734x + 27.497 |
| R: TTGGGTTACACCGAAGACG | |||||
| β-Tub | F: GCGGGAAACAACTGGGCTAA | 104 | 108.95 | 0.9987 | y = −3.1246x + 30.147 |
| R: CCCTGAAGGCAATCGCAACC | |||||
| EF1-α | F: TGACAAAGGCTGCCGAGAA | 132 | 103.07 | 0.9999 | y = −3.2506x + 25.485 |
| R: TGGAAACACGGCTGGAGAA | |||||
| RPS10 | F: AGAAATGCCTCCAAGCGAACT | 119 | 107.24 | 0.9995 | y = −3.1598x + 28.862 |
| R: CTACAACGAGCCCAAACACCC | |||||
| RPS15 | F: CACTGCCGTGCTAGGAGGA | 107 | 105.48 | 0.9994 | y = −3.1973x + 28.448 |
| R: CGTTGGGCGGAGTTTCTT | |||||
| RPL6 | F: ATGGTTGCTACGGCTGTGAA | 145 | 109.02 | 0.9992 | y = −3.1231x + 29.159 |
| R: GGAACATCTGGCTTCTGCTAT | |||||
| RPL13 | F: ACCTTTGCCAATCCTTGTG | 109 | 105.61 | 0.9982 | y = −3.1944x + 28.732 |
| R: GGAAACCGATCCGACTTTT | |||||
| RPL32 | F: TCTGCGAAAGGATCACCATG | 148 | 106.00 | 0.9986 | y = −3.186x + 27.886 |
| R: CCTTCGGTTTACGCCAGTTT | |||||
| HSP90 | F: ATTCGCCGTGCTGTATCGTAA | 93 | 102.63 | 0.9999 | y = −3.2604x + 27.998 |
| R: TTCTTGGCAGCCATGTATCCC |
| Conditions | CRGs * | geNorm | NormFinder | BestKeeper | ∆Ct | Recommendation | ||||
|---|---|---|---|---|---|---|---|---|---|---|
| Stability | Rank | Stability | Rank | Stability | Rank | Stability | Rank | |||
| Developmental stage | Act | 0.792 | 10 | 2.205 | 10 | 1.827 | 10 | 2.225 | 10 | RPS10, RPL32 |
| GAPDH | 0.433 | 9 | 0.443 | 9 | 0.474 | 6 | 0.811 | 9 | ||
| β-Tub | 0.341 | 7 | 0.44 | 8 | 0.515 | 8 | 0.674 | 8 | ||
| EF1-α | 0.254 | 4 | 0.29 | 4 | 0.451 | 5 | 0.59 | 4 | ||
| RPS10 | 0.154 | 1 | 0.115 | 1 | 0.297 | 1 | 0.55 | 2 | ||
| RPS15 | 0.285 | 5 | 0.388 | 7 | 0.421 | 4 | 0.635 | 5 | ||
| RPL6 | 0.216 | 3 | 0.311 | 5 | 0.413 | 3 | 0.59 | 3 | ||
| RPL13 | 0.317 | 6 | 0.362 | 6 | 0.518 | 9 | 0.649 | 7 | ||
| RPL32 | 0.154 | 1 | 0.197 | 2 | 0.348 | 2 | 0.548 | 1 | ||
| HSP90 | 0.366 | 8 | 0.205 | 3 | 0.493 | 7 | 0.646 | 6 | ||
| Tissue | Act | 1.42 | 10 | 1.836 | 10 | 1.36 | 10 | 2.056 | 10 | RPS10, RPS15 |
| GAPDH | 0.792 | 6 | 0.548 | 1 | 0.644 | 2 | 1.236 | 4 | ||
| β-Tub | 0.515 | 5 | 1.294 | 8 | 1.02 | 9 | 1.476 | 8 | ||
| EF1-α | 0.982 | 7 | 0.763 | 3 | 0.703 | 5 | 1.279 | 5 | ||
| RPS10 | 0.235 | 1 | 0.758 | 2 | 0.621 | 1 | 1.197 | 1 | ||
| RPS15 | 0.235 | 1 | 0.796 | 5 | 0.654 | 3 | 1.205 | 3 | ||
| RPL6 | 1.117 | 8 | 1.097 | 6 | 0.825 | 6 | 1.44 | 7 | ||
| RPL13 | 0.46 | 4 | 1.165 | 7 | 0.862 | 7 | 1.373 | 6 | ||
| RPL32 | 0.406 | 3 | 0.773 | 4 | 0.701 | 4 | 1.203 | 2 | ||
| HSP90 | 1.261 | 9 | 1.515 | 9 | 1.007 | 8 | 1.737 | 9 | ||
| Host | Act | 0.721 | 10 | 1.534 | 10 | 1.411 | 10 | 1.564 | 10 | RPS10, RPS15 |
| GAPDH | 0.455 | 8 | 0.397 | 7 | 0.385 | 8 | 0.684 | 7 | ||
| β-Tub | 0.211 | 1 | 0.388 | 6 | 0.224 | 4 | 0.61 | 6 | ||
| EF1-α | 0.364 | 5 | 0.118 | 1 | 0.245 | 5 | 0.555 | 3 | ||
| RPS10 | 0.322 | 4 | 0.123 | 2 | 0.165 | 1 | 0.54 | 1 | ||
| RPS15 | 0.298 | 3 | 0.143 | 3 | 0.166 | 2 | 0.548 | 2 | ||
| RPL6 | 0.421 | 7 | 0.507 | 8 | 0.269 | 7 | 0.687 | 7 | ||
| RPL13 | 0.211 | 1 | 0.35 | 5 | 0.223 | 3 | 0.578 | 4 | ||
| RPL32 | 0.385 | 6 | 0.248 | 4 | 0.262 | 6 | 0.597 | 5 | ||
| HSP90 | 0.511 | 9 | 0.748 | 9 | 0.489 | 9 | 0.849 | 9 | ||
| Temperature | Act | 0.374 | 10 | 0.443 | 10 | 0.531 | 7 | 0.499 | 10 | EF1-α, RPL6 |
| GAPDH | 0.224 | 4 | 0.249 | 5 | 0.518 | 6 | 0.359 | 5 | ||
| β-Tub | 0.312 | 8 | 0.32 | 8 | 0.298 | 1 | 0.417 | 8 | ||
| EF1-α | 0.166 | 1 | 0.032 | 1 | 0.477 | 4 | 0.279 | 1 | ||
| RPS10 | 0.28 | 7 | 0.255 | 6 | 0.458 | 3 | 0.365 | 6 | ||
| RPS15 | 0.267 | 6 | 0.295 | 7 | 0.49 | 5 | 0.382 | 7 | ||
| RPL6 | 0.166 | 1 | 0.164 | 2 | 0.545 | 10 | 0.316 | 2 | ||
| RPL13 | 0.25 | 5 | 0.233 | 4 | 0.437 | 2 | 0.352 | 4 | ||
| RPL32 | 0.197 | 3 | 0.223 | 3 | 0.532 | 8 | 0.342 | 3 | ||
| HSP90 | 0.343 | 9 | 0.347 | 9 | 0.537 | 9 | 0.429 | 9 | ||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, C.; Yin, Z.; Wang, Y.; Liu, Y.; Zhao, S.; Dai, X.; Wang, R.; Su, L.; Chen, H.; Zheng, L.; et al. The Selection and Validation of Reference Genes for RT-qPCR Analysis of the Predatory Natural Enemy Orius nagaii (Hemiptera: Anthocoridae). Insects 2024, 15, 936. https://doi.org/10.3390/insects15120936
Wang C, Yin Z, Wang Y, Liu Y, Zhao S, Dai X, Wang R, Su L, Chen H, Zheng L, et al. The Selection and Validation of Reference Genes for RT-qPCR Analysis of the Predatory Natural Enemy Orius nagaii (Hemiptera: Anthocoridae). Insects. 2024; 15(12):936. https://doi.org/10.3390/insects15120936
Chicago/Turabian StyleWang, Chengxing, Zhenjuan Yin, Yu Wang, Yan Liu, Shan Zhao, Xiaoyan Dai, Ruijuan Wang, Long Su, Hao Chen, Li Zheng, and et al. 2024. "The Selection and Validation of Reference Genes for RT-qPCR Analysis of the Predatory Natural Enemy Orius nagaii (Hemiptera: Anthocoridae)" Insects 15, no. 12: 936. https://doi.org/10.3390/insects15120936
APA StyleWang, C., Yin, Z., Wang, Y., Liu, Y., Zhao, S., Dai, X., Wang, R., Su, L., Chen, H., Zheng, L., & Zhai, Y. (2024). The Selection and Validation of Reference Genes for RT-qPCR Analysis of the Predatory Natural Enemy Orius nagaii (Hemiptera: Anthocoridae). Insects, 15(12), 936. https://doi.org/10.3390/insects15120936





