Identification and Functional Analysis of Three NlCstF Genes in Nilaparvata lugens
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Insects
2.2. Total RNA Extraction, cDNA Synthesis and Cloning of Three NlCstF Genes
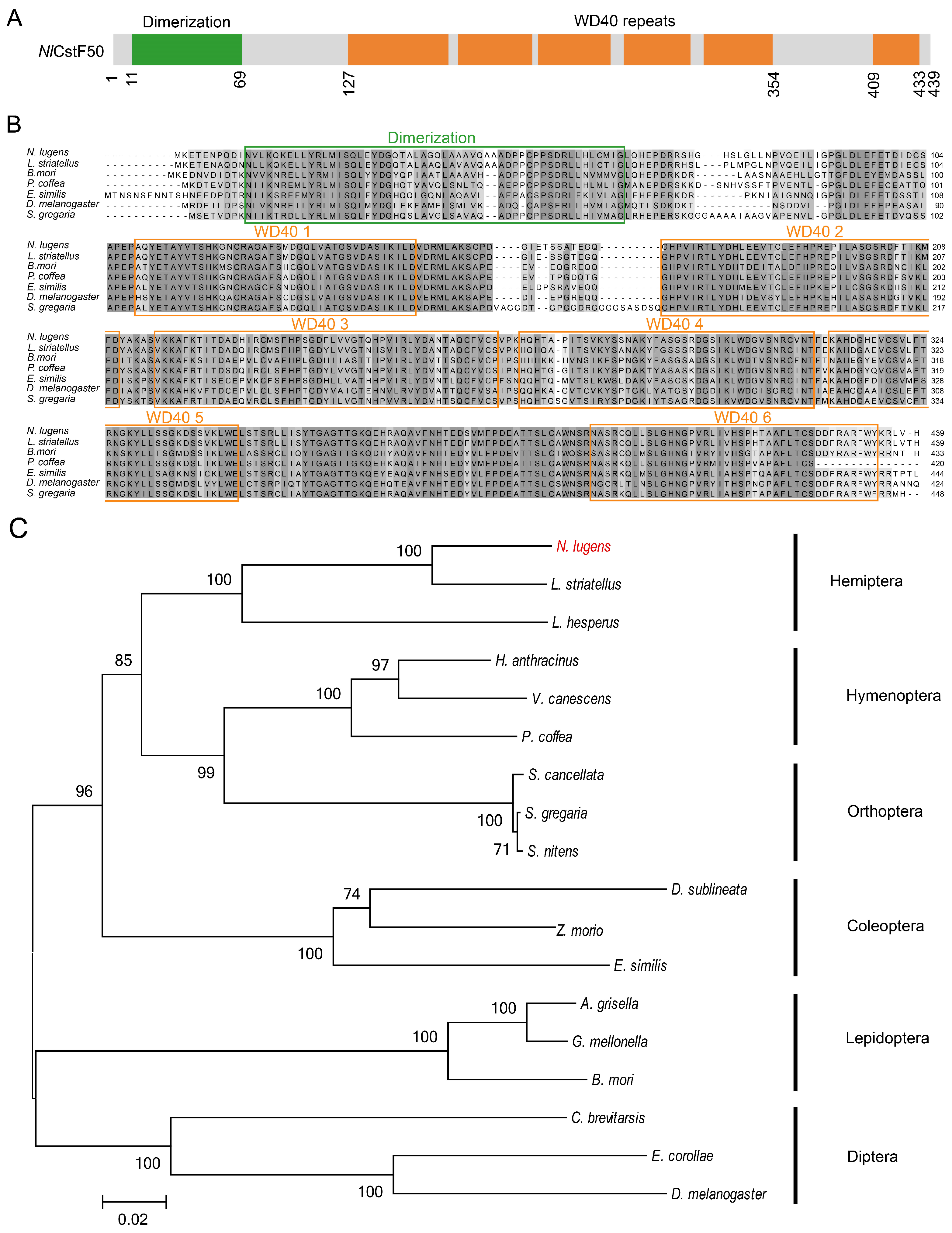
2.3. Sequence and Phylogenetic Analysis
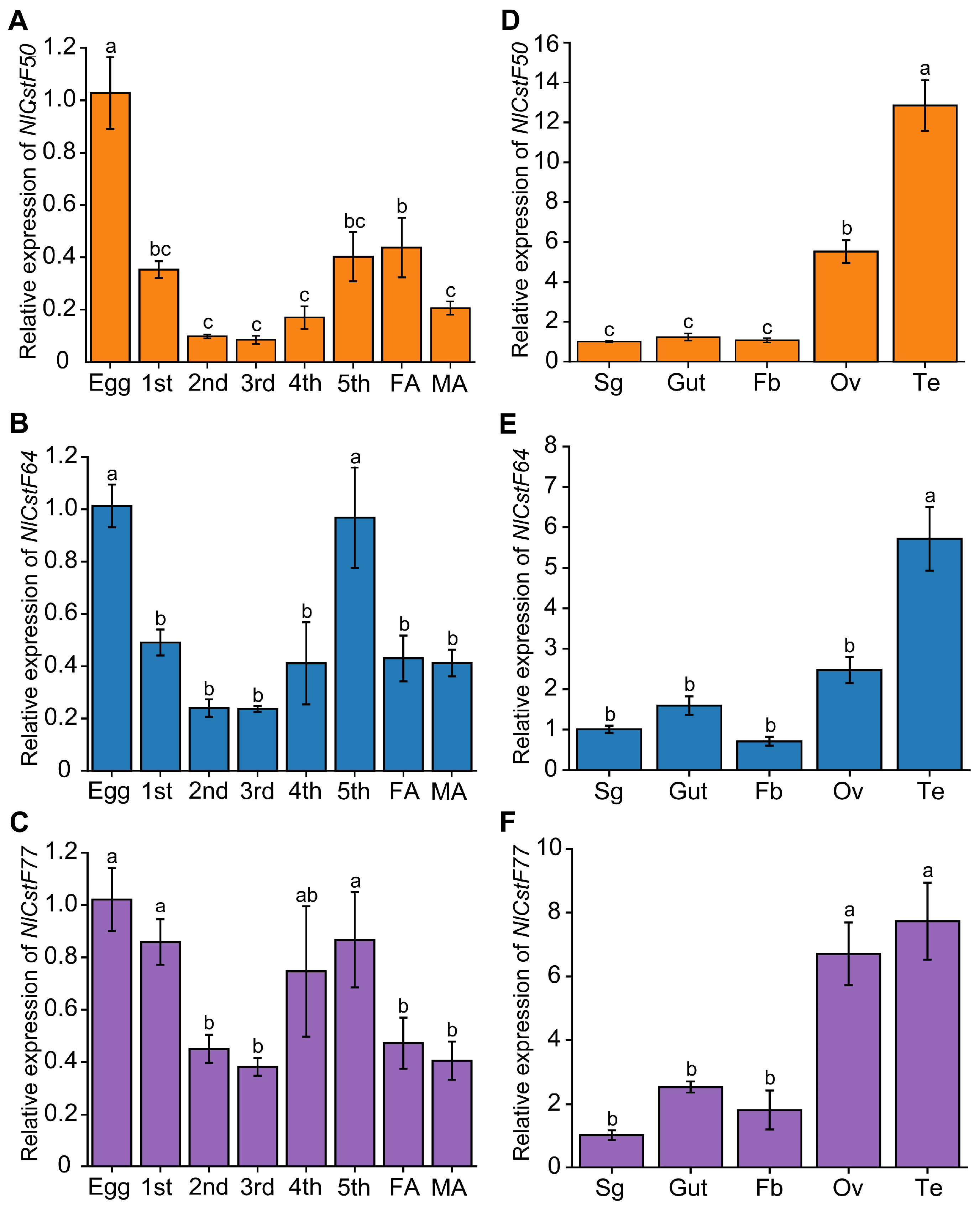
2.4. Expression Pattern Analysis of NlCstFs and qRT-PCR
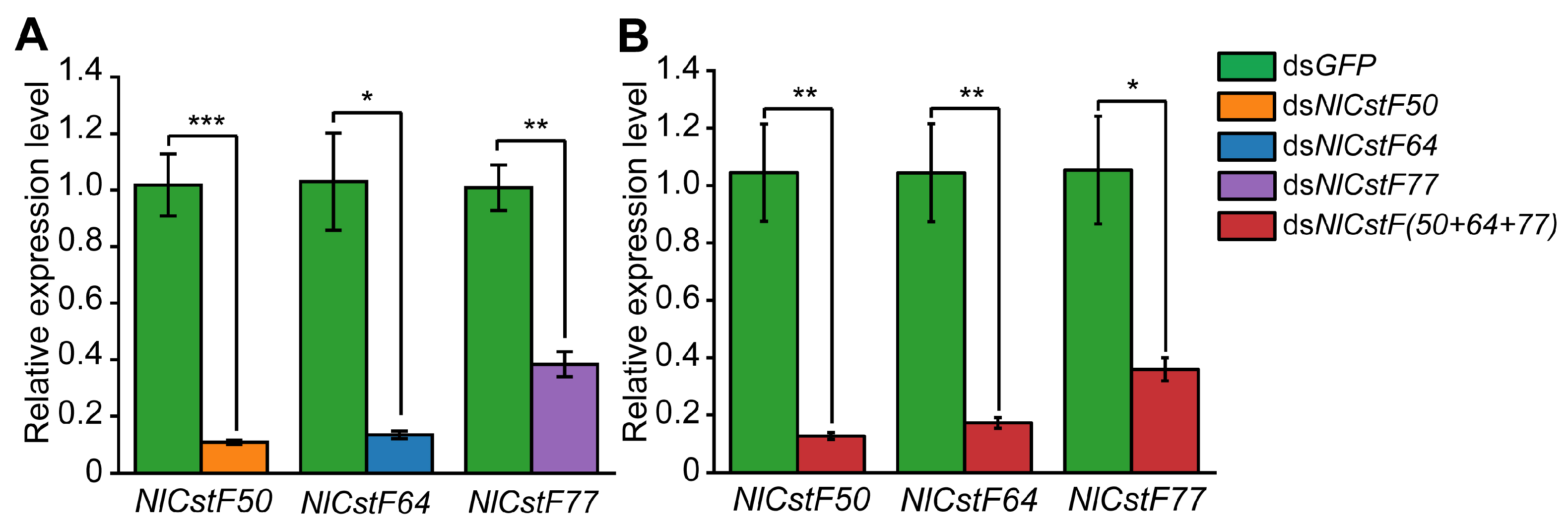
2.5. Double-Stranded RNA (dsRNA) Synthesis and RNAi
2.6. Effect of RNAi on the Survival of BPH
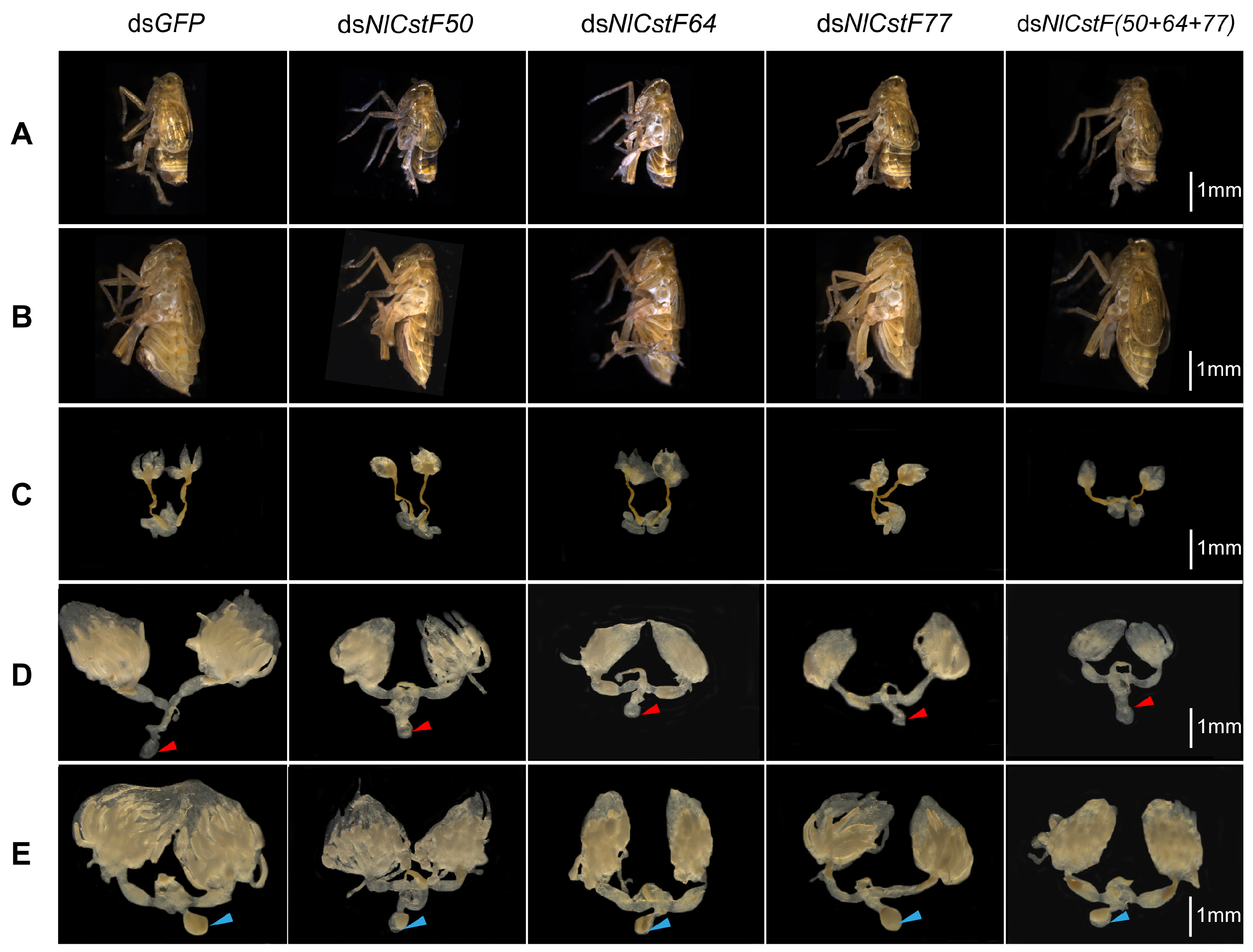
2.7. Effect of RNAi on Reproduction of BPH
2.8. Data Analysis
3. Results
3.1. Sequence and Phylogenetic Analysis of Three NlCstF Genes
3.2. Spatiotemporal Expression Patterns of Three NlCstF Genes
3.3. RNAi of NlCstFs Impacted Nymphal Survival
3.4. Effects of NlCstFs on the Reproduction of N. lugens
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Rosonina, E.; Kaneko, S.; Manley, J.L. Terminating the transcript: Breaking up is hard to do. Genes. Dev. 2006, 20, 1050–1056. [Google Scholar] [CrossRef] [PubMed]
- Xiang, K.; Tong, L.; Manley, J.L. Delineating the structural blueprint of the pre-mRNA 3′-end processing machinery. Mol. Cell Biol. 2014, 34, 1894–1910. [Google Scholar] [CrossRef] [PubMed]
- Christofori, G.; Keller, W. 3′ cleavage and polyadenylation of mRNA precursors in vitro requires a poly(A) polymerase, a cleavage factor, and a snRNP. Cell 1988, 54, 875–889. [Google Scholar] [CrossRef]
- Gilmartin, G.M.; Nevins, J.R. An ordered pathway of assembly of components required for polyadenylation site recognition and processing. Genes. Dev. 1989, 3, 2180–2190. [Google Scholar] [CrossRef]
- Takagaki, Y.; Ryner, L.C.; Manley, J.L. Four factors are required for 3′-end cleavage of pre-mRNAs. Genes. Dev. 1989, 3, 1711–1724. [Google Scholar] [CrossRef]
- Takagaki, Y.; Manley, J.L.; MacDonald, C.C.; Wilusz, J.; Shenk, T. A multisubunit factor, CstF, is required for polyadenylation of mammalian pre-mRNAs. Genes. Dev. 1990, 4, 2112–2120. [Google Scholar] [CrossRef]
- Takagaki, Y.; Manley, J.L. RNA recognition by the human polyadenylation factor CstF. Mol. Cell. Biol. 1997, 17, 3907–3914. [Google Scholar] [CrossRef]
- Yang, W.; Hsu, P.L.; Yang, F.; Song, J.-E.; Varani, G. Reconstitution of the CstF complex unveils a regulatory role for CstF-50 in recognition of 3′-end processing signals. Nucleic. Acids. Res. 2018, 46, 493–503. [Google Scholar] [CrossRef]
- Moreno-Morcillo, M.; Minvielle-Sébastia, L.; Mackereth, C.; Fribourg, S. Hexameric architecture of CstF supported by CstF-50 homodimerization domain structure. RNA 2011, 17, 412–418. [Google Scholar] [CrossRef][Green Version]
- Takagaki, Y.; Manley, J.L. A human polyadenylation factor is a G protein beta-subunit homologue. J. Biol. Chem. 1992, 267, 23471–23474. [Google Scholar] [CrossRef]
- Takagaki, Y.; Manley, J.L. Complex protein interactions within the human polyadenylation machinery identify a novel component. Mol. Cell. Biol. 2000, 20, 1515–1525. [Google Scholar] [CrossRef] [PubMed]
- Fong, N.; Bird, G.; Vigneron, M.; Bentley, D.L. A 10 residue motif at the C-terminus of the RNA pol II CTD is required for transcription, splicing and 3′ end processing. EMBO J. 2003, 22, 4274–4282. [Google Scholar] [CrossRef] [PubMed]
- Takagaki, Y.; MacDonald, C.C.; Shenk, T.; Manley, J.L. The human 64-kDa polyadenylylation factor contains a ribonucleoprotein-type RNA binding domain and unusual auxiliary motifs. Proc. Natl. Acad. Sci. USA 1992, 89, 1403–1407. [Google Scholar] [CrossRef] [PubMed]
- MacDonald, C.C.; Wilusz, J.; Shenk, T. The 64-kilodalton subunit of the CstF polyadenylation factor binds to pre-mRNAs downstream of the cleavage site and influences cleavage site location. Mol. Cell. Biol. 1994, 14, 6647–6654. [Google Scholar]
- Hatton, L.S.; Eloranta, J.J.; Figueiredo, L.M.; Takagaki, Y.; Manley, J.L.; O’Hare, K. The Drosophila homologue of the 64 kDa subunit of cleavage stimulation factor interacts with the 77 kDa subunit encoded by the suppressor of forked gene. Nucleic. Acids. Res. 2000, 28, 520–526. [Google Scholar] [CrossRef][Green Version]
- Qu, X.; Perez-Canadillas, J.-M.; Agrawal, S.; De Baecke, J.; Cheng, H.; Varani, G.; Moore, C. The c-terminal domains of vertebrate CstF-64 and its yeast orthologue rna15 form a new structure critical for mRNA 3′-end processing. J. Biol. Chem. 2007, 282, 2101–2115. [Google Scholar] [CrossRef]
- Preker, P.J.; Keller, W. The HAT helix, a repetitive motif implicated in RNA processing. Trends. Biochem. Sci. 1998, 23, 15–16. [Google Scholar] [CrossRef]
- Bai, Y.; Auperin, T.C.; Chou, C.-Y.; Chang, G.-G.; Manley, J.L.; Tong, L. Crystal structure of murine CstF-77: Dimeric association and implications for polyadenylation of mRNA precursors. Mol. Cell 2007, 25, 863–875. [Google Scholar] [CrossRef]
- Legrand, P.; Pinaud, N.; Minvielle-Sebastia, L.; Fribourg, S. The structure of the CstF-77 homodimer provides insights into CstF assembly. Nucleic. Acids. Res. 2007, 35, 4515–4522. [Google Scholar] [CrossRef][Green Version]
- Simonelig, M.; Elliott, K.; Mitchelson, A.; O’Hare, K. Interallelic complementation at the suppressor of forked locus of Drosophila reveals complementation between suppressor of forked proteins mutated in different regions. Genetics 1996, 142, 1225–1235. [Google Scholar] [CrossRef]
- Benoit, B.; Juge, F.; Iral, F.; Audibert, A.; Simonelig, M. Chimeric human CstF-77/Drosophila suppressor of forked proteins rescue suppressor of forked mutant lethality and mRNA 3′ end processing in Drosophila. Proc. Natl. Acad. Sci. USA 2002, 99, 10593–10598. [Google Scholar] [CrossRef] [PubMed]
- Mandart, E.; Parker, R. Effects of mutations in the Saccharomyces cerevisiae RNA14, RNA15, and PAP1 genes on polyadenylation in vivo. Mol. Cell. Biol. 1995, 15, 6979–6986. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Yao, Y.; Song, L.; Katz, Y.; Galili, G. Cloning and characterization of Arabidopsis homologues of the animal CstF complex that regulates 3′ mRNA cleavage and polyadenylation. J. Exp. Bot. 2002, 53, 2277–2278. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Wilson, T.G. Studies on the female-sterile phenotype of 1(1)su(f)ts76a, a temperature-sensitive allele of the suppressor of forked mutation in Drosophila melanogaster. Development 1980, 55, 247–256. [Google Scholar] [CrossRef]
- Audibert, A.; Simonelig, M. The suppressor of forked gene of Drosophila, which encodes a homologue of human CstF-77K involved in mRNA 3′-end processing, is required for progression through mitosis. Mech. Dev. 1999, 82, 41–50. [Google Scholar] [CrossRef]
- Minvielle-Sebastia, L.; Winsor, B.; Bonneaud, N.; Lacroute, F. Mutations in the yeast RNA14 and RNA15 genes result in an abnormal mRNA decay rate; sequence analysis reveals an RNA-binding domain in the RNA15 protein. Mol. Cell. Biol. 1991, 11, 3075–3087. [Google Scholar]
- Liu, F.; Marquardt, S.; Lister, C.; Swiezewski, S.; Dean, C. Targeted 3′ processing of antisense transcripts triggers Arabidopsis FLC chromatin silencing. Science 2010, 327, 94–97. [Google Scholar] [CrossRef]
- Zeng, W.; Dai, X.; Sun, J.; Hou, Y.; Ma, X.; Cao, X.; Zhao, Y.; Cheng, Y. Modulation of auxin signaling and development by polyadenylation machinery. Plant. Physiol. 2019, 179, 686–699. [Google Scholar] [CrossRef]
- Jing, S.; Zhao, Y.; Du, B.; Chen, R.; Zhu, L.; He, G. Genomics of interaction between the brown planthopper and rice. Curr. Opin. Insect Sci. 2017, 19, 82–87. [Google Scholar] [CrossRef]
- Tamura, K.; Stecher, G.; Peterson, D.; Filipski, A.; Kumar, S. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol. Biol. Evol. 2013, 30, 2725–2729. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Shangguan, X.; Zhang, J.; Liu, B.; Zhao, Y.; Wang, H.; Wang, Z.; Guo, J.; Rao, W.; Jing, S.; Guan, W.; et al. A mucin-like protein of planthopper is required for feeding and induces immunity response in plants. Plant. Physiol. 2018, 176, 552–565. [Google Scholar] [CrossRef] [PubMed]
- Mandel, C.R.; Bai, Y.; Tong, L. Protein factors in pre-mRNA 3′-end processing. Cell. Mol. Life Sci. 2008, 65, 1099–1122. [Google Scholar] [CrossRef] [PubMed]
- Xi, Y.; Pan, P.L.; Ye, Y.X.; Yu, B.; Xu, H.J.; Zhang, C.X. Chitinase-like gene family in the brown planthopper, Nilaparvata lugens. Insect. Mol. Biol. 2014, 24, 29–40. [Google Scholar] [CrossRef]
- Xi, Y.; Pan, P.L.; Zhang, C.X. The β-N-acetylhexosaminidase gene family in the brown planthopper, Nilaparvata lugens. Insect. Mol. Biol. 2015, 24, 601–610. [Google Scholar] [CrossRef]
- Xi, Y.; Pan, P.L.; Ye, Y.X.; Yu, B.; Zhang, C.X. Chitin deacetylase family genes in the brown planthopper, Nilaparvata lugens (Hemiptera: Delphacidae). Insect. Mol. Biol. 2014, 23, 695–705. [Google Scholar] [CrossRef]
- Audibert, A.; Simonelig, M. Autoregulation at the level of mRNA 3′ end formation of the suppressor of forked gene of Drosophila melanogaster is conserved in Drosophila virilis. Proc. Natl. Acad. Sci. USA 1998, 95, 14302–14307. [Google Scholar] [CrossRef]
- Wallace, A.M.; Dass, B.; Ravnik, S.E.; Tonk, V.; Jenkins, N.A.; Gilbert, D.J.; Copeland, N.G.; MacDonald, C.C. Two distinct forms of the 64,000 Mr protein of the cleavage stimulation factor are expressed in mouse male germ cells. Proc. Natl. Acad. Sci. USA 1999, 96, 6763–6768. [Google Scholar] [CrossRef]
- Dass, B.; Tardif, S.; Park, J.Y.; Tian, B.; Weitlauf, H.M.; Hess, R.A.; Carnes, K.; Griswold, M.D.; Small, C.L.; Macdonald, C.C. Loss of polyadenylation protein tauCstF-64 causes spermatogenic defects and male infertility. Proc. Natl. Acad. Sci. USA 2007, 104, 20374–20379. [Google Scholar] [CrossRef]
- Audibert, A.; Juge, F.; Simonelig, M. The suppressor of forked protein of Drosophila, a homologue of the human 77K protein required for mRNA 3′-end formation, accumulates in mitotically-active cells. Mech. Dev. 1998, 72, 53–63. [Google Scholar] [CrossRef]




Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jing, S.; Wang, F.; Ren, A.; Zheng, F.; Yu, B.; Xu, J.; Liu, Y.; Yang, J.; Chen, R.; Zeng, W.; et al. Identification and Functional Analysis of Three NlCstF Genes in Nilaparvata lugens. Insects 2024, 15, 867. https://doi.org/10.3390/insects15110867
Jing S, Wang F, Ren A, Zheng F, Yu B, Xu J, Liu Y, Yang J, Chen R, Zeng W, et al. Identification and Functional Analysis of Three NlCstF Genes in Nilaparvata lugens. Insects. 2024; 15(11):867. https://doi.org/10.3390/insects15110867
Chicago/Turabian StyleJing, Shengli, Feifei Wang, Aobo Ren, Fang Zheng, Bingbing Yu, Jingang Xu, Yali Liu, Jing Yang, Ruixian Chen, Wei Zeng, and et al. 2024. "Identification and Functional Analysis of Three NlCstF Genes in Nilaparvata lugens" Insects 15, no. 11: 867. https://doi.org/10.3390/insects15110867
APA StyleJing, S., Wang, F., Ren, A., Zheng, F., Yu, B., Xu, J., Liu, Y., Yang, J., Chen, R., Zeng, W., Zhang, Y., Ke, D., Ma, X., Tang, H., Liu, Q., & Yu, B. (2024). Identification and Functional Analysis of Three NlCstF Genes in Nilaparvata lugens. Insects, 15(11), 867. https://doi.org/10.3390/insects15110867





