The First Two Complete Mitochondrial Genomes for the Subfamily Meligethinae (Coleoptera: Nitidulidae) and Implications for the Higher Phylogeny of Nitidulidae
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Sample Preparation and DNA Extraction
2.2. Sequence Analysis
2.3. Phylogenetic Analysis
3. Results and Discussion
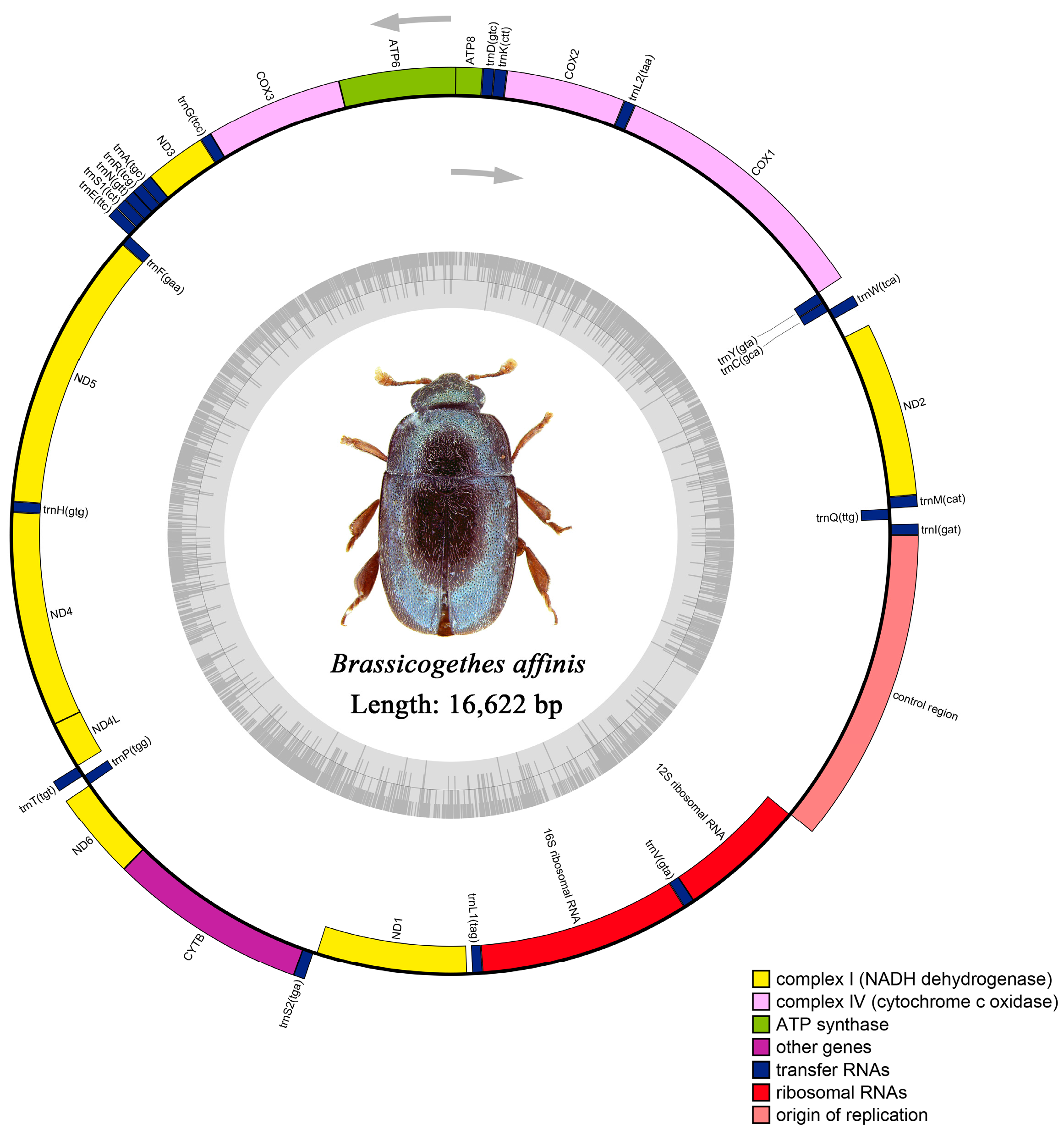
3.1. Genome Structure and Base Composition
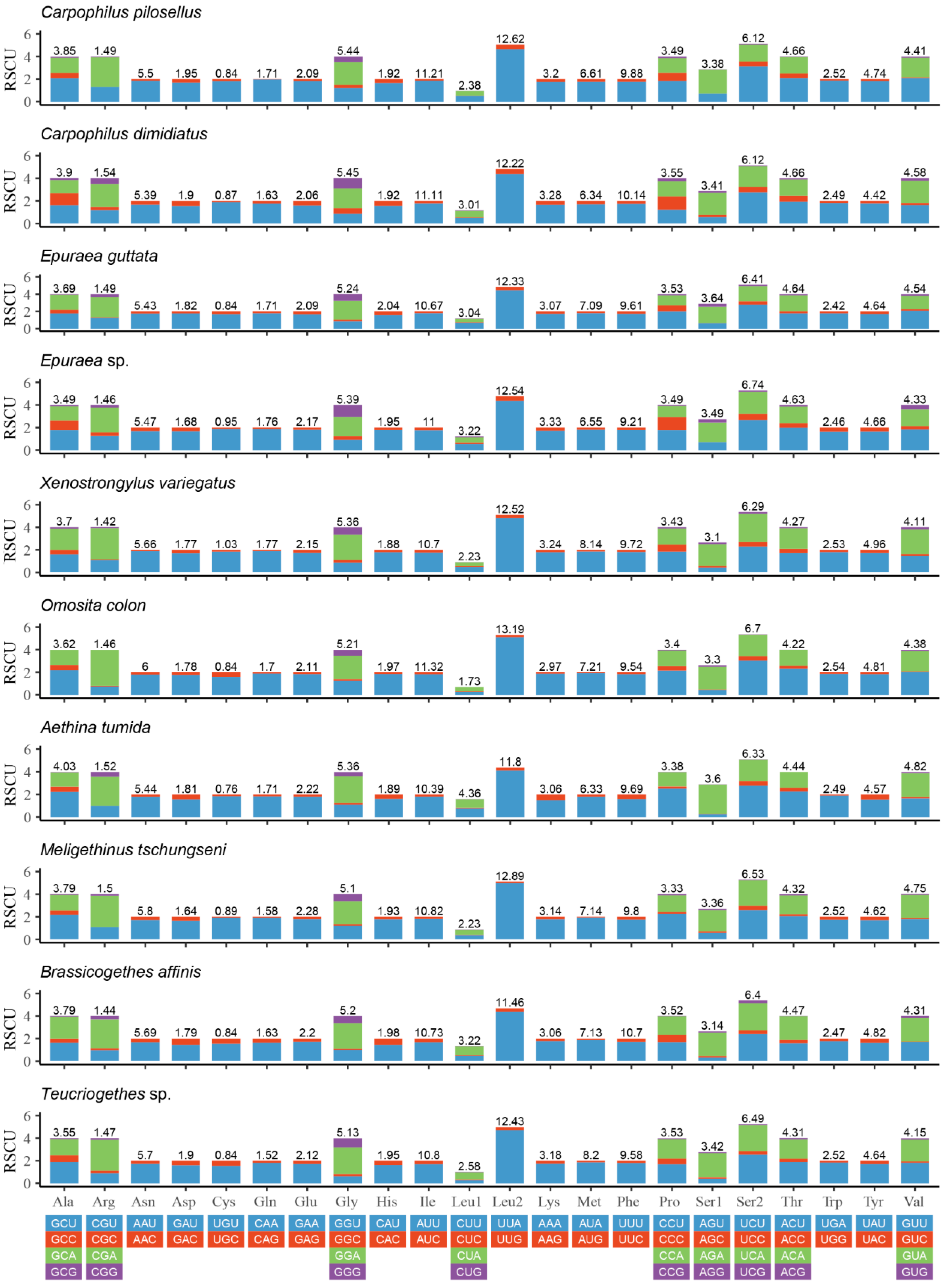
3.2. Protein-Coding Genes (PCGs) and Codon Usage
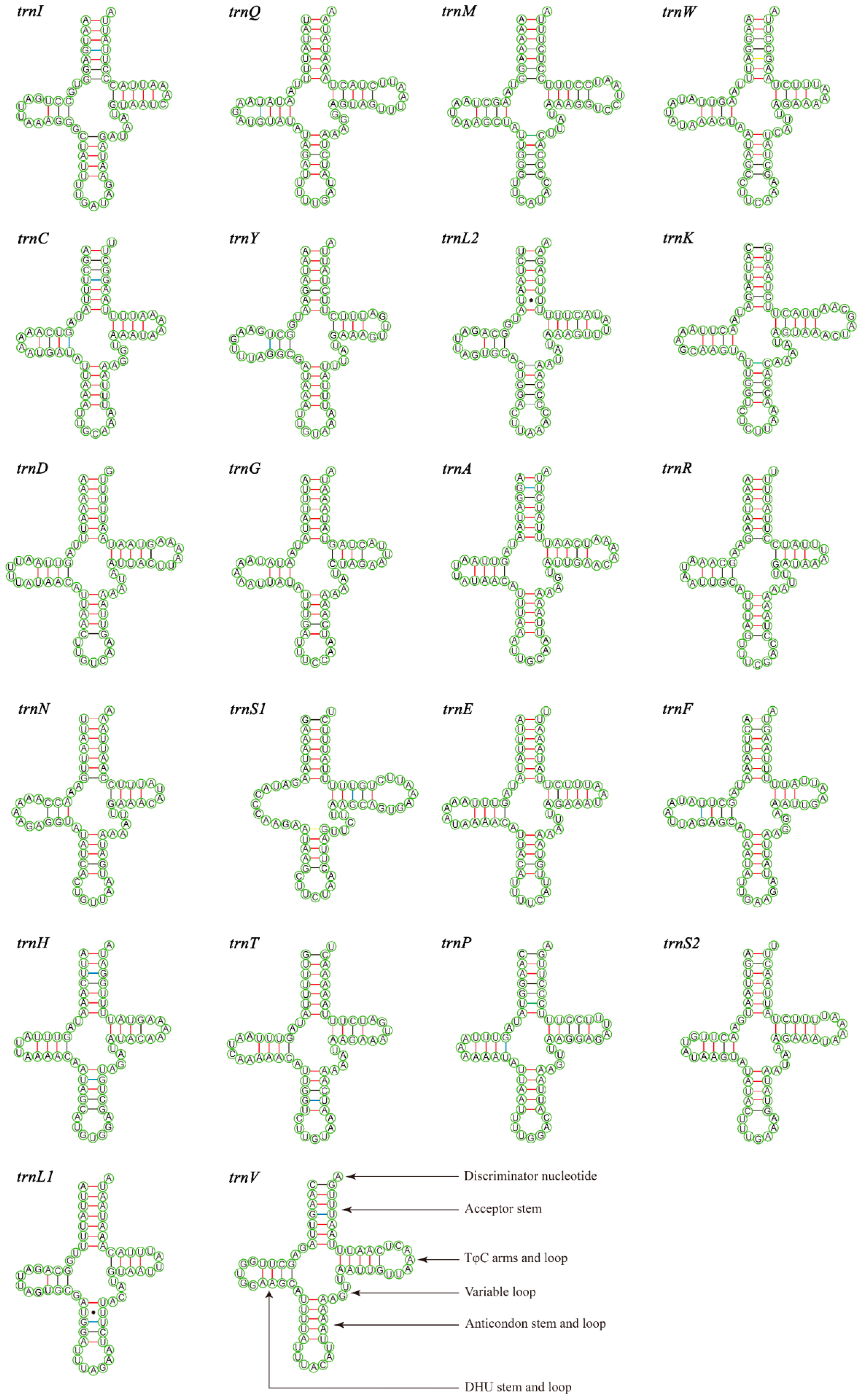
3.3. Transfer and Ribosomal RNA Genes
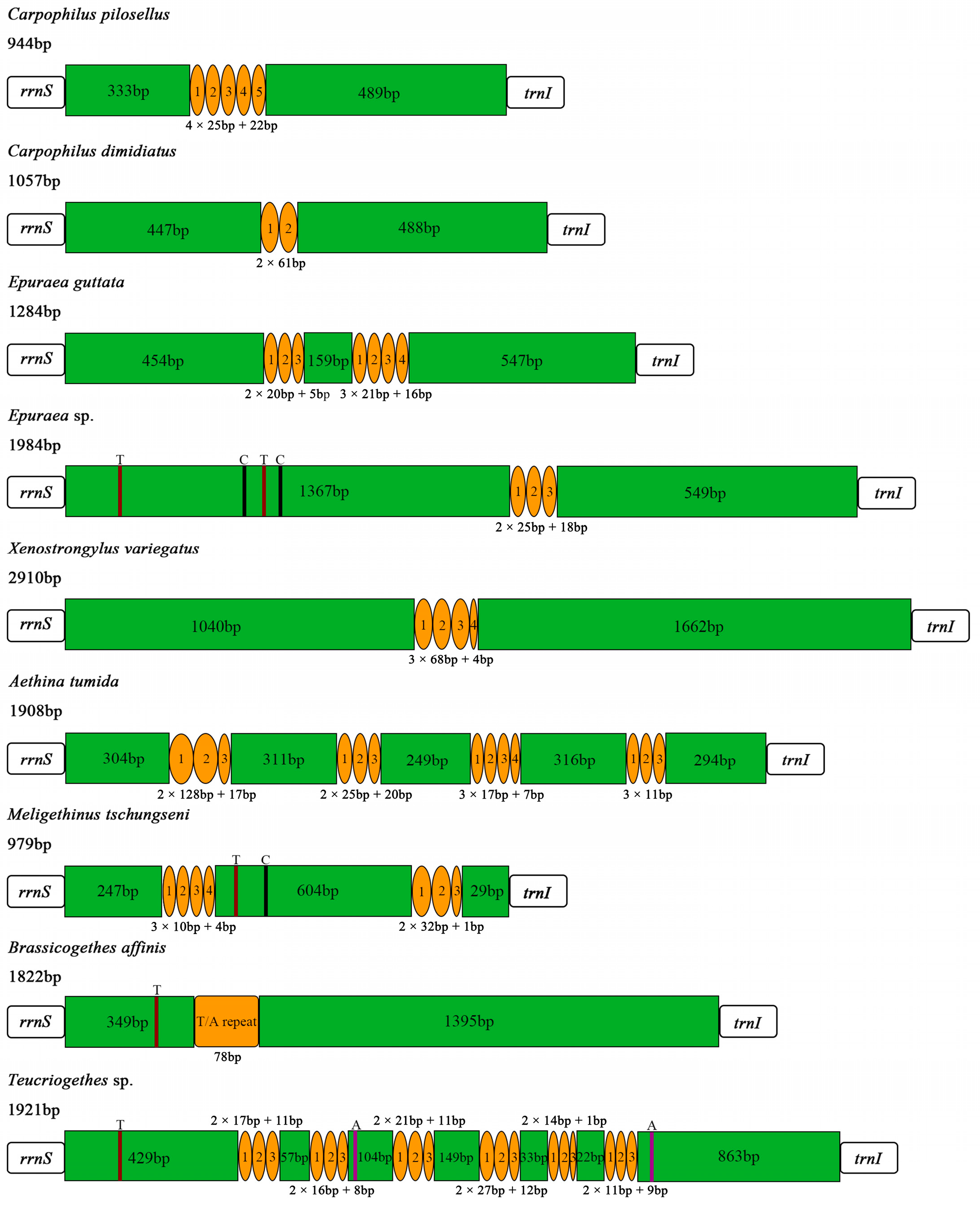
3.4. Control Region
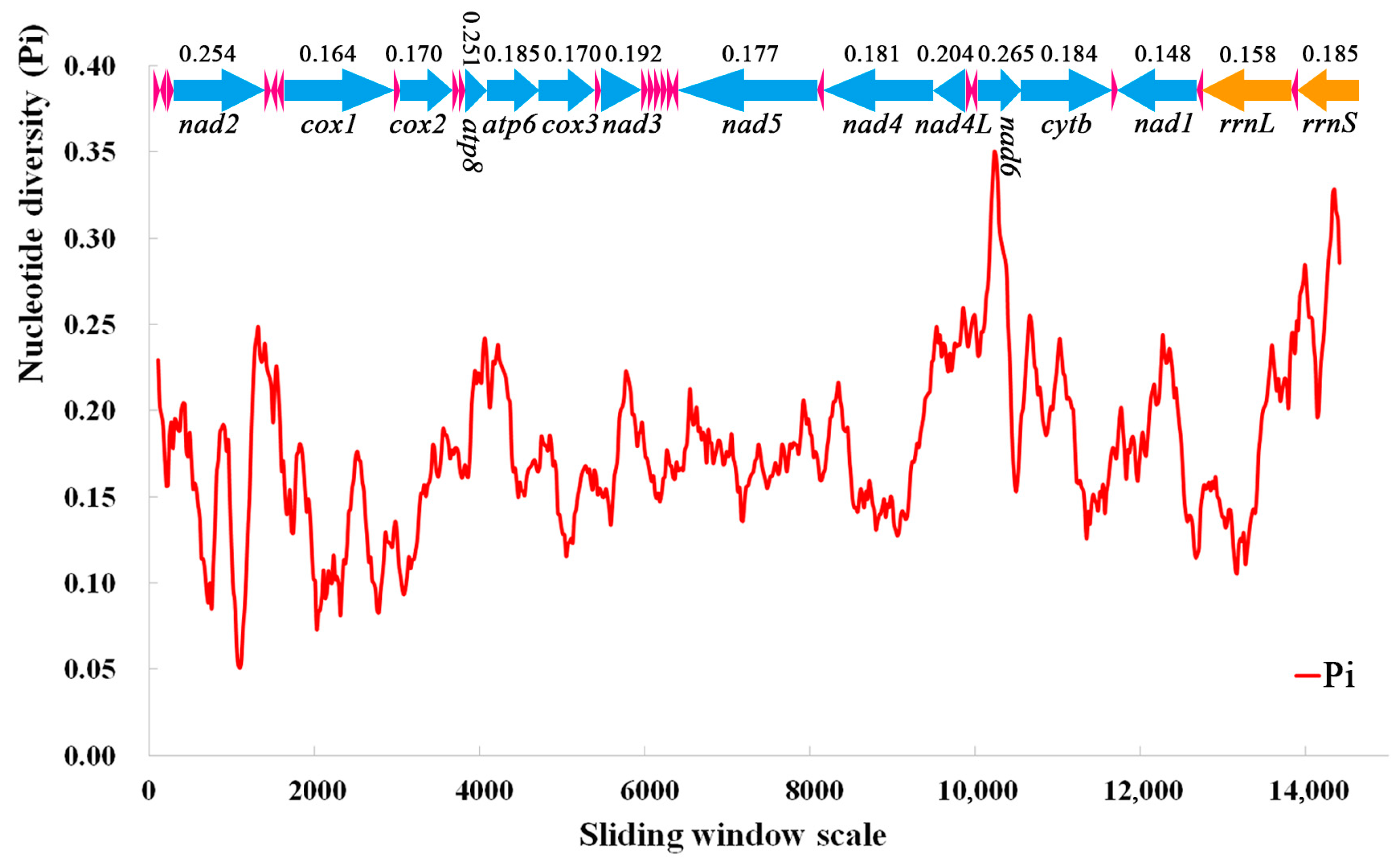
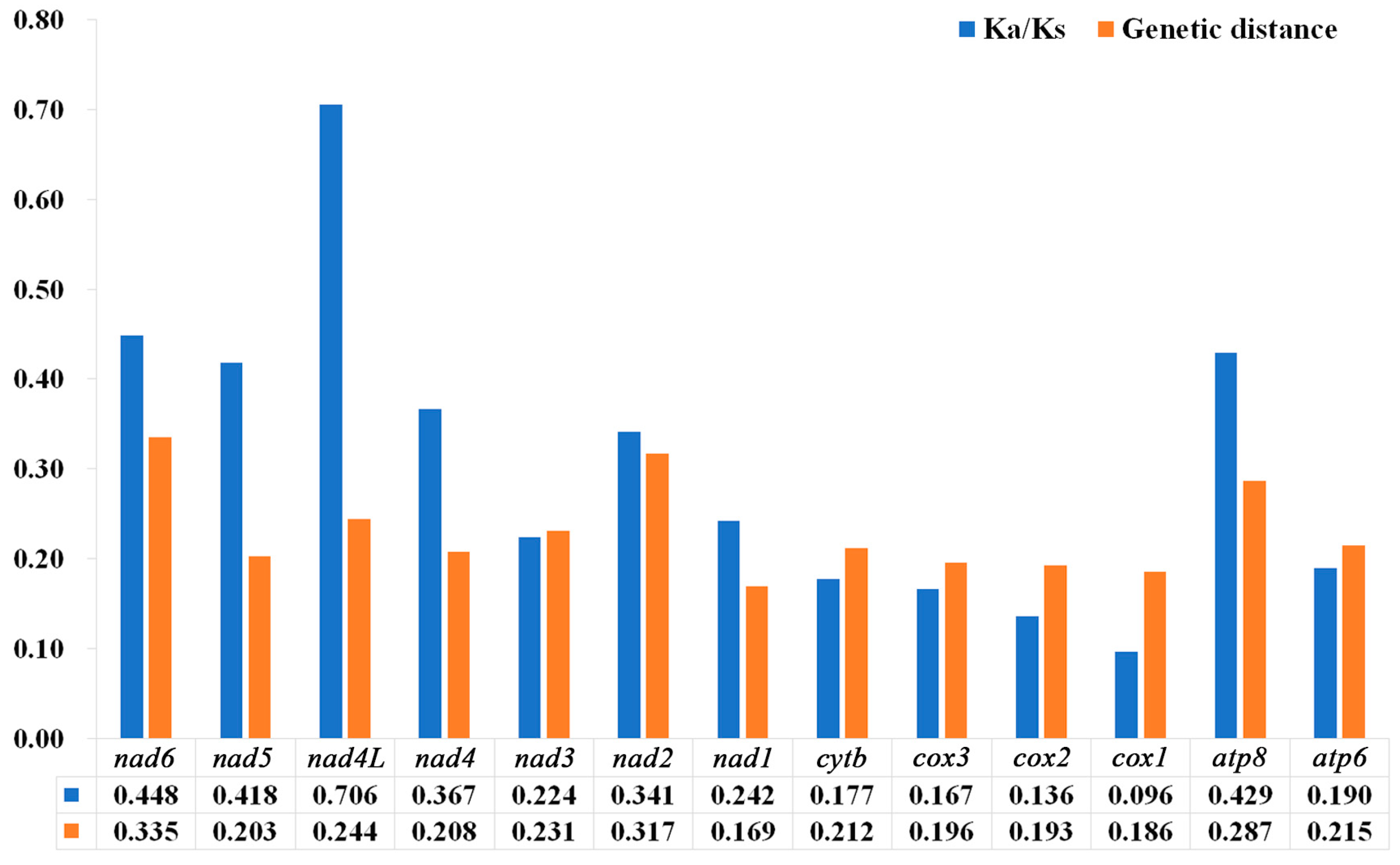
3.5. Nucleotide Diversity and Genetic Distance
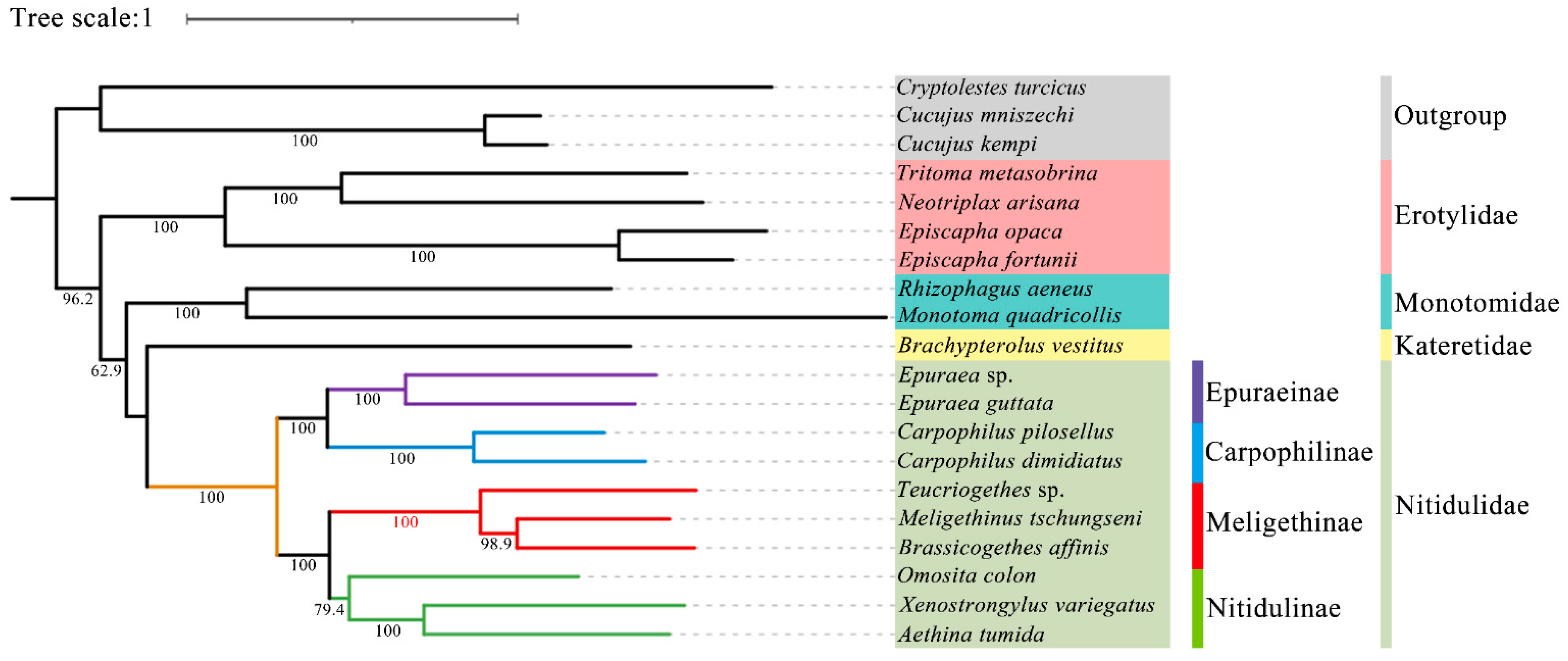
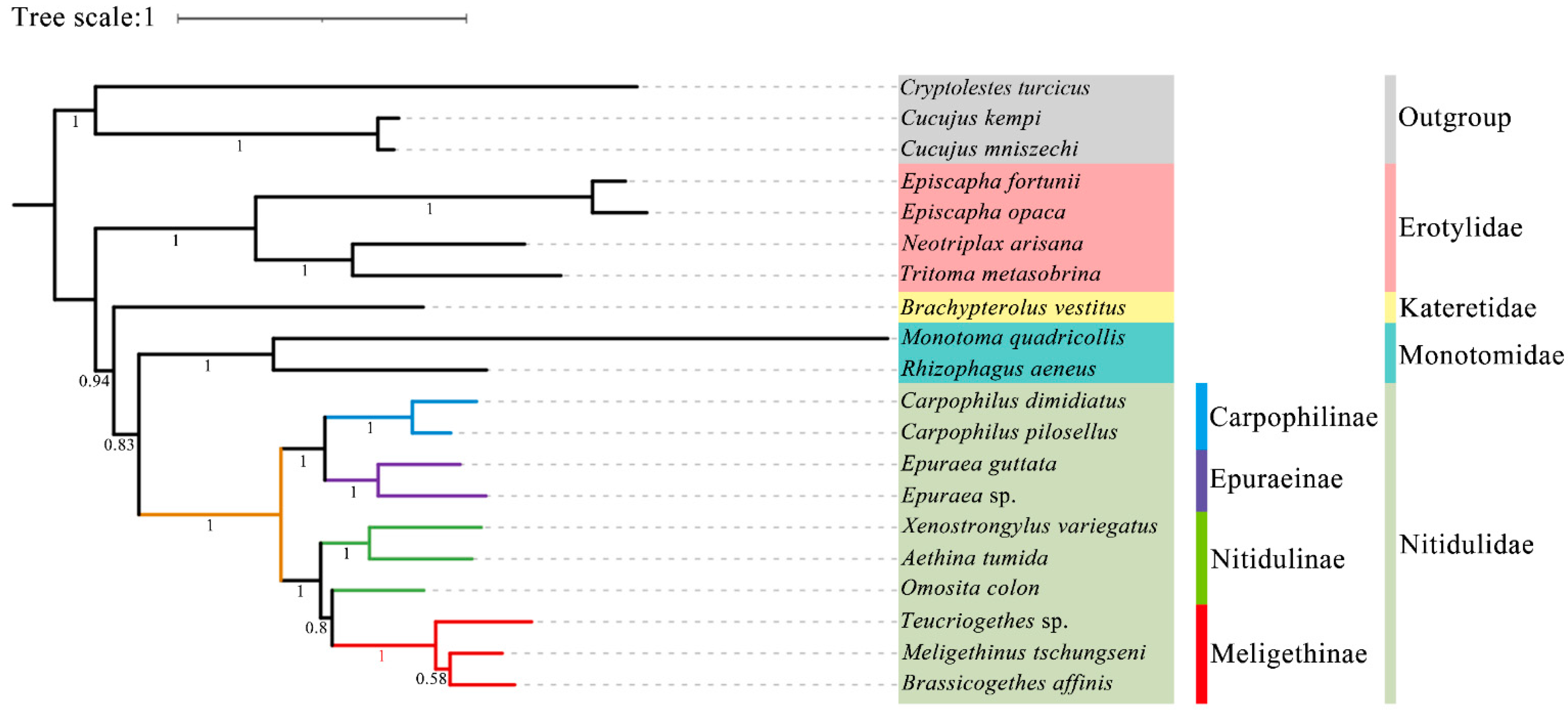
3.6. Phylogenetic Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Lee, M.H.; Lee, S.; Leschen, R.A.B.; Lee, S. Evolution of feeding habits of sap beetles (Coleoptera: Nitidulidae) and placement of Calonecrinae. Syst. Entomol. 2020, 45, 911–923. [Google Scholar] [CrossRef]
- Robertson, J.A.; Ślipiński, A.; Moulton, M.; Shockley, F.W.; Giorgi, A.; Lord, N.P.; Mckenna, D.D.; Tomaszewska, W.; Forrester, J.; Miller, K.B. Phylogeny and classification of Cucujoidea and the recognition of a new superfamily Coccinelloidea (Coleoptera: Cucujiformia). Syst. Entomol. 2015, 40, 745–778. [Google Scholar] [CrossRef]
- Cai, C.; Tihelka, E.; Giacomelli, M.; Lawrence, J.F.; Kundrata, R.; Yamamoto, S.; Thayer, M.K.; Newton, A.F.; Leschen, R.A.B.; Gimmel, M.L.; et al. Integrated phylogenomics and fossil data illuminate the evolution of beetles. R. Soc. Open Sci. 2022, 9, 211771. [Google Scholar] [CrossRef]
- Audisio, P. Fauna d’Italia 32. Coleoptera, Nitidulidae–Kateridae; Edizioni Calderini: Roma, Italy, 1993; Volume 32, p. 971. [Google Scholar]
- Audisio, P.; Cline, A.R.; De Biase, A.; Antonini, G.; Mancini, E.; Trizzino, M.; Costantini, L.; Strika, S.; Lamanna, F.; Cerretti, P. Preliminary re-examination of genus-level taxonomy of the pollen beetle subfamily Meligethinae (Coleoptera: Nitidulidae). Acta Entomol. Musei Natl. Pragae 2009, 49, 341–504. [Google Scholar]
- Audisio, P.; Sabatelli, S.; Jelínek, J. Revision of the pollen beetle genus Meligethes (Coleoptera: Nitidulidae). Fragm. Entomol. 2014, 46, 19–112. [Google Scholar] [CrossRef][Green Version]
- Jelínek, J.; Carlton, C.E.; Cline, A.R.; Leschen, R.A.B. Nitidulidae Latreille. In Handbook of Zoology. Arthropoda: Insecta. Coleoptera, 2 Morphology and Systematics (Elateroidea, Bostrichiformia, Cucujiformia Partim); Beutel, R.G., Leschen, R.A.B., Lawrence, J.F., Eds.; Walter De Gruyter: Berlin, Germany, 2010; pp. 390–407. [Google Scholar]
- Liu, M.; Yang, X.; Huang, M.; Jelínek, J.; Audisio, P. Four new species of Meligethes Stephens from China and additional data on other species of the genus (Coleoptera: Nitidulidae: Meligethinae). Zootaxa 2016, 4121, 101–116. [Google Scholar] [CrossRef] [PubMed]
- Liu, M.; Huang, M.; Cline, A.R.; Audisio, P. Two new Lamiogethes Audisio & Cline from China (Coleoptera: Nitidulidae, Meligethinae). Fragm. Entomol. 2017, 49, 145–150. [Google Scholar]
- Liu, M.; Huang, M.; Cline, A.R.; Sabatelli, S.; Audisio, P. A new species of Meligethes Stephens from China and additional data on members of the M. chinensis species-complex (Coleoptera: Nitidulidae, Meligethinae). Fragm. Entomol. 2017, 49, 79–84. [Google Scholar] [CrossRef][Green Version]
- Liu, M.; Huang, M.; Cline, A.R.; Audisio, P. New and poorly known Meligethes Stephens from China, with bionomical data on some species (Coleoptera: Nitidulidae: Meligethinae). Zootaxa 2018, 4392, 546–566. [Google Scholar] [CrossRef]
- Liu, M.; Huang, M.; Cline, A.R.; Cardoli, P.; Audisio, P.; Sabatelli, S. Re-examination of the genus-level taxonomy of the pollen beetle subfamily Meligethinae—Part 1. Sagittogethes Audisio & Cline 2009 and allied genera; with description of a new genus (Coleoptera: Nitidulidae). Fragm. Entomol. 2020, 52, 119–135. [Google Scholar]
- Liu, M.; Yang, X.K.; Huang, M.; Cline, A.R.; Sabatelli, S.; Audisio, P. Five new species of Lamiogethes Audisio & Cline from China (Coleoptera: Nitidulidae: Meligethinae). Zootaxa 2020, 4728, 63–76. [Google Scholar]
- Liu, M.; Huang, M.; Cline, A.R.; Mancini, E.; Scaramuzzi, A.; Paradisi, S.; Audisio, P.; Badano, D.; Sabatelli, S. Rosaceae, Brassicaceae and pollen beetles: Exploring relationships and evolution in an anthophilous beetle lineage (Nitidulidae, Meligethes-complex of genera) using an integrative approach. Front. Zool. 2021, 18, 9. [Google Scholar] [CrossRef] [PubMed]
- Sabatelli, S.; Liu, M.; Badano, D.; Mancini, E.; Trizzino, M.; Cline, A.R.; Endrestol, A.; Huang, M.; Audisio, P. Molecular phylogeny and host-plant use (Lamiaceae) of the Thymogethes pollen beetles (Coleoptera). Zool. Scr. 2020, 49, 28–46. [Google Scholar] [CrossRef]
- Kirejtshuk, A.G. On recent knowledge on the sap-beetles (Coleoptera, Nitidulidae) of India. Zool. Inst. Russ. Acad. Sci. 1999, 1, 21–32. [Google Scholar]
- Krishnan, K.T.; Neumann, P.; Ahmad, A.H.; Pimid, M. A scientific note on the association of Haptoncus luteolus (Coleoptera: Nitidulidae) with colonies of multiple stingless bee species. Apidologie 2015, 46, 262–264. [Google Scholar] [CrossRef]
- Deng, G.R.; Zeng, Q.D.; Li, W.Q.; Huang, D.X.; Zhou, Z.H. The harm of Tricanus japonicus on Dictyophora indusiata and its control. Plant Prot. 2006, 32, 117–118. [Google Scholar]
- Lounsberry, Z.; Spiewok, S.; Pernal, S.F.; Sonstegard, T.S.; Hood, W.M.; Pettis, J.; Neumann, P.; Evans, J.D. Worldwide Diaspora of Aethina tumida (Coleoptera: Nitidulidae), a Nest Parasite of Honey Bees. Ann. Entomol. Soc. Am. 2010, 103, 671–677. [Google Scholar] [CrossRef]
- Bai, X.G.; Cao, Y.; Zhou, Y.X. Identification of seven species of larvae of Nitidulidae from China. J. Zhengzhou Grain Coll. 1992, 50–56. [Google Scholar] [CrossRef]
- Dobson, R.M. The Species of Carpophilus Stephens (Col. Nitidulidae) associated with Stored Products. Bull. Entomol. Res. 1954, 45, 389–402. [Google Scholar] [CrossRef]
- Nadel, H.; Pena, J.E. Identity, Behavior, and Efficacy of Nitidulid Beetles (Coleoptera: Nitidulidae) Pollinating Commercial Annona Species in Florida. Environ. Entomol. 1994, 23, 878–886. [Google Scholar] [CrossRef]
- Yang, Q.P.; Liu, W.C.; Huang, C. Statistics and analysis on oilseed rape losses caused by main diseases and insect pests in recent ten years. Plant Prot. 2018, 44, 24–30. [Google Scholar]
- He, C.G.; Wang, G.L.; Fan, Y.H.; Zou, Y.X.; Deng, Y.X.; Deng, H.Y. Study on a new pest of oilseed-Xenostrongylus variegatus. Acta Agric. Boreali Occident. Sin. 1998, 7, 18–23. [Google Scholar]
- Bocak, L.; Barton, C.; Crampton-Platt, A.; Chesters, D.; Ahrens, D.; Vogler, A.P. Building the Coleoptera tree-of-life for >8000 species: Composition of public DNA data and fit with Linnaean classification. Syst. Entomol. 2014, 39, 97–110. [Google Scholar] [CrossRef]
- Tang, P.; Li, M.; Feng, R.; Wang, J.; Liu, M.; Wang, Y.; Yuan, M. Phylogenetic relationships among superfamilies of Cucujiformia (Coleoptera: Polyphaga) inferred from mitogenomic data. Sci. Sinca 2019, 49, 163–171. [Google Scholar] [CrossRef]
- Lawrence, J.F.; Ślipiński, A.; Seago, A.E.; Thayer, M.K.; Newton, A.F.; Marvaldi, A.E. Phylogeny of the Coleoptera Based on Morphological Characters of Adults and Larvae. Ann. Zool. 2011, 61, 1–217. [Google Scholar] [CrossRef]
- Cline, A.R.; Smith, T.R.; Miller, K.; Moulton, M.; Whiting, M.; Audisio, P. Molecular phylogeny of Nitidulidae: Assessment of subfamilial and tribal classification and formalization of the family Cybocephalidae (Coleoptera: Cucujoidea). Syst. Entomol. 2014, 39, 758–772. [Google Scholar] [CrossRef]
- Chen, X.; Song, Q.; Huang, M. Characterization of the Complete Mitochondrial Genomes from Two Nitidulid Pests with Phylogenetic Implications. Insects 2020, 11, 779. [Google Scholar] [CrossRef]
- McKenna, D.D.; Shin, S.; Ahrens, D.; Balke, M.; Beza-Beza, C.; Clarke, D.J.; Donath, A.; Escalona, H.E.; Friedrich, F.; Letsch, H.; et al. The evolution and genomic basis of beetle diversity. Proc. Natl. Acad. Sci. USA 2019, 116, 24729–24737. [Google Scholar] [CrossRef]
- Leschen, R.A.; Lawrence, J.F.; Ślipiński, S. Classification of basal Cucujoidea (Coleoptera: Polyphaga): Cladistic analysis, keys and review of new families. Invertebr. Syst. 2005, 19, 17–73. [Google Scholar] [CrossRef]
- Cline, A.R.; Slipinski, S.A. Discolomatidae Horn, 1878. In Handbuch der Zoologie/Handbook of Zoology; Leschen, R.A.B., Beutel, R.G., Lawrence, J.F., Eds.; Band. Arthropoda: Insecta, Teilband; W. DeGruyter: Berlin, Germany, 2010; Volume IV, pp. 435–442. [Google Scholar]
- Li, Y.W.; Yu, L.; Zhang, Y.P. “Long-branch Attraction” artifact in phylogenetic reconstruction. Hereditas 2020, 29, 659–667. [Google Scholar] [CrossRef]
- Kirejtshuk, A.G. English translation in Entomological Review. Systematic position of the genus Calonecrus J. Thomas and notes on the phylogeny of the family Nitidulidae (Coleoptera). Entomologicheskoye Obozreniye. Russ. Engl. Transl. Entomol. Rev. 1982, 61, 117–129. [Google Scholar]
- Kirejtshuk, A.G. On polyphyly of the Carpophilinae with description of a new subfamily, Cillaeinae (Coleoptera: Nitidulidae). Coleopt. Bull. 1986, 40, 217–221. [Google Scholar]
- Kirejtshuk, A.G. New taxa of the Nitidulidae (Coleoptera) of the eastern hemisphere. Trudy Zool. Inst. Ross. Akad. Nauk 1995, 258, 3–50. [Google Scholar]
- Kirejtshuk, A.G. A current generic classification of sap beetles (Coleoptera, Nitidulidae). Zoosyst. Ross. 2008, 17, 107–122. [Google Scholar] [CrossRef]
- Trizzino, M.; Audisio, P.; Antonini, G.; De Biase, A.; Mancini, E. Comparative analysis of sequences and secondary structures of the rRNA internal transcribed spacer 2 (ITS2) in pollen beetles of the subfamily Meligethinae (Coleoptera, Nitidulidae): Potential use of slippage-derived sequences in molecular systematics. Mol. Phylogenet. Evol. 2009, 51, 215–226. [Google Scholar] [CrossRef]
- Audisio, P.; De Biase, A.; Trizzino, M.; Mancini, E.; Antini, G. A new species of Meligethes (Coleoptera: Nitidulidae: Meligethinae) of the M. lugubris complex from Sardinia. Zootaxa 2009, 2318, 386–393. [Google Scholar] [CrossRef]
- Song, F.; Li, H.; Jiang, P.; Zhou, X.; Liu, J.; Sun, C.; onVogler, A.P.; Cai, W. Capturing the Phylogeny of Holometabola with Mitochondrial Genome Data and Bayesian Site-Heterogeneous Mixture Models. Genome Biol. Evol. 2016, 8, 1411–1426. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Li, H.; Song, F.; Zhao, Y.; Wilson, J.J.; Cai, W. Higher-level phylogeny and evolutionary history of Pentatomomorpha (Hemiptera: Heteroptera) inferred from mitochondrial genome sequences. Syst. Entomol. 2019, 44, 810–819. [Google Scholar] [CrossRef]
- Li, Q.; Yang, L.; Xiang, D.; Wan, Y.; Wu, Q.; Huang, W.; Zhao, G. The complete mitochondrial genomes of two model ectomycorrhizal fungi (Laccaria): Features, intron dynamics and phylogenetic implications. Int. J. Biol. Macromol. 2020, 145, 974–984. [Google Scholar] [CrossRef]
- Du, Z.; Hasegawa, H.; Cooley, J.R.; Simon, C.; Yoshimura, J.; Cai, W.; Sota, T.; Li, H. Mitochondrial Genomics Reveals Shared Phylogeographic Patterns and Demographic History among Three Periodical Cicada Species Groups. Mol. Biol. Evol. 2019, 36, 1187–1200. [Google Scholar] [CrossRef]
- Dowton, M.; Cameron, S.L.; Dowavic, J.I.; Austin, A.D.; Whiting, M.F. Characterization of 67 Mitochondrial tRNA Gene Rearrangements in the Hymenoptera Suggests That Mitochondrial tRNA Gene Position Is Selectively Neutral. Mol. Biol. Evol. 2009, 26, 1607–1617. [Google Scholar] [CrossRef] [PubMed]
- Song, S.N.; Tang, P.; Wei, S.J.; Chen, X.X. Comparative and phylogenetic analysis of the mitochondrial genomes in basal hymenopterans. Sci. Rep. 2016, 6, 20972. [Google Scholar] [CrossRef] [PubMed]
- Mao, M.; Gibson, T.; Dowton, M. Higher-level phylogeny of the Hymenoptera inferred from mitochondrial genomes. Mol. Biol. Evol. 2015, 84, 34–43. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Lan, Y.; Xia, L.; Cui, M.; Sun, W.; Dong, Z.; Cao, Y. The First Complete Mitochondrial Genomes of Two Sibling Species from Nitidulid Beetles Pests. Insects 2020, 11, 24. [Google Scholar] [CrossRef] [PubMed]
- Xu, W.; Wang, Y.; Wang, M.; Wang, Y.; Zhang, Y.; Wang, J. Characterization of the complete mitochondrial genome of Omosita colon (Coleoptera: Nitidulidae). Mitochondrial DNA B 2021, 6, 1547–1553. [Google Scholar] [CrossRef]
- Duquesne, V.; Delcont, A.; Huleux, A.; Beven, V.; Touzain, F.; Ribiere-Chabert, M. Complete Mitochondrial Genome Sequence of Aethina tumida (Coleoptera: Nitidulidae), a Beekeeping Pest. Genome Announc. 2017, 5, e01165-17. [Google Scholar] [CrossRef] [PubMed]
- Kearse, M.; Moir, R.; Wilson, A.; Stones-Havas, S.; Cheung, M.; Sturrock, S.; Buxton, S.; Cooper, A.; Markowitz, S.; Duran, C.; et al. Geneious Basic: An integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 2012, 28, 1647–1649. [Google Scholar] [CrossRef]
- Bernt, M.; Donath, A.; Juhling, F.; Externbrink, F.; Florentz, C.; Fritzsch, G.; Putz, J.; Middendorf, M.; Stadler, P.F. MITOS: Improved de novo metazoan mitochondrial genome annotation. Mol. Biol. Evol. 2013, 69, 313–319. [Google Scholar] [CrossRef]
- Lowe, T.M.; Chan, P.P. tRNAscan-SE On-line: Integrating search and context for analysis of transfer RNA genes. Nucleic Acids Res. 2016, 44, W54–W57. [Google Scholar] [CrossRef]
- Benson, G. Tandem repeats finder: A program to analyze DNA sequences. Nucleic Acids Res. 1999, 27, 573–580. [Google Scholar] [CrossRef]
- Lohse, M.; Drechsel, O.; Kahlau, S.; Bock, R. OrganellarGenomeDRAW—A suite of tools for generating physical maps of plastid and mitochondrial genomes and visualizing expression data sets. Nucleic Acids Res. 2013, 41, W575–W581. [Google Scholar] [CrossRef] [PubMed]
- Zhang, D.; Gao, F.; Jakovlic, I.; Zou, H.; Zhang, J.; Li, W.X.; Wang, G.T. PhyloSuite: An integrated and scalable desktop platform for streamlined molecular sequence data management and evolutionary phylogenetics studies. Mol. Ecol. Resour. 2020, 20, 348–355. [Google Scholar] [CrossRef] [PubMed]
- Rozas, J.; Ferrer-Mata, A.; Sanchez-DelBarrio, J.C.; Guirao-Rico, S.; Librado, P.; Ramos-Onsins, S.E.; Sanchez-Gracia, A. DnaSP 6: DNA Sequence Polymorphism Analysis of Large Data Sets. Mol. Biol. Evol. 2017, 34, 3299–3302. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef] [PubMed]
- Sun, T.Y.; Liu, G.; Li, L.; Xin, T.; Lei, K.; Xia, B. The complete mitochondrial genome of Cryptolestes turcicus (Grouvelle) (Coleoptera: Laemophloeidae). Mitochondrial DNA A 2016, 27, 3701–3702. [Google Scholar] [CrossRef] [PubMed]
- Jin, M.; Zwick, A.; Ślipiński, A.; Marris, J.W.M.; Thomas, M.C.; Pang, H. A comprehensive phylogeny of flat bark beetles (Coleoptera: Cucujidae) with a revised classification and a new South American genus. Syst. Entomol. 2020, 45, 248–268. [Google Scholar] [CrossRef]
- Liu, J.; Wang, Y.; Zhang, R.; Shi, C.; Lu, W.; Li, J.; Bai, M. Three Complete Mitochondrial Genomes of Erotylidae (Coleoptera: Cucujoidea) with Higher Phylogenetic Analysis. Insects 2021, 12, 524. [Google Scholar] [CrossRef]
- Lam-Tung, N.; Schmidt, H.A.; von Haeseler, A.; Bui Quang, M. IQ-TREE: A Fast and Effective Stochastic Algorithm for Estimating Maximum-Likelihood Phylogenies. Mol. Biol. Evol. 2015, 32, 268–274. [Google Scholar] [CrossRef]
- Letunic, I.; Bork, P. Interactive tree of life (iTOL) v3: An online tool for the display and annotation of phylogenetic and other trees. Nucleic Acids Res. 2016, 44, W242–W245. [Google Scholar] [CrossRef]
- Ojala, D.; Montoya, J.; Attardi, G. tRNA punctuation model of RNA processing in human mitochondria. Nature 1981, 290, 470–474. [Google Scholar] [CrossRef]
- Mancini, E.; De Biase, A.; Mariottini, P.; Bellini, A.; Audisio, P. Structure and evolution of the mitochondrial control region of the pollen beetle Meligethes thalassophilus (Coleoptera: Nitidulidae). Genome 2008, 51, 196–207. [Google Scholar] [CrossRef] [PubMed]
- Ye, F.; Easy, R.H.; King, S.D.; Cone, D.K.; You, P. Comparative analyses within Gyrodactylus (Platyhelminthes: Monogenea) mitochondrial genomes and conserved polymerase chain reaction primers for gyrodactylid mitochondrial DNA. J. Fish Dis. 2017, 40, 541–555. [Google Scholar] [CrossRef] [PubMed]
- Xie, Y.; Zhang, Z.; Niu, L.; Wang, Q.; Wang, C.; Lan, J.; Deng, J.; Fu, Y.; Nie, H.; Yan, N.; et al. The mitochondrial genome of Baylisascaris procyonis. PLoS ONE 2011, 6, e27066. [Google Scholar] [CrossRef] [PubMed]
- Jia, W.Z.; Yan, H.B.; Guo, A.J.; Zhu, X.Q.; Wang, Y.C.; Shi, W.G.; Chen, H.T.; Zhan, F.; Zhang, S.H.; Fu, B.Q.; et al. Complete mitochondrial genomes of Taenia multiceps, T. hydatigena and T. pisiformis: Additional molecular markers for a tapeworm genus of human and animal health significance. BMC Genom. 2010, 11, 447. [Google Scholar] [CrossRef] [PubMed]
- Hebert, P.D.N.; Ratnasingham, S.; deWaard, J.R. Barcoding animal life: Cytochrome c oxidase subunit 1 divergences among closely related species. R. Soc. 2003, 270 (Suppl. S1), S96–S99. [Google Scholar] [CrossRef]
- Cai, C.Y.; Wang, Y.L.; Liang, L.; Yin, Z.W.; Thayer, M.K.; Newton, A.F.; Zhou, Y.L. Congruence of morphological and molecular phylogenies of the rove beetle subfamily Staphylininae (Coleoptera: Staphylinidae). Sci. Rep. 2019, 9, 15137. [Google Scholar] [CrossRef]
- Cao, Y.K.; Huang, M. A SEM study of the antenna and mouthparts of Omosita colon (Linnaeus) (Coleoptera: Nitidulidae). Microsc. Res. Tech. 2016, 79, 1152–1164. [Google Scholar] [CrossRef]
- Crowson, R.A. A new genus of Boganiidae (Coleoptera) from Australia, with observations on glandular openings, cycad associations and geographical distribution in the family. Aust. J. Entomol. 1990, 29, 91–99. [Google Scholar] [CrossRef]

| Family | Subfamily | Species | Accession Number | Reference |
|---|---|---|---|---|
| Laemophloeidae | Cryptolestes turcicus | KT070712.1 | [58] | |
| Cucujidae | Cucujus kempi | NC_051939.1 | [59] | |
| Cucujus mniszechi | NC_051938.1 | [59] | ||
| Erotylidae | Tritoma metasobrina | MZ014622.1 | [60] | |
| Episcapha opaca | MZ014623.1 | [60] | ||
| Neotriplax arisana | MZ014624.1 | [60] | ||
| Episcapha fortunii | NC_067051.1 | Unpublished | ||
| Monotomidae | Monotoma quadricollis | NC_036266.1 | Unpublished | |
| Rhizophagus aeneus | KX087340.1 | Unpublished | ||
| Kateretidae | Brachypterolus vestitus | KX087245.1 | Unpublished | |
| Nitidulidae | Carpophilinae | Carpophilus pilosellus | NC_046035.1 | [47] |
| Carpophilus dimidiatus | NC_046036.1 | [47] | ||
| Epuraeinae | Epuraea guttata | KX087289.1 | Unpublished | |
| Epuraea sp. | MW044619.1 | [29] | ||
| Nitidulinae | Xenostrongylus variegatus | MW044620.1 | [29] | |
| Omosita colon | NC_050852.1 | [48] | ||
| Aethina tumida | NC_036104.1 | [49] | ||
| Meligethinae | Meligethinus tschungseni | ON782471 | This study | |
| Brassicogethes affinis | ON782472 | This study | ||
| Teucriogethes sp. | OR387485 | Unpublished |
| Gene | Position | Size (bp) | Intergenic Nucleotides | Codon | Strand | ||
|---|---|---|---|---|---|---|---|
| From | To | Start | Stop | ||||
| Meligethinus tschungseni/Brassicogethes affinis | |||||||
| trnI | 1/1 | 65/65 | 65/65 | +/+ | |||
| trnQ | 94/91 | 162/159 | 69/69 | 28/25 | −/− | ||
| trnM | 162/159 | 230/227 | 69/69 | −1/−1 | +/+ | ||
| nad2 | 231/228 | 1364/1232 | 1134/1005 | ATT/ATT | TAA/TAA | +/+ | |
| trnW | 1363/1355 | 1428/1420 | 66/66 | −2/122 | +/+ | ||
| trnC | 1421/1413 | 1483/1474 | 63/62 | −8/−8 | −/− | ||
| trnY | 1489/1457 | 1554/1540 | 66/66 | 5 | −/− | ||
| cox1 | 1596/1538 | 3093/3083 | 1498/1546 | 41/−3 | ATC/ATT | T/T | +/+ |
| trnL2 | 3094/3084 | 3158/3148 | 65/65 | +/+ | |||
| cox2 | 3159/3149 | 3846/3836 | 688/688 | ATC/ATC | T/T | +/+ | |
| trnK | 3847/3837 | 3916/3907 | 70/71 | +/+ | |||
| trnD | 3917/3908 | 3985/3973 | 69/66 | +/+ | |||
| atp8 | 3986/3974 | 4141/4129 | 156/156 | ATC/ATT | TAG/TAG | +/+ | |
| atp6 | 4138/4126 | 4809/4797 | 672/672 | −4/−4 | ATA/ATA | TAA/TAA | +/+ |
| cox3 | 4809/4797 | 5597/5585 | 789/789 | −1/−1 | ATG/ATG | TAA/TAA | +/+ |
| trnG | 5598/5585 | 5662/5649 | 65/65 | /−1 | +/+ | ||
| nad3 | 5663/5650 | 6016/6003 | 354/354 | ATT/ATT | TAG/TAG | +/+ | |
| trnA | 6015/6002 | 6081/6068 | 67/67 | −2/−2 | +/+ | ||
| trnR | 6081/6069 | 6144/6132 | 64/64 | −1 | +/+ | ||
| trnN | 6144/6132 | 6208/6195 | 65/64 | −1/−1 | +/+ | ||
| trnS1 | 6209/6196 | 6275/6262 | 6767 | +/+ | |||
| trnE | 6277/6264 | 6342/6329 | 66/66 | 1/1 | +/+ | ||
| trnF | 6342/6329 | 6407/6394 | 66/66 | −1/−1 | −/− | ||
| nad5 | 6408/6395 | 8121/8108 | 1714/1714 | ATC/ATC | T/T | −/− | |
| trnH | 8122/8109 | 8185/8172 | 64/64 | −/− | |||
| nad4 | 8186/8173 | 9509/9490 | 1324/1318 | ATA/ATT | T/T | −/− | |
| nad4L | 9506/9490 | 9802/9777 | 297/288 | −4/−1 | ATA/ATG | TAA/TAA | −/− |
| trnT | 9795/9780 | 9860/9845 | 66/66 | −8/2 | +/+ | ||
| trnP | 9861/9846 | 9924/9909 | 64/64 | −/− | |||
| nad6 | 9926/9911 | 10,429/10,417 | 504/507 | 1/1 | ATT/ATT | TAA/TAA | +/+ |
| cytb | 10,429/10,417 | 11,565/11,559 | 1137/1143 | −1/−1 | ATG/ATG | TAG/TAG | +/+ |
| trnS2 | 11,564/11,558 | 11,630/11,625 | 67/68 | −2/−2 | +/+ | ||
| nad1 | 11,649/11,644 | 12,581/12,564 | 933/921 | 18/18 | ATT/ATT | TAG/TAG | −/− |
| trnL1 | 12,601/12,596 | 12,663/12,658 | 63/63 | 19/31 | −/− | ||
| rrnL | 12,664/12,659 | 13,955/13,950 | 1292/1292 | −/− | |||
| trnV | 13,956/13,951 | 14,024/14,018 | 69/68 | −/− | |||
| rrnS | 14,024/14,018 | 14,804/14,800 | 781/783 | −/− | |||
| control region | 14,805/14,801 | 15,783/16,622 | 979/1822 | ||||
| Species | Whole Genome | AT Skew | GC Skew | PCGs | tRNAs | rRNAs | Control Region | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Size (bp) | AT (%) | Size (bp) | AT (%) | Size (bp) | AT (%) | Size (bp) | AT (%) | Size (bp) | AT (%) | |||
| C1. | 15,686 | 77.2 | 0.027 | −0.177 | 11,103 | 76.5 | 1442 | 76.3 | 2079 | 77.5 | 944 | 86.8 |
| C2. | 15,717 | 75.2 | 0.038 | −0.202 | 11,094 | 74.5 | 1441 | 74.9 | 2061 | 75 | 1057 | 83.5 |
| E1. | 16,021 | 76.5 | 0.043 | −0.19 | 11,073 | 75.7 | 1451 | 75.7 | 2081 | 76.4 | 1284 | 85.0 |
| E2. | 16,641 | 76.4 | −0.015 | −0.216 | 11,100 | 74.9 | 1445 | 75.7 | 2081 | 78.8 | 1984 | 82.6 |
| X. | 17,657 | 77.2 | 0.021 | −0.141 | 11,040 | 77 | 1454 | 78.2 | 2079 | 81.3 | 2910 | 74.7 |
| O. | 16,544 | 79.3 | 0.029 | −0.178 | 11,127 | 77.9 | 1453 | 79.4 | 2083 | 82.2 | 645 | 86.1 |
| A. | 16,576 | 76.9 | 0.034 | −0.223 | 11,109 | 75.4 | 1460 | 77.2 | 2064 | 79.5 | 1908 | 82.4 |
| M. | 15,783 | 77 | 0.029 | −0.236 | 11,196 | 77.2 | 1455 | 78 | 2073 | 81.5 | 979 | 62.5 |
| B. | 16,622 | 76.7 | 0.061 | −0.175 | 11,097 | 75.8 | 1451 | 79 | 2075 | 79.3 | 1822 | 76.1 |
| T. | 16,737 | 79.9 | 0.102 | −0.165 | 11,082 | 76.5 | 1461 | 79.1 | 2099 | 81.2 | 1921 | 97.7 |
| Regions | Size (bp) | T(U) | C | A | G | AT(%) | GC(%) | AT Skew | GC Skew |
|---|---|---|---|---|---|---|---|---|---|
| Meligethinus tschungseni | |||||||||
| Full genome | 15,783 | 37.4 | 14.2 | 39.6 | 8.8 | 77 | 23 | 0.029 | −0.236 |
| PCGs | 11,196 | 43.7 | 11.4 | 33.5 | 11.3 | 77.2 | 22.7 | −0.132 | −0.006 |
| tRNAs | 1455 | 36.8 | 9.8 | 41.2 | 12.2 | 78 | 22 | 0.056 | 0.106 |
| rRNAs | 2073 | 42.5 | 6.2 | 39 | 12.3 | 81.5 | 18.5 | −0.042 | 0.328 |
| 1st codon position | 3732 | 37.4 | 10.6 | 34.5 | 17.5 | 71.9 | 28.1 | −0.04 | 0.248 |
| 2nd codon position | 3732 | 47.5 | 17.9 | 21.2 | 13.3 | 68.7 | 31.2 | −0.383 | −0.147 |
| 3rd codon position | 3732 | 46.3 | 5.8 | 44.8 | 3 | 91.1 | 8.8 | −0.016 | −0.317 |
| Control region | 979 | 41.2 | 34.4 | 21.3 | 3.1 | 62.5 | 37.5 | −0.317 | −0.837 |
| Brassicogethes affinis | |||||||||
| Full genome | 16,622 | 36 | 13.7 | 40.7 | 9.6 | 76.7 | 23.3 | 0.061 | −0.175 |
| PCGs | 11,097 | 42.4 | 12.8 | 33.4 | 11.5 | 75.8 | 24.3 | −0.119 | −0.052 |
| tRNAs | 1451 | 37.6 | 9.2 | 41.4 | 11.9 | 79 | 21.1 | 0.047 | 0.128 |
| rRNAs | 2075 | 42 | 7.3 | 37.3 | 13.3 | 79.3 | 20.6 | −0.058 | 0.291 |
| 1st codon position | 3699 | 36.8 | 11.8 | 34.1 | 17.2 | 70.9 | 29 | −0.038 | 0.189 |
| 2nd codon position | 3699 | 47.4 | 18.1 | 21.4 | 13.1 | 68.8 | 31.2 | −0.379 | −0.163 |
| 3rd codon position | 3699 | 42.8 | 8.4 | 44.6 | 4.2 | 87.4 | 12.6 | 0.02 | −0.332 |
| Control region | 1822 | 44.1 | 12.2 | 32.0 | 11.7 | 76.1 | 23.9 | −0.159 | −0.021 |
| Gene | Start Codon/Stop Codon | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| C1. | C2. | E1. | E2. | X. | O. | A. | M. | B. | T. | |
| atp6 | ATA/TAA | ATG/TAA | ATG/TAA | ATG/TAA | ATA/TAA | ATG/TAA | ATA/TAA | ATA/TAA | ATA/TAA | ATA/TAA |
| atp8 | ATC/TAG | ATC/TAG | ATT/TAG | ATC/TAG | ATC/T | ATT/TAG | ATT/TAG | ATC/TAG | ATT/TAG | ATC/TAG |
| cox1 | ATT/T | ATT/T | ATT/T | ATC/T | ATT/T | ATT/T | ATT/T | ATC/T | ATT/T | ATA/T |
| cox2 | ATT/T | ATC/T | ATA/T | ATT/T | ATT/T | ATT/T | ATT/T | ATC/T | ATC/T | ATT/T |
| cox3 | ATG/T | ATG/T | ATG/T | ATG/T | ATG/T | ATG/T | ATG/T | ATG/TAA | ATG/TAA | ATG/TAA |
| cytb | ATG/TAG | ATG/TAG | ATA/TAG | ATG/TAG | ATG/TAG | ATG/TAG | ATG/TAA | ATG/TAG | ATG/TAG | ATG/TAA |
| nad1 | ATG/TAG | ATA/TAG | TTG/TAG | ATT/TAG | ATT/TAG | TTG/TAG | TTG/TAG | ATT/TAG | ATT/TAG | ATA/TAG |
| nad2 | ATT/TAA | ATT/TAA | ATT/TAA | ATT/TAA | ATT/TAA | ATT/TAA | ATT/TAA | ATT/TAA | ATT/TAA | ATT/TAG |
| nad3 | ATT/TAG | ATT/TAG | ATA/TAG | ATT/TAG | ATT/TAG | ATT/TAA | ATA/TAG | ATT/TAG | ATT/TAG | ATT/TAG |
| nad4 | ATG/T | ATG/T | ATG/TAA | ATA/T | ATT/T | ATG/T | ATG/T | ATA/T | ATT/T | ATA/T |
| nad4L | ATG/TAA | ATG/TAA | ATG/TAA | ATG/TAA | ATG/TAA | ATG/TAA | ATG/TAA | ATA/TAA | ATG/TAA | ATG/TAA |
| nad5 | ATT/T | ATT/T | ATA/T | ATT/T | ATT/T | ATT/T | ATA/T | ATC/T | ATC/T | ATC/T |
| nad6 | ATA/TAA | ATA/TAA | ATC/TAA | ATA/TAA | ATA/TAA | ATT/TAA | ATA/TAA | ATT/TAA | ATT/TAA | ATT/TAA |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dai, J.; Liu, M.; Di Giulio, A.; Sabatelli, S.; Wang, W.; Audisio, P. The First Two Complete Mitochondrial Genomes for the Subfamily Meligethinae (Coleoptera: Nitidulidae) and Implications for the Higher Phylogeny of Nitidulidae. Insects 2024, 15, 57. https://doi.org/10.3390/insects15010057
Dai J, Liu M, Di Giulio A, Sabatelli S, Wang W, Audisio P. The First Two Complete Mitochondrial Genomes for the Subfamily Meligethinae (Coleoptera: Nitidulidae) and Implications for the Higher Phylogeny of Nitidulidae. Insects. 2024; 15(1):57. https://doi.org/10.3390/insects15010057
Chicago/Turabian StyleDai, Jiaqi, Meike Liu, Andrea Di Giulio, Simone Sabatelli, Wenkai Wang, and Paolo Audisio. 2024. "The First Two Complete Mitochondrial Genomes for the Subfamily Meligethinae (Coleoptera: Nitidulidae) and Implications for the Higher Phylogeny of Nitidulidae" Insects 15, no. 1: 57. https://doi.org/10.3390/insects15010057
APA StyleDai, J., Liu, M., Di Giulio, A., Sabatelli, S., Wang, W., & Audisio, P. (2024). The First Two Complete Mitochondrial Genomes for the Subfamily Meligethinae (Coleoptera: Nitidulidae) and Implications for the Higher Phylogeny of Nitidulidae. Insects, 15(1), 57. https://doi.org/10.3390/insects15010057








