Host-Encoded Aminotransferase Import into the Endosymbiotic Bacteria Nardonella of Red Palm Weevil
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Weevil Rearing and Tissue Collection
2.2. Characterization of Aminotransferase Genes
2.3. RNA Isolation and cDNA Synthesis
2.4. Relative Transcript Analysis
2.5. RNA Interference of RfGOT1 and RfGOT2A Genes
2.6. Tyrosine Detection
2.7. Histology
3. Results
3.1. Identification of Tyrosine Transaminase Genes in RPW
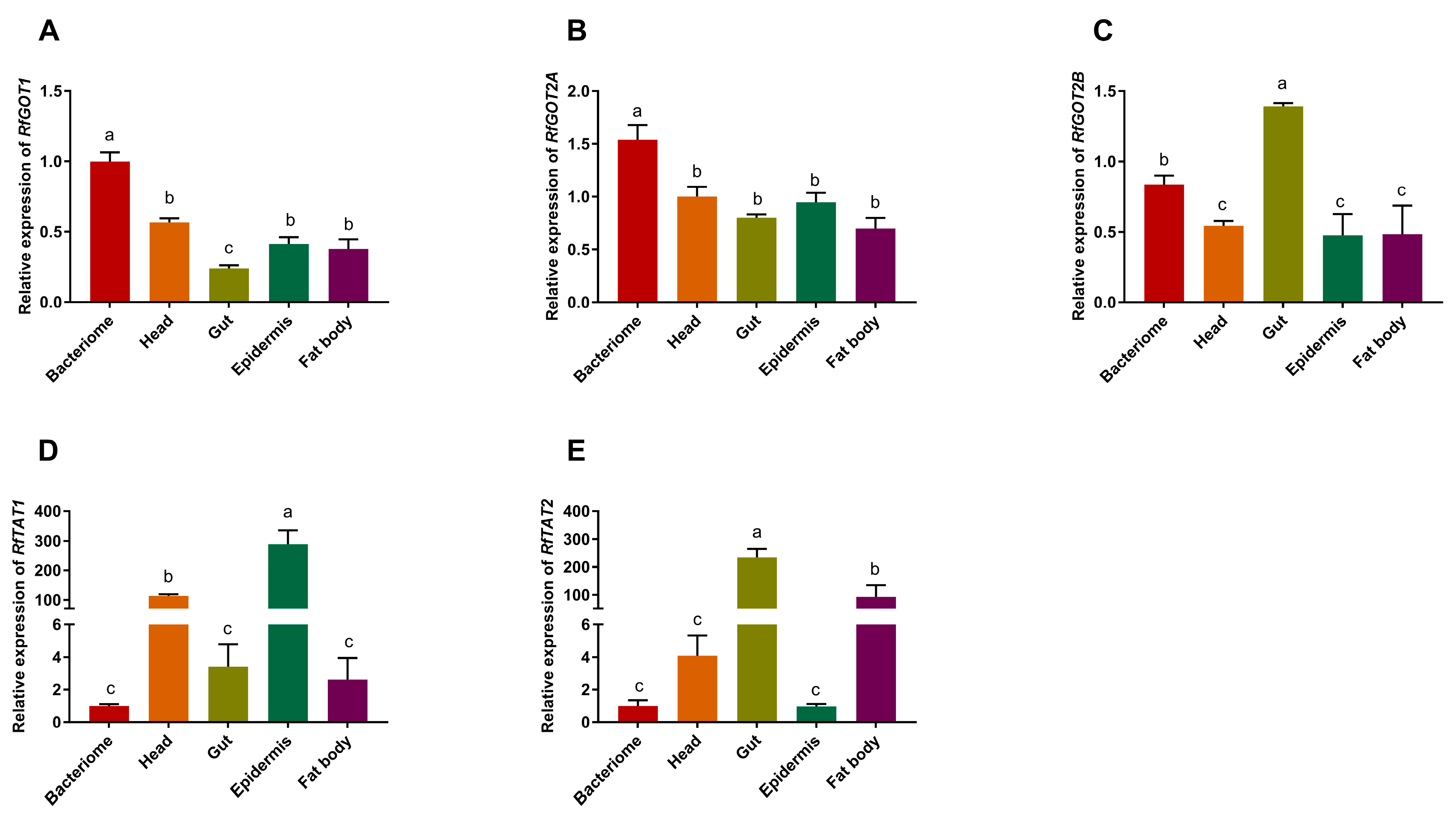
3.2. Analysis of the Relative Expression of RPWTyrosine Transaminase Genes
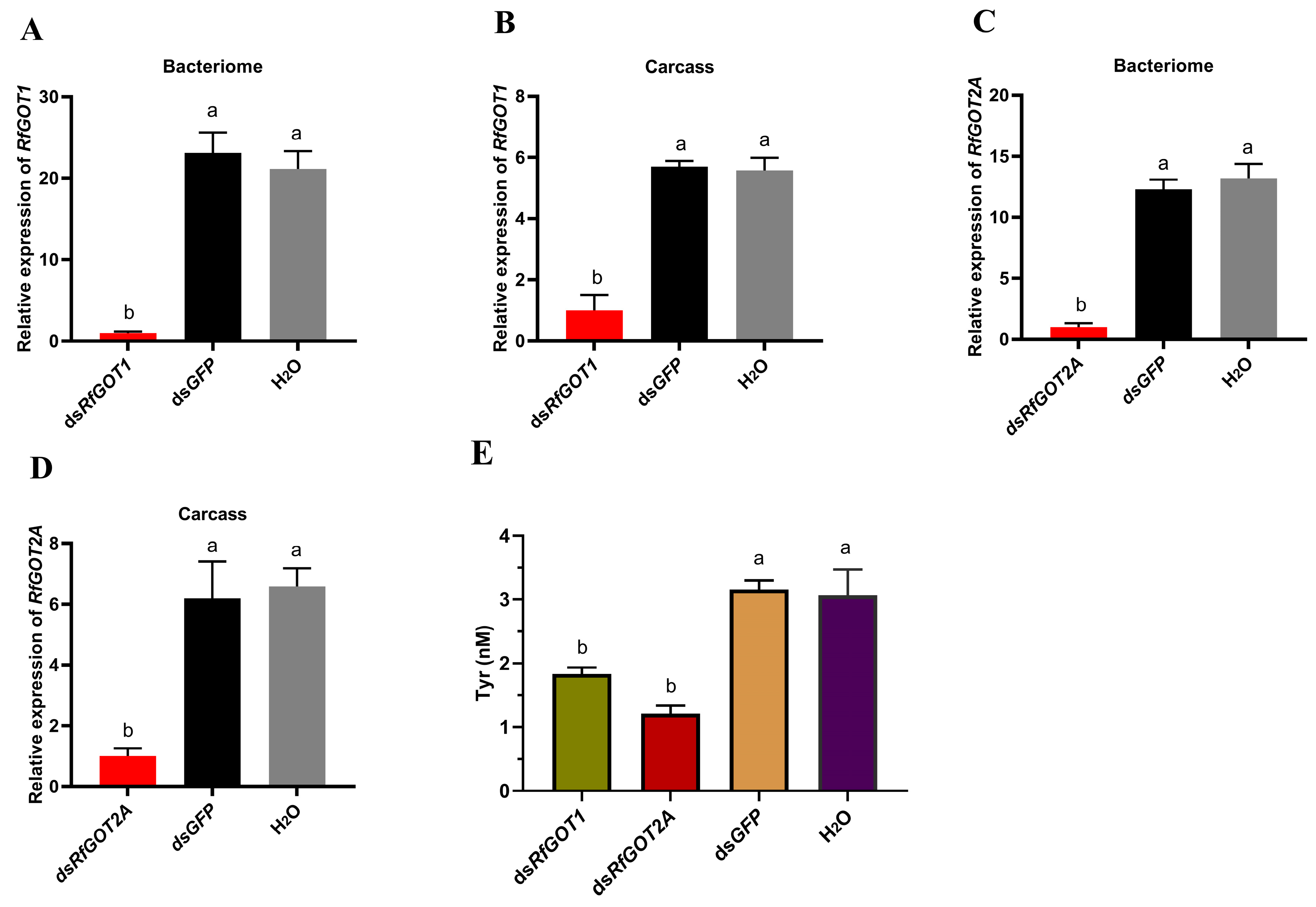
3.3. Effect of RfGOT1 and RfGOT2A Silencing by RNAi in RPW
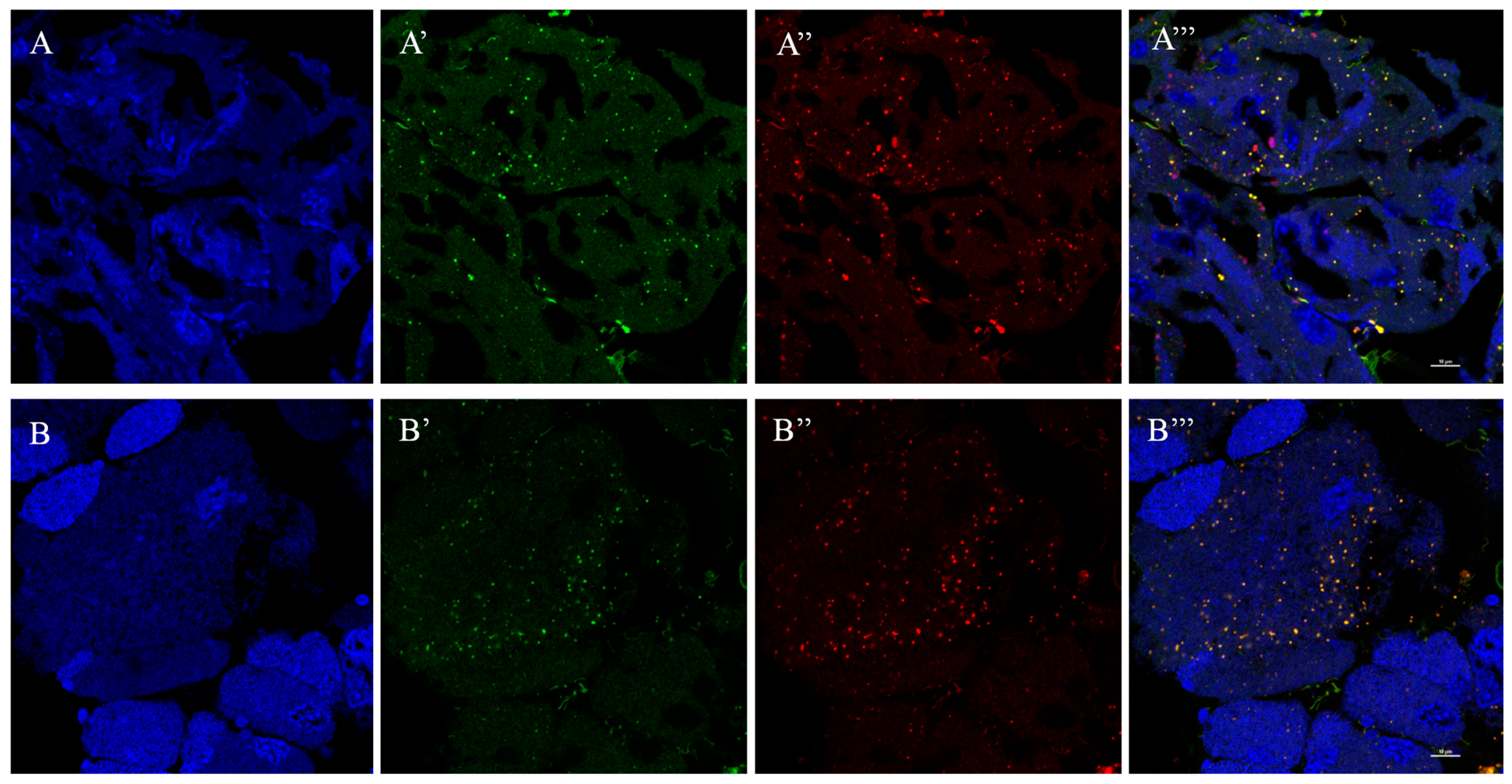
3.4. Localization of RfGOT1 and RfGOT2A in the Bacteriocyte
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Douglas, A.E. Housing microbial symbionts: Evolutionary origins and diversification of symbiotic organs in animals. Philos. Trans. R. Soc. B-Biol. Sci. 2020, 375, 20190603. [Google Scholar] [CrossRef] [PubMed]
- Alarcón, M.E.; Polo, P.G.; Akyüz, S.N.; Rafiqi, A.M. Evolution and ontogeny of bacteriocytes in insects. Front. Physiol. 2022, 13, 1034066. [Google Scholar] [CrossRef] [PubMed]
- Rio, R.; Attardo, G.M.; Weiss, B.L. Grandeur alliances: Symbiont metabolic integration and obligate arthropod hematophagy. Trends Parasitol. 2016, 32, 739–749. [Google Scholar] [CrossRef]
- Gupta, A.; Nair, S. Dynamics of insect-microbiome interaction influence host and microbial symbiont. Front. Microbiol. 2020, 11, 1357. [Google Scholar] [CrossRef] [PubMed]
- Schwartzman, J.A.; Koch, E.; Heath-Heckman, E.A.C.; Zhou, L.; Kremer, N.; Mcfall-Ngai, M.J.; Ruby, E.G. The chemistry of negotiation: Rhythmic, glycan-driven acidification in a symbiotic conversation. Proc. Natl. Acad. Sci. USA 2015, 112, 566–571. [Google Scholar] [CrossRef] [PubMed]
- Mckenna, D.D.; Sequeira, A.S.; Marvaldi, A.E.; Farrell, B.D. Temporal lags and overlap in the diversification of weevils and flowering plants. Proc. Natl. Acad. Sci. USA 2009, 106, 7083–7088. [Google Scholar] [CrossRef] [PubMed]
- Xu, J.; Tan, J.B.; Li, Y.D.; Xu, Y.H.; Tang, A.; Zhou, H.K.; Shi, P.Q. Diversity and dynamics of endosymbionts in a single population of sweet potato weevil, Cylas formicarius (Coleoptera: Brentidae): A preliminary study. J. Insect Sci. 2023, 23, 13. [Google Scholar] [CrossRef] [PubMed]
- Maire, J.; Chouaia, B.; Zaidman-Rémy, A.; Heddi, A. Endosymbiosis morphological reorganization during metamorphosis diverges in weevils. Commun. Integr. Biol. 2020, 13, 184–188. [Google Scholar] [CrossRef]
- Hosokawa, T.; Koga, R.; Tanaka, K.; Moriyama, M.; Anbutsu, H.; Fukatsu, T. Nardonella endosymbionts of Japanese pest and non-pest weevils (Coleoptera: Curculionidae). Appl. Entomol. Zoolog. 2015, 50, 223–229. [Google Scholar] [CrossRef]
- Palmieri, L.; Pavarini, R.; Sharma, P.P. Draft genome sequence of “Candidatus Nardonella dryophthoridicola” strain narmhe1, endosymbiont of Metamasius hemipterus (Coleoptera, Curculionidae, Dryophthorinae). Microbiol. Resour. Ann. 2022, 11, e73822. [Google Scholar] [CrossRef]
- Anbutsu, H.; Moriyama, M.; Nikoh, N.; Hosokawa, T.; Futahashi, R.; Tanahashi, M.; Meng, X.; Kuriwada, T.; Mori, N.; Oshima, K.; et al. Small genome symbiont underlies cuticle hardness in beetles. Proc. Natl. Acad. Sci. USA 2017, 114, e8382–e8391. [Google Scholar] [CrossRef] [PubMed]
- Kanyile, S.N.; Engl, T.; Heddi, A.; Kaltenpoth, M. Endosymbiosis allows Sitophilus oryzae to persist in dry conditions. Front. Microbiol. 2023, 14, 1199370. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Rajabi, H.; Ghoroubi, N.; Lin, C.; Gorb, S.N. Biomechanical strategies underlying the robust body armour of an aposematic weevil. Front. Physiol. 2018, 9, 1410. [Google Scholar] [CrossRef] [PubMed]
- Wang, G.; Zhou, Y.; Tang, B.; Ali, H.; Hou, Y. Immune function differences between two color morphs of the red palm weevil Rhynchophorus ferrugineus (Coleoptera: Curculionidae) at different life stages. Ecol. Evol. 2021, 11, 5702–5712. [Google Scholar] [CrossRef] [PubMed]
- Douglas, A.E. Phloem-sap feeding by animals: Problems and solutions. J. Exp. Bot. 2006, 57, 747–754. [Google Scholar] [CrossRef] [PubMed]
- Douglas, A.E. Molecular dissection of nutrient exchange at the insect-microbial interface. Curr. Opin. Insect Sci. 2014, 4, 23–28. [Google Scholar] [CrossRef] [PubMed]
- Luan, J.B.; Chen, W.; Hasegawa, D.K.; Simmons, A.M.; Wintermantel, W.M.; Ling, K.S.; Fei, Z.; Liu, S.S.; Douglas, A.E. Metabolic coevolution in the bacterial symbiosis of whiteflies and related plant sap-feeding insects. Genome Biol. Evol. 2015, 7, 2635–2647. [Google Scholar] [CrossRef] [PubMed]
- Szabo, G.; Schulz, F.; Toenshoff, E.R.; Volland, J.M.; Finkel, O.M.; Belkin, S.; Horn, M. Convergent patterns in the evolution of mealybug symbioses involving different intrabacterial symbionts. ISME J. 2017, 11, 715–726. [Google Scholar] [CrossRef]
- Klein, A.; Schrader, L.; Gil, R.; Manzano-Marín, A.; Flórez, L.; Wheeler, D.; Werren, J.H.; Latorre, A.; Heinze, J.; Kaltenpoth, M.; et al. A novel intracellular mutualistic bacterium in the invasive ant Cardiocondyla obscurior. ISME J. 2016, 10, 376–388. [Google Scholar] [CrossRef]
- Simonet, P.; Gaget, K.; Parisot, N.; Duport, G.; Rey, M.; Febvay, G.; Charles, H.; Callaerts, P.; Colella, S.; Calevro, F. Disruption of phenylalanine hydroxylase reduces adult lifespan and fecundity, and impairs embryonic development in parthenogenetic pea aphids. Sci. Rep. 2016, 6, 34321. [Google Scholar] [CrossRef]
- Russell, C.W.; Bouvaine, S.; Newell, P.D.; Douglas, A.E. Shared metabolic pathways in a coevolved insect-bacterial symbiosis. Appl. Environ. Microbiol. 2013, 79, 6117–6123. [Google Scholar] [CrossRef] [PubMed]
- Mccutcheon, J.P.; von Dohlen, C.D. An interdependent metabolic patchwork in the nested symbiosis of mealybugs. Curr. Biol. 2011, 21, 1366–1372. [Google Scholar] [CrossRef] [PubMed]
- Husnik, F.; Nikoh, N.; Koga, R.; Ross, L.; Duncan, R.P.; Fujie, M.; Tanaka, M.; Satoh, N.; Bachtrog, D.; Wilson, A.C.; et al. Horizontal gene transfer from diverse bacteria to an insect genome enables a tripartite nested mealybug symbiosis. Cell 2013, 153, 1567–1578. [Google Scholar] [CrossRef] [PubMed]
- Duncan, R.P.; Husnik, F.; Van Leuven, J.T.; Gilbert, D.G.; Davalos, L.M.; Mccutcheon, J.P.; Wilson, A.C. Dynamic recruitment of amino acid transporters to the insect/symbiont interface. Mol. Ecol. 2014, 23, 1608–1623. [Google Scholar] [CrossRef] [PubMed]
- Feng, H.; Edwards, N.; Anderson, C.M.H.; Althaus, M.; Duncan, R.P.; Hsu, Y.; Luetje, C.W.; Price, D.R.G.; Wilson, A.C.C.; Thwaites, D.T. Trading amino acids at the aphid-buchnera symbiotic interface. Proc. Natl. Acad. Sci. USA 2019, 116, 16003–16011. [Google Scholar] [CrossRef] [PubMed]
- Ribeiro, C.W.; Baldacci-Cresp, F.; Pierre, O.; Larousse, M.; Benyamina, S.; Lambert, A.; Hopkins, J.; Castella, C.; Cazareth, J.; Alloing, G.; et al. Regulation of differentiation of nitrogen-fixing bacteria by microsymbiont targeting of plant thioredoxin s1. Curr. Biol. 2017, 27, 250–256. [Google Scholar] [CrossRef] [PubMed]
- Nakabachi, A.; Ishida, K.; Hongoh, Y.; Ohkuma, M.; Miyagishima, S.Y. Aphid gene of bacterial origin encodes a protein transported to an obligate endosymbiont. Curr. Biol. 2014, 24, R640–R641. [Google Scholar] [CrossRef]
- Bublitz, D.C.; Chadwick, G.L.; Magyar, J.S.; Sandoz, K.M.; Brooks, D.M.; Mesnage, S.; Ladinsky, M.S.; Garber, A.I.; Bjorkman, P.J.; Orphan, V.J.; et al. Peptidoglycan production by an insect-bacterial mosaic. Cell 2019, 179, 703–712. [Google Scholar] [CrossRef]
- Nurashikin-Khairuddin, W.; Abdul-Hamid, S.N.A.; Mansor, M.S.; Bharudin, I.; Othman, Z.; Jalinas, J. A review of entomopathogenic nematodes as a biological control agent for red palm weevil, Rhynchophorus ferrugineus (Coleoptera: Curculionidae). Insects 2022, 13, 245. [Google Scholar] [CrossRef]
- Hazzouri, K.M.; Sudalaimuthuasari, N.; Kundu, B.; Nelson, D.; Al-Deeb, M.A.; Le Mansour, A.; Spencer, J.J.; Desplan, C.; Amiri, K.M.A. The genome of pest Rhynchophorus ferrugineus reveals gene families important at the plant-beetle interface. Commun. Biol. 2020, 3, 323. [Google Scholar] [CrossRef]
- Chouaia, B.; Montagna, M.; Suma, P.; Faoro, F. Complete genome sequence of Rhynchophorus ferrugineus endocytobiont “Candidatus Nardonella dryophthoridicola” strain nardrf. Microbiol. Resour. Ann. 2021, 10, e35521. [Google Scholar] [CrossRef] [PubMed]
- Edgar, R.C. Muscle: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004, 32, 1792–1797. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Tamura, K. Mega7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Zimmerman, S.G.; Peters, N.C.; Altaras, A.E.; Berg, C.A. Optimized RNA ISH, RNA FISH and protein-RNA double labeling (IF/FISH) in Drosophila ovaries. Nat. Protoc. 2013, 8, 2158–2179. [Google Scholar] [CrossRef] [PubMed]
- Wei, T.; Kikuchi, A.; Moriyasu, Y.; Suzuki, N.; Shimizu, T.; Hagiwara, K.; Chen, H.; Takahashi, M.; Ichiki-Uehara, T.; Omura, T. The spread of rice dwarf virus among cells of its insect vector exploits virus-induced tubular structures. J. Virol. 2006, 80, 8593–8602. [Google Scholar] [CrossRef] [PubMed]
- Perreau, J.; Moran, N.A. Genetic innovations in animal-microbe symbioses. Nat. Rev. Genet. 2022, 23, 23–39. [Google Scholar] [CrossRef] [PubMed]
- Van de Velde, W.; Zehirov, G.; Szatmari, A.; Debreczeny, M.; Ishihara, H.; Kevei, Z.; Farkas, A.; Mikulass, K.; Nagy, A.; Tiricz, H.; et al. Plant peptides govern terminal differentiation of bacteria in symbiosis. Science 2010, 327, 1122–1126. [Google Scholar] [CrossRef]
- Guefrachi, I.; Pierre, O.; Timchenko, T.; Alunni, B.; Barrière, Q.; Czernic, P.; Villaécija-Aguilar, J.A.; Verly, C.; Bourge, M.; Fardoux, J.; et al. Bradyrhizobium Bcla is a peptide transporter required for bacterial differentiation in symbiosis with Aeschynomene legumes. Mol. Plant-Microbe Interact. 2015, 28, 1155–1166. [Google Scholar] [CrossRef]
- Haag, A.F.; Baloban, M.; Sani, M.; Kerscher, B.; Pierre, O.; Farkas, A.; Longhi, R.; Boncompagni, E.; Hérouart, D.; Dall’Angelo, S.; et al. Protection of Sinorhizobium against host cysteine-rich antimicrobial peptides is critical for symbiosis. PLoS Biol. 2011, 9, e1001169. [Google Scholar] [CrossRef]
- Mattiuzzo, M.; Bandiera, A.; Gennaro, R.; Benincasa, M.; Pacor, S.; Antcheva, N.; Scocchi, M. Role of the Escherichia coli sbma in the antimicrobial activity of proline-rich peptides. Mol. Microbiol. 2007, 66, 151–163. [Google Scholar] [CrossRef]
- Poliakov, A.; Russell, C.W.; Ponnala, L.; Hoops, H.J.; Sun, Q.; Douglas, A.E.; van Wijk, K.J. Large-scale label-free quantitative proteomics of the pea aphid-buchnera symbiosis. Mol. Cell. Proteom. 2011, 10, M110.007039. [Google Scholar] [CrossRef] [PubMed]
- Nowack, E.C.; Grossman, A.R. Trafficking of protein into the recently established photosynthetic organelles of Paulinella chromatophora. Proc. Natl. Acad. Sci. USA 2012, 109, 5340–5345. [Google Scholar] [CrossRef] [PubMed]
- Singer, A.; Poschmann, G.; Mühlich, C.; Valadez-Cano, C.; Hänsch, S.; Hüren, V.; Rensing, S.A.; Stühler, K.; Nowack, E. Massive protein import into the early-evolutionary-stage photosynthetic organelle of the amoeba Paulinella chromatophora. Curr. Biol. 2017, 27, 2763–2773. [Google Scholar] [CrossRef] [PubMed]
- Oberleitner, L.; Poschmann, G.; Macorano, L.; Schott-Verdugo, S.; Gohlke, H.; Stühler, K.; Nowack, E. The puzzle of metabolite exchange and identification of putative octotrico peptide repeat expression regulators in the nascent photosynthetic organelles of Paulinella chromatophora. Front. Microbiol. 2020, 11, 607182. [Google Scholar] [CrossRef]
- Wilson, A.C.; Duncan, R.P. Signatures of host/symbiont genome coevolution in insect nutritional endosymbioses. Proc. Natl. Acad. Sci. USA 2015, 112, 10255–10261. [Google Scholar] [CrossRef]
- Duncan, R.P.; Anderson, C.M.H.; Thwaites, D.T.; Luetje, C.W.; Wilson, A.C.C. Co-option of a conserved host glutamine transporter facilitates aphid/buchnera metabolic integration. Proc. Natl. Acad. Sci. USA 2023, 120, e2308448120. [Google Scholar] [CrossRef]


| Primer | Sequence (5′–3′) | Products (bp) |
|---|---|---|
| For RT-qPCR | ||
| RfGOT1-F | CTTCTGGAGACCTGGATAA | 155 |
| RfGOT1-R | CACTTCACTTGGGTTGTTC | |
| RfGOT2A-F | GGTTCAACTAAGAGTTGGG | 180 |
| RfGOT2A-R | TACAGCATGGGCCAGATAG | |
| RfGOT2B-F | CTTTCACGCAGTTGCTCAC | 164 |
| RfGOT2B-R | ATCGTACCGCCCATACATC | |
| RfTAT1-F | CAAAGCGGTTGCAGAATAT | 151 |
| RfTAT1-R | GGGTCTGGGTAGCAGAATG | |
| RfTAT2-F | AGGACTGACGGCGTATCTG | 181 |
| RfTAT2-R | TGGCGTGATTACGATTCTG | |
| RfActin-F | CCAAGGGAGCCAAGCAATT | 163 |
| RfActin-R | CGCTGATGCCCCTATGTATGT | |
| For dsRNA synthesis | ||
| dsRfGOT1-F | taatacgactcactatagggAGGAGTTGGAGCTTATCGCA | |
| dsRfGOT1-R | taatacgactcactatagggGTTGGATCACATCCGGTAGG | |
| dsRfGOT2A-F | taatacgactcactatagggGTGGGAGCCTATCGAGATGA | |
| dsRfGOT2A-R | taatacgactcactatagggTGCTGGGATCAACACCTGTA | |
| dsGFP-F | taatacgactcactatagggCAGTGCTTCAGCCGCTAC | |
| dsGFP-R | taatacgactcactatagggGTTCACCTGCCGTTCTTGA | |
| For fluorescence in situ hybridization | ||
| NarYahi933R | AATCTTGCGATCGTACTTCT | |
| NarYahi1308R | TTCTCGCGAAATTGCTTCT | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Huang, Y.; Feng, Z.-F.; Li, F.; Hou, Y.-M. Host-Encoded Aminotransferase Import into the Endosymbiotic Bacteria Nardonella of Red Palm Weevil. Insects 2024, 15, 35. https://doi.org/10.3390/insects15010035
Huang Y, Feng Z-F, Li F, Hou Y-M. Host-Encoded Aminotransferase Import into the Endosymbiotic Bacteria Nardonella of Red Palm Weevil. Insects. 2024; 15(1):35. https://doi.org/10.3390/insects15010035
Chicago/Turabian StyleHuang, Ying, Zhen-Feng Feng, Fan Li, and You-Ming Hou. 2024. "Host-Encoded Aminotransferase Import into the Endosymbiotic Bacteria Nardonella of Red Palm Weevil" Insects 15, no. 1: 35. https://doi.org/10.3390/insects15010035
APA StyleHuang, Y., Feng, Z.-F., Li, F., & Hou, Y.-M. (2024). Host-Encoded Aminotransferase Import into the Endosymbiotic Bacteria Nardonella of Red Palm Weevil. Insects, 15(1), 35. https://doi.org/10.3390/insects15010035






