Functional Analysis of Amino Acid Transporter Genes ACYPI000536 and ACYPI004320 in Acyrthosiphon pisum
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Insect Culture
2.2. Plant Cultivation
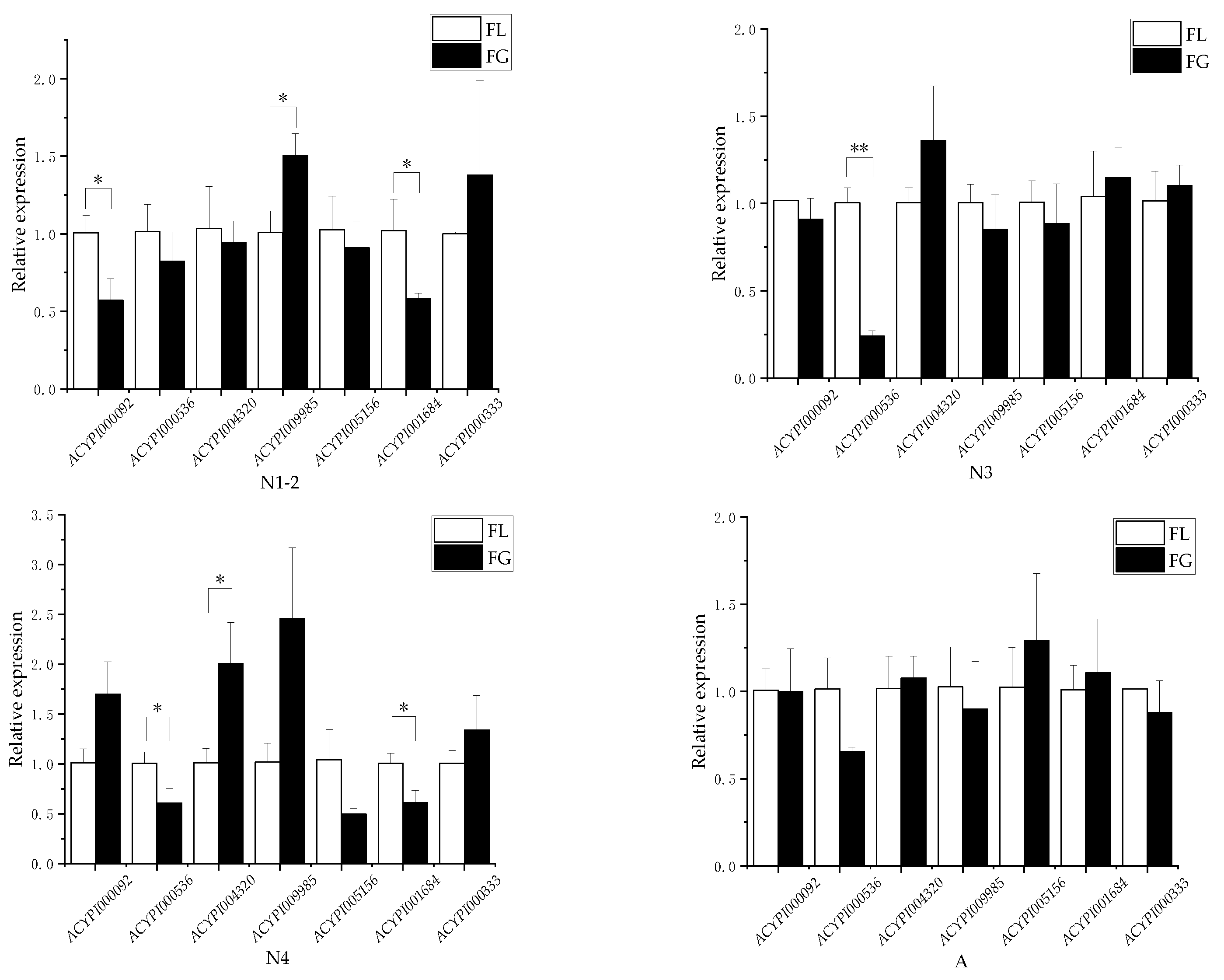
2.3. Differential Gene Expression Analysis of Amino Acid Transporter in Pea Aphids
2.4. Quantitative Real-Time PCR Analysis of Relative Expression Levels
2.5. Synthetic dsRNA
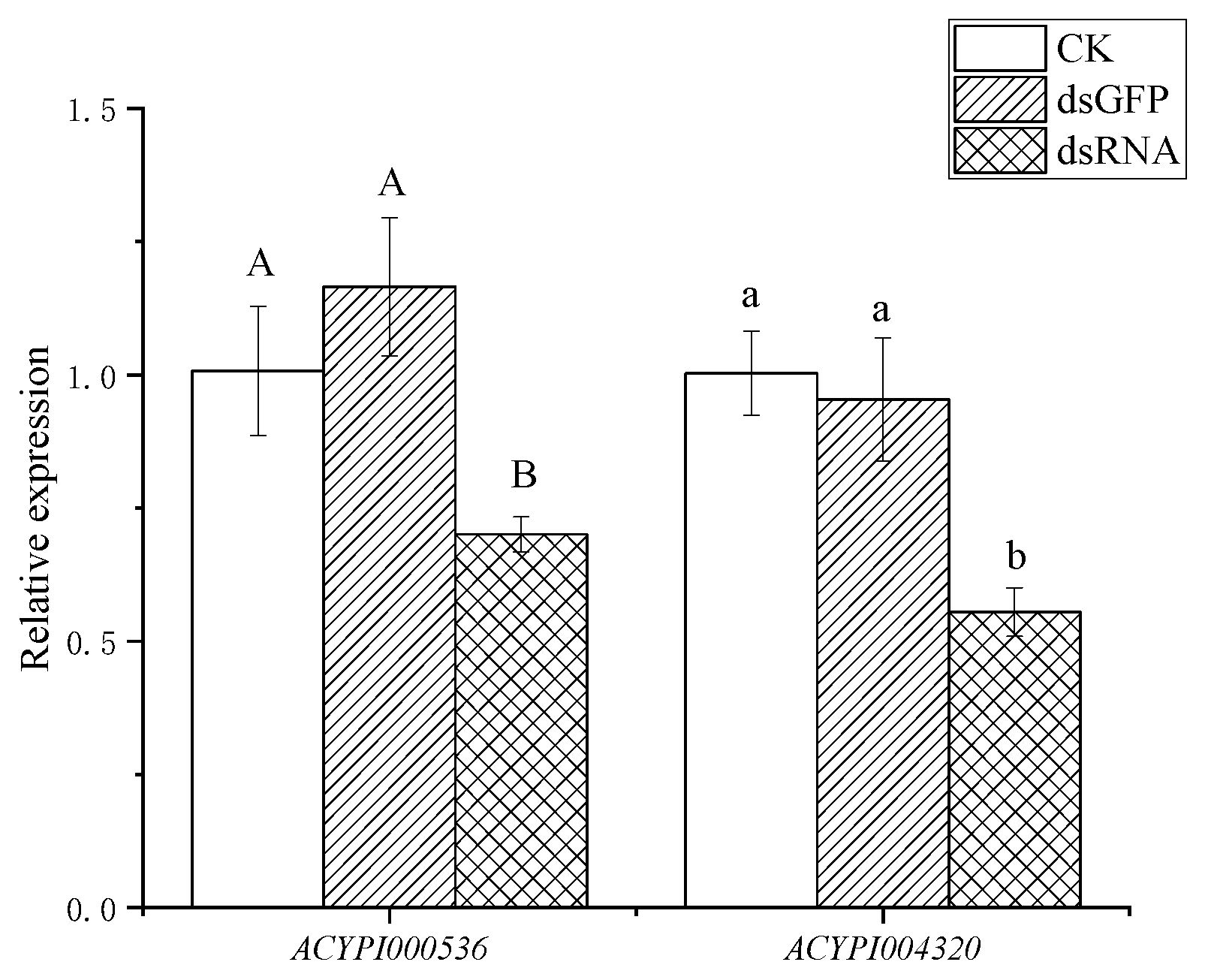
2.6. RNA Interference
2.7. Determination of the Sensitivity of Pea Aphids to “Gannong 5” and “Lie Renhe” Alfalfa after RNA Interference
2.8. Determination of Free Amino Acid Content of Pea Aphids after RNA Interference
2.8.1. Sample Preparation
2.8.2. Determination of Free Amino Acids
Flow Phase Configuration
Instrument Conditions
- (1)
- Chromatographic column: HILIC-Z, 2.7 μm, 2.1 mm × 150 mm, or chromatographic column with the same performance;
- (2)
- The specific elution procedure of mobile phase is shown in the Table 3.
- (3)
- Flow rate: 0.5 mL/min;
- (4)
- Column temperature: 25 °C; injection volume: 1 μL.
- (1)
- Scanning mode: positive ion mode;
- (2)
- Detection method: multiple reaction monitoring (MRM);
- (3)
- Drying gas (N2) temperature: 230 °C;
- (4)
- Dry gas flow rate: 11.0 L/min;
- (5)
- Sheath gas temperature: 390 °C;
- (6)
- Sheath gas flow rate: 12.0 L/min;
- (7)
- Atomizing gas (N2) pressure: 0.14 MPa (20 psi);
- (8)
- Capillary voltage: 1500 V.
3. Results
3.1. Effects of Resistant and Susceptible Alfalfa Varieties on Differential Genes of Amino Acid Transporters in Pea Aphids
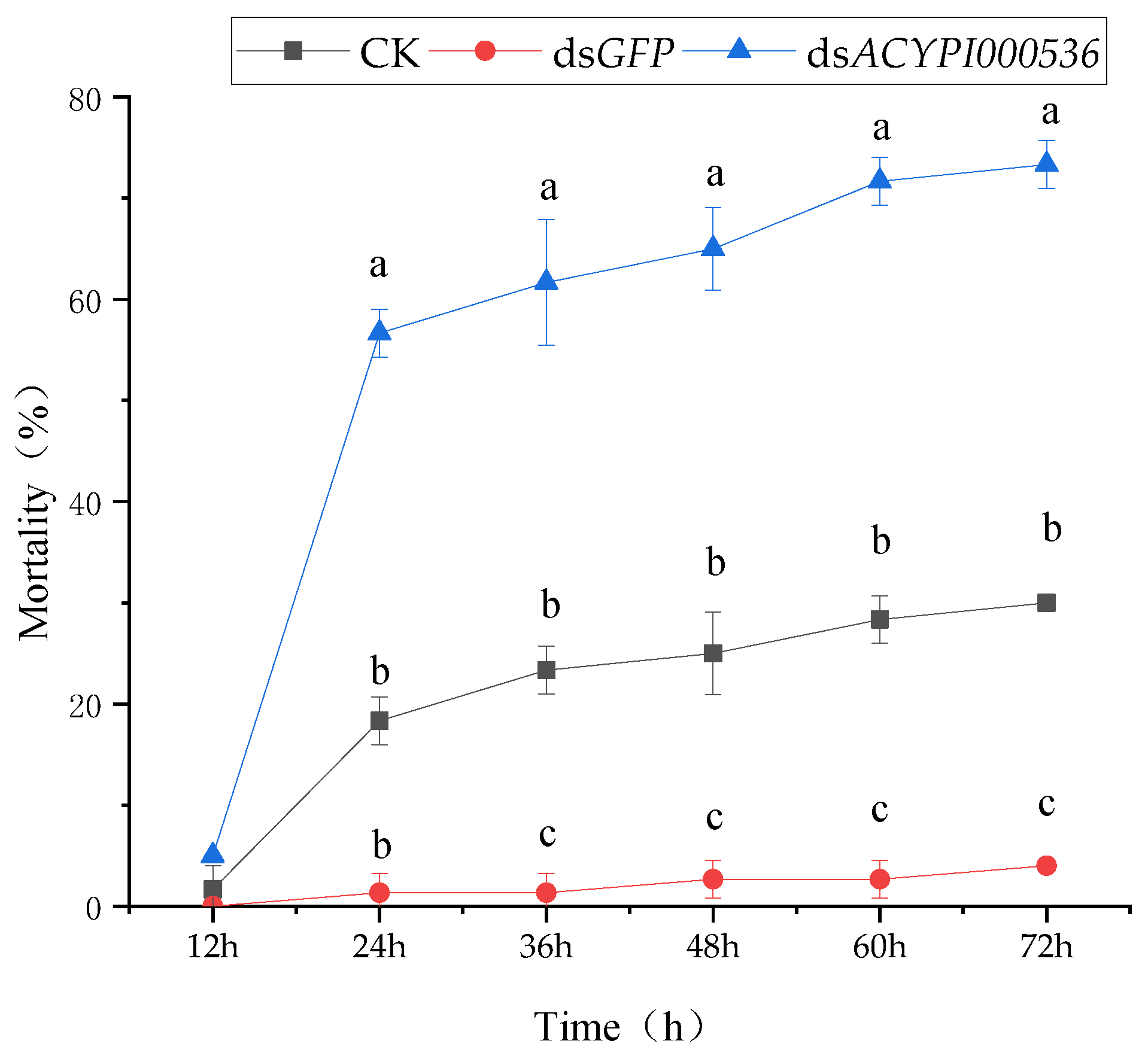
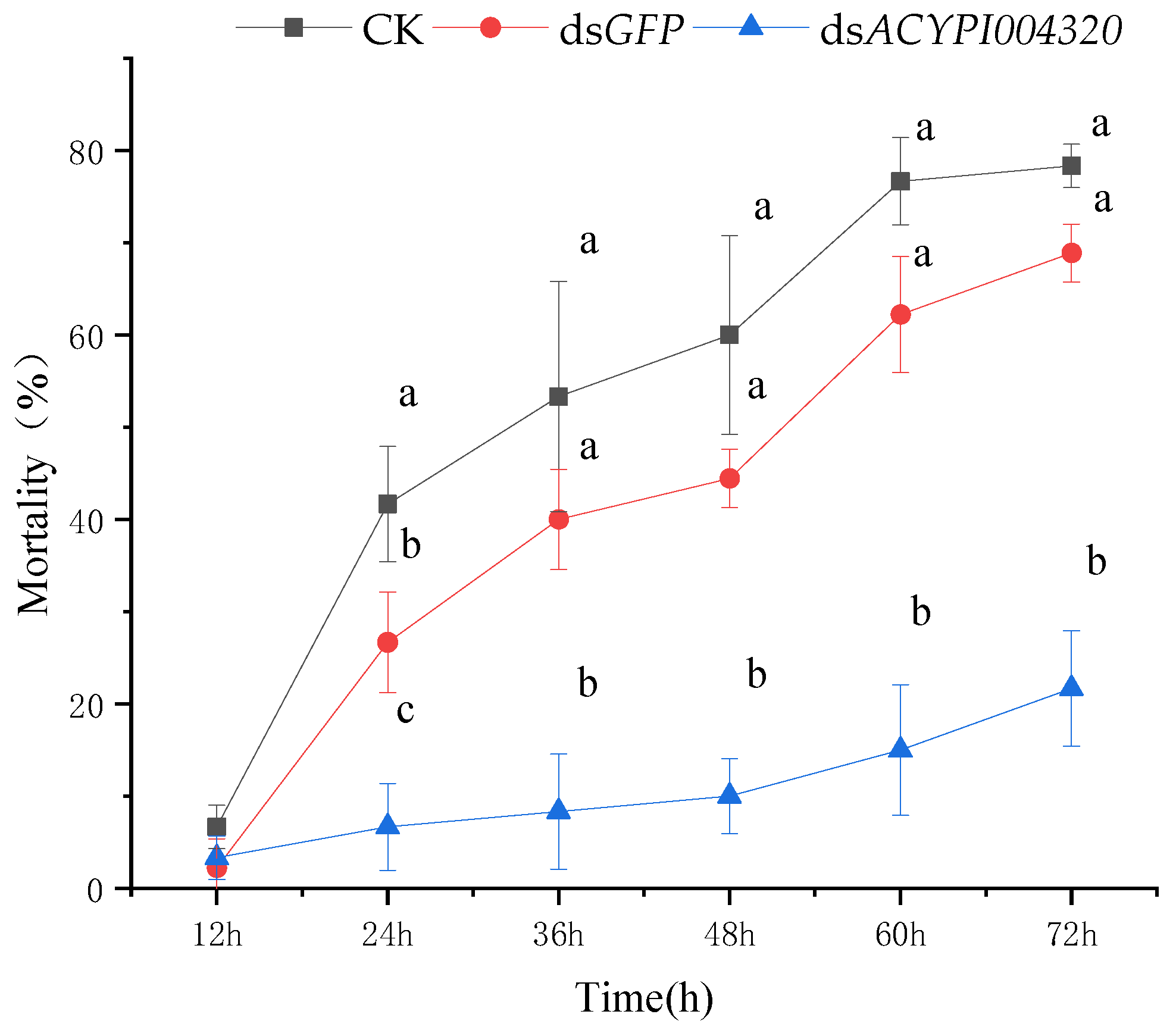
3.2. Effect of RNA Interference on the Survival of Pea Aphids
3.3. Determination of Free Amino Acid Content of Pea Aphids after RNA Interference
3.4. Determination of Pea Aphid Sensitivity to Susceptible Alfalfa Varieties after RNA Interference
3.4.1. The Sensitivity of Pea Aphids to “Lie Renhe” Alfalfa after Interfering with ACYPI000536 Was Determined
3.4.2. The Sensitivity of Pea Aphids to “Gannong 5” Alfalfa after Interfering with ACYPI004320 Was Determined
4. Discussion
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Harmon, J.P.; Moran, N.A.; Ives, A.R. Species response to environmental change: Impacts of food web interactions and evolution. Science 2009, 323, 1347–1350. [Google Scholar] [CrossRef] [PubMed]
- Ma, Y.L.; Liu, C.Z. Review on ecological characteristics and control of aphids. Pratacult. Sci. 2014, 31, 519–525. [Google Scholar]
- Gamalath, N.S.; Tufail, M.; Sharma, P.N.; Mori, N.; Takeda, M.; Nakamura, C. Differential expression of vitellogenin mRNA and protein in response to rice resistance genes in two strains of Nilaparvata lugens (Hemiptera: Delphacidae) with different levels of virulence. Appl. Entomol. Zool. 2012, 47, 9–16. [Google Scholar] [CrossRef]
- Wang, S.S.; Zhu, Y.L.; Song, L.W.; Da, L.T. Field Evaluation of resistance of different alfalfa varieties to Acyrthosiphon pisum Harris. Acta Agrestia Sin. 2014, 22, 1139–1142. [Google Scholar]
- Wei, J.W.; Liu, L.; Wang, S.S.; Chen, L.X.; Jiang, M.J.; Wang, G.H.; Xu, H.H. Effects of resistant and susceptible alfalfa varieties on adaptability and enzyme activity of pea aphid. Acta Agrestia Sin. 2022, 30, 1171–1177. [Google Scholar]
- Hansen, A.K.; Moran, N.A. Aphid genome expression reveals host–symbiont cooperation in the production of amino acids. Proc. Natl. Acad. Sci. USA 2011, 108, 2849–2854. [Google Scholar] [CrossRef] [PubMed]
- Boudko, D.Y. Molecular basis of essential amino acid transport from studies of insect nutrient amino acid transporters of the SLC6 family (NAT-SLC6). J. Insect Physiol. 2012, 58, 433–449. [Google Scholar] [CrossRef] [PubMed]
- Douglas, A.E. The nutritional physiology of aphids. Adv. Insect Physiol. 2003, 31, 73–140. [Google Scholar]
- Castagna, M.; Shayakul, C.; Trotti, D.; Sacchi, V.F.; Hediger, M.A. Molecular characteristics of mammalian and insect amino acid transporters: Implications for amino acid homeostasis. J. Exp. Biol. 1997, 200 Pt 2, 269–286. [Google Scholar] [CrossRef]
- Wieczorek, H.; Putzenlechner, M.; Zeiske, W.; Klein, U. A vacuolar-type proton pump energizes K+/H+ antiport in an animal plasma membrane. J. Biol. Chem. 1991, 266, 15340. [Google Scholar] [CrossRef]
- Boudko, D.Y.; Tsujimoto, H.; Rodriguez, S.D.; Meleshkevitch, E.A.; Price, D.P.; Drake, L.L.; Hansen, I.A. Substrate specificity and transport mechanism of amino-acid transceptor Slimfast from Aedes aegypti. Nat. Commun. 2015, 6, 8546. [Google Scholar] [CrossRef]
- Goberdhan, D.C.; Meredith, D.; Boyd, C.R.; Wilson, C. PAT-related amino acid transporters regulate growth via a novel mechanism that does not require bulk transport of amino acids. Development 2005, 132, 2365–2375. [Google Scholar] [CrossRef] [PubMed]
- Attardo, G.M.; Hansen, I.A.; Shiao, S.H.; Raikhel, A.S. Identification of two cationic amino acid transporters required for nutritional signaling during mosquito reproduction. J. Exp. Biol. 2006, 209, 3071. [Google Scholar] [CrossRef] [PubMed]
- Evans, A.M.; Aimanova, K.G.; Gill, S.S. Characterization of a blood-meal-responsive proton-dependent amino acid transporter in the disease vector, Aedes aegypti. J. Exp. Biol. 2009, 212 Pt 20, 3263–3271. [Google Scholar] [CrossRef]
- Wilson, A. Genome expansion and differential expression of amino acid transporters at the aphid/Buchnera symbiotic interface. Mol. Biol. Evol. 2011, 28, 3113. [Google Scholar]
- Duncan, R.P.; Nathanson, L.; Wilson, A.C. Novel male-biased expression in paralogs of the aphid slimfast nutrient amino acid transporter expansion. BMC Evol. Biol. 2011, 11, 253. [Google Scholar] [CrossRef] [PubMed]
- Duncan, R.P.; Husnik, F.; Van Leuven, J.T.; Gilbert, D.G.; Dávalos, L.M.; McCutcheon, J.P.; Wilson, A.C. Dynamic recruitment of amino acid transporters to the insect/symbiont interface. Mol. Ecol. 2014, 23, 1608–1623. [Google Scholar] [CrossRef] [PubMed]
- Price, D.R.; Feng, H.; Baker, J.D.; Bavan, S.; Luetje, C.W.; Wilson, A.C. Aphid amino acid transporter regulates glutamine supply to intracellular bacterial symbionts. Proc. Natl. Acad. Sci. USA 2014, 111, 320–325. [Google Scholar] [CrossRef] [PubMed]
- Fu, K.Y.; Guo, W.C.; Ahmat, T.; Li, G.Q. Knockdown of a nutrient amino acid transporter gene ldnat1 reduces free neutral amino acid contents and impairs Leptinotarsa decemlineata pupation. Sci. Rep. 2015, 5, 18124. [Google Scholar] [CrossRef] [PubMed]
- Price, D.R.; Wilson, A.C.; Luetje, C.W. Proton-dependent glutamine uptake by aphid bacteriocyte amino acid transporter apglnt1. Biochim. Biophys. Acta Biomembr. 2015, 1848, 2085–2091. [Google Scholar] [CrossRef][Green Version]
- Dahan, R.A.; Duncan, R.P.; Wilson, A.C.; Dávalos, L.M. Amino acid transporter expansions associated with the evolution of obligate endosymbiosis in sap-feeding insects (Hemiptera: Sternorrhyncha). BMC Evol. Biol. 2015, 15, 52. [Google Scholar] [CrossRef] [PubMed]
- Saier, M.H. Families of transmembrane transporters selective for amino acids and their derivatives. Microbiology 2000, 146, 1775–1795. [Google Scholar] [PubMed]
- He, C. Breeding of a new aphid-resistant alfalfa variety Gannong No.5. Chin. Sci. Technol. Achiev. 2010. [Google Scholar]
- Ye, C.; Jiang, Y.D.; An, X.; Yang, L.; Shang, F.; Niu, J.; Wang, J.J. Effects of RNAi-based silencing of chitin synthase gene on moulting and fecundity in pea aphids (Acyrthosiphon pisum). Sci. Rep. 2019, 9, 3694. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Y. Study on the Interrelation between Pea Aphid and Host Plant. Master’s Thesis, Shihezi University, Shihezi, China, 2014. [Google Scholar]
- GB/T 30987-2020; Determination of Free Amino Acids in Plants. National Standardization Committee (China): Beijing, China, 2020.
- Min, K.; Tatar, M. Restriction of amino acids extends lifespan in Drosophila melanogaster. Mech. Ageing Dev. 2006, 127, 643–646. [Google Scholar] [CrossRef] [PubMed]
- Vrzal, E.; Allan, S.; Hahn, D. Amino acids in nectar enhance longevity of female Culex quinquefasciatus mosquitoes. J. Insect Physiol. 2010, 56, 1659–1664. [Google Scholar] [CrossRef] [PubMed]
- Paoli, P.P.; Donley, D.; Stabler, D.; Saseendranath, A.; Nicolson, S.W.; Simpson, S.J.; Wright, G.A. Nutritional balance of essential amino acids and carbohydrates of the adult worker honeybee depends on age. Amino Acids 2014, 46, 1449–1458. [Google Scholar] [CrossRef]
| Primer Name | Forward Primer | Reverse Primer |
|---|---|---|
| RPS20 | AAGTGTGTGCTCCGAGATGA | CAGCAATGACACCGGGTTC |
| RPL 7 | TTGAAGAGCGTAAGGGAACT | TATTGGTGATTGGAATGCGTTG |
| ACYPI004320 | GTTAGTACTAGCCGGTAAC | GTACTATCTCGGTAAGTGC |
| ACYPI000333 | CACTCTTGTCCTCCTAAC | GATATCAGAGGGTAGACAG |
| ACYPI000536 | GCTTCACTCGCTATATTTGC | TTTATCAACAGGAACGCTGA |
| ACYPI000092 | GTCTTACCATTAGAGCAGG | CCGGTAAGAACTATGAGAC |
| ACYPI001684 | CAAATGAGTCAGAGTCTCG | GGATGACAAGTCCAAGATG |
| ACYPI009985 | CACCACTACTACTACCAC | CAGTCAATCTAGTCTTCCC |
| ACYPI005156 | GCTGATCCTCAGGTATAG | GAGAGTCCCAGTTTACTAC |
| Primer Name | Forward Primer | Reverse Primer |
|---|---|---|
| dsGFP | TAATACGACTCACTATAGGG CAGTTCTTGTTGAATTAGATG | TAATACGACTCACTATAGGG GATTTTGGTTTGTCTCCCATG |
| dsACYPI000536 | TAATACGACTCACTATAGGG TAGCCTTACCGATTCTTC | TAATACGACTCACTATAGGG GAACCTTCAGTGGAATTG |
| dsACYPI04320 | TAATACGACTCACTATAGGG CCGTGTTCTACTTCATTG | TAATACGACTCACTATAGGG ATTATGGAGCCGAGTATC |
| Time/min | Mobile Phase | |
|---|---|---|
| A/% | B/% | |
| 0 | 0 | 100 |
| 11.5 | 30 | 70 |
| 12 | 0 | 100 |
| 30 | 0 | 100 |
| Sample | Mortality/% | |||
|---|---|---|---|---|
| 12 h | 24 h | 36 h | 48 h | |
| CK | 1.67% ± 0.0136a | 2.78% ± 0.0079b | 2.78% ± 0.0079c | 6.11% ± 0.0079a |
| dsGFP | 2.22% ± 0.0079a | 3.33% ± 0.0000b | 3.33% ± 0.0000c | 5.56% ± 0.0079a |
| dsACYPI000536 | 1.11% ± 0.0079a | 2.22% ± 0.0079b | 5.56% ± 0.0079b | 8.33% ± 0.0136a |
| dsACYPI004320 | 2.78% ± 0.0079a | 5.56% ± 0.0079a | 7.22% ± 0.0079a | 8.89% ± 0.0157a |
| Amino Acid | CK (μg/mg) | dsGFP (μg/mg) | dsACYPI000536 (μg/mg) |
|---|---|---|---|
| Thr | 0.3707 ± 0.0233 a | 0.3860 ± 0.0097 a | 0.3614 ± 0.0183 a |
| Phe | 0.3084 ± 0.0121 a | 0.2984 ± 0.0231 a | 0.3057 ± 0.0085 a |
| Leu | 0.2781 ± 0.0044 ab | 0.2986 ± 0.0150 a | 0.2526 ± 0.0067 b |
| Ile | 0.4834 ± 0.0218 a | 0.4497 ± 0.0416 a | 0.4507 ± 0.0286 a |
| Asn | 0.2001 ± 0.0089 b | 0.2107 ± 0.0162 b | 0.3357 ± 0.0181 a |
| Trp | 0.3655 ± 0.0071 a | 0.3550 ± 0.0317 a | 0.3510 ± 0.0182 a |
| Met | 0.3902 ± 0.0144 a | 0.3570 ± 0.0236 a | 0.3605 ± 0.3989 a |
| Pro | 1.2001 ± 0.0144 a | 1.1147 ± 0.1288 a | 1.2603 ± 0.1244 a |
| Val | 0.6750 ± 0.0155 a | 0.6458 ± 0.0307 a | 0.6823 ± 0.0487 a |
| Tyr | 5.2146 ± 0.3226 a | 4.9761 ± 0.1097 a | 3.5015 ± 0.0047 b |
| Cys | 0.3382 ± 0.0273 a | 0.3371 ± 0.0392 a | 0.2933 ± 0.0162 a |
| Ala | 0.7501 ±0.0109 a | 0.7188 ± 0.0217 a | 0.5700 ± 0.0783 b |
| Gly | 0.1348 ± 0.0099 a | 0.1215 ± 0.0113 a | 0.1560 ± 0.0317 a |
| Ser | 0.5416 ± 0.0344 a | 0.5403 ± 0.0336 a | 0.2419 ± 0.0191 b |
| Glu | 0.1961 ± 0.0058 a | 0.1900 ± 0.0215 a | 0.1842 ± 0.0180 a |
| Asp | 0.2669 ± 0.0196 a | 0.2529 ± 0.0057 a | 0.2442 ± 0.0060 a |
| His | 0.3702 ± 0.0090 b | 0.3643 ± 0.0184 b | 0.6279 ± 0.0328 a |
| Cys-Cys | 0.2714 ± 0.0424 a | 0.2716 ± 0.0048 a | 0.2827 ± 0.0280 a |
| Arg | 1.0935 ± 0.1458 a | 0.9939 ± 0.1436 a | 0.9531 ± 0.0722 a |
| Lys | 0.3850 ± 0.0183 b | 0.3538 ± 0.0102 b | 0.5002 ± 0.0395 a |
| Total Content | 13.8340 ± 0.4266 a | 13.1694 ± 0.4213 a | 11.9152 ± 0.5598 b |
| Amino Acid | CK (μg/mg) | dsGFP (μg/mg) | dsACYPI004320 (μg/mg) |
|---|---|---|---|
| Thr | 0.3707 ± 0.0233 a | 0.3860 ± 0.0097 a | 0.3956 ± 0.0204 a |
| Phe | 0.3084 ± 0.0121 b | 0.2984 ± 0.0231 b | 0.3643 ± 0.0172 a |
| Leu | 0.2781 ± 0.0044 a | 0.2986 ± 0.0150 a | 0.2921 ± 0.0091 a |
| Ile | 0.4834 ± 0.0218 a | 0.4497 ± 0.0416 a | 0.4926 ± 0.0055 a |
| Asn | 0.2001 ± 0.0089 b | 0.2107 ± 0.0162 b | 0.2744 ± 0.0323 a |
| Trp | 0.3655 ± 0.0071 a | 0.3550 ± 0.0317 a | 0.3491 ± 0.0129 a |
| Met | 0.3902 ± 0.0144 a | 0.3570 ± 0.0236 a | 0.3497 ± 0.0269 a |
| Pro | 1.2001 ± 0.0144 a | 1.1147 ± 0.1288 a | 1.2373 ± 0.0709 a |
| Val | 0.6750 ± 0.0155 a | 0.6458 ± 0.0307 a | 0.7482 ± 0.0965 a |
| Tyr | 5.2146 ± 0.3226 a | 4.9761 ± 0.1097 a | 3.8781 ± 0.1699 b |
| Cys | 0.3382 ± 0.0273 a | 0.3371 ± 0.0392 a | 0.3374 ± 0.0074 a |
| Ala | 0.7501 ±0.0109 a | 0.7188 ± 0.0217 a | 0.2300 ± 0.0182 b |
| Gly | 0.1348 ± 0.0099 a | 0.1215 ± 0.0113 a | 0.1339 ± 0.0160 a |
| Ser | 0.5416 ± 0.0344 a | 0.5403 ± 0.0336 a | 0.2426 ± 0.0152 b |
| Glu | 0.1961 ± 0.0058 a | 0.1900 ± 0.0215 a | 0.1935 ± 0.0005 a |
| Asp | 0.2669 ± 0.0196 a | 0.2529 ± 0.0057 a | 0.3112 ± 0.0580 a |
| His | 0.3702 ± 0.0090 a | 0.3643 ± 0.0184 a | 0.3755 ± 0.0026 a |
| Cys-Cys | 0.2714 ± 0.0424 a | 0.2716 ± 0.0048 a | 0.3507 ± 0.0533 a |
| Arg | 1.0935 ± 0.1458 a | 0.9939 ± 0.1436 a | 0.7504 ± 0.0133 a |
| Lys | 0.3850 ± 0.0183 a | 0.3538 ± 0.0102 a | 0.3737 ± 0.0238 a |
| Total Content | 13.8340 ± 0.4266 a | 13.1694 ± 0.4213 a | 11.6803 ± 0.2118 b |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yao, L.; Wang, S.; Ma, R.; Wei, J.; Song, L.; Liu, L. Functional Analysis of Amino Acid Transporter Genes ACYPI000536 and ACYPI004320 in Acyrthosiphon pisum. Insects 2024, 15, 20. https://doi.org/10.3390/insects15010020
Yao L, Wang S, Ma R, Wei J, Song L, Liu L. Functional Analysis of Amino Acid Transporter Genes ACYPI000536 and ACYPI004320 in Acyrthosiphon pisum. Insects. 2024; 15(1):20. https://doi.org/10.3390/insects15010020
Chicago/Turabian StyleYao, Lu, Senshan Wang, Rui Ma, Jiangwen Wei, Liwen Song, and Lei Liu. 2024. "Functional Analysis of Amino Acid Transporter Genes ACYPI000536 and ACYPI004320 in Acyrthosiphon pisum" Insects 15, no. 1: 20. https://doi.org/10.3390/insects15010020
APA StyleYao, L., Wang, S., Ma, R., Wei, J., Song, L., & Liu, L. (2024). Functional Analysis of Amino Acid Transporter Genes ACYPI000536 and ACYPI004320 in Acyrthosiphon pisum. Insects, 15(1), 20. https://doi.org/10.3390/insects15010020




