The Effect of Chromosomes on Courtship Behavior in Sibling Species of the Drosophila virilis Group
Abstract
Simple Summary
Abstract
1. Introduction
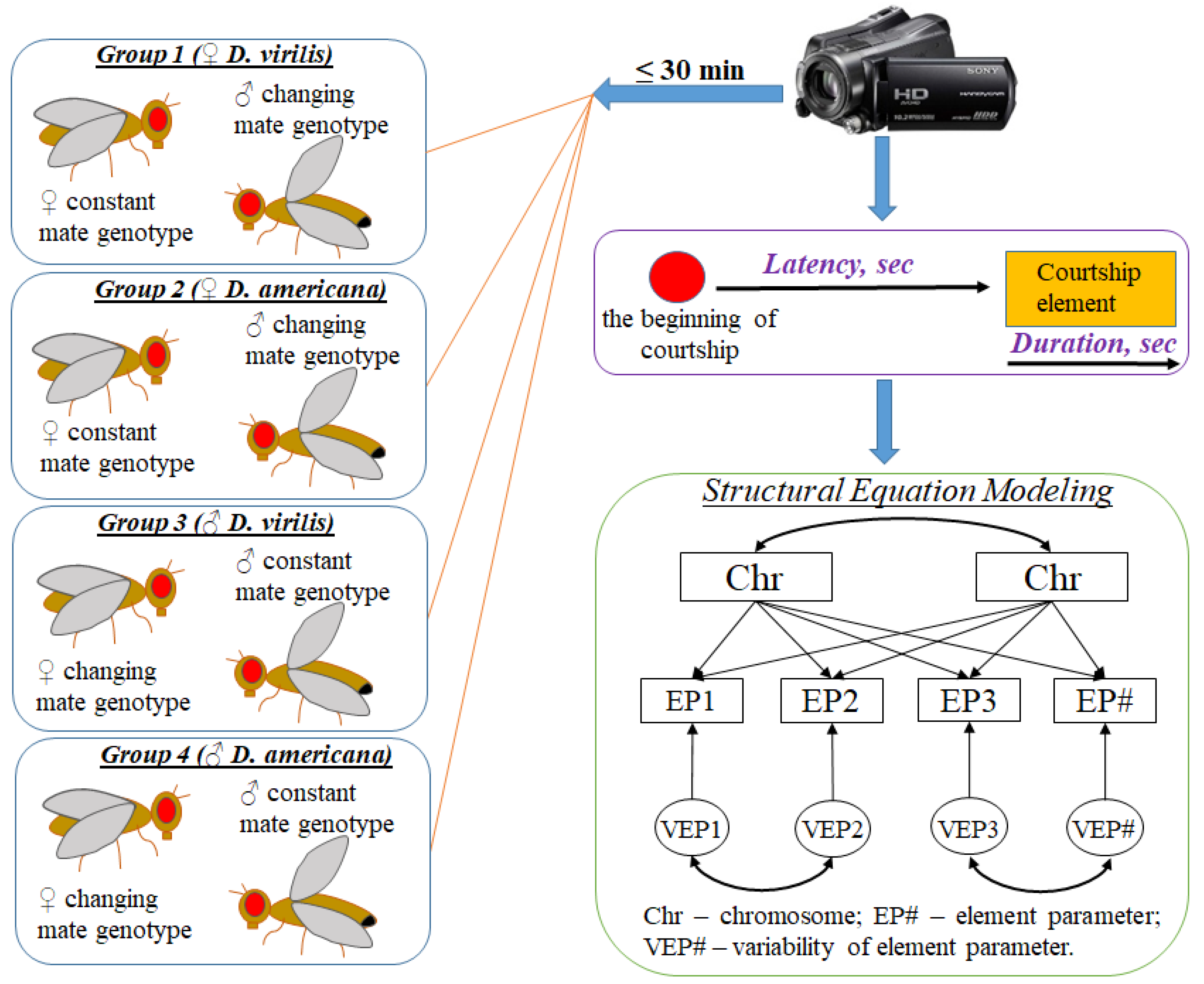
2. Materials and Methods
2.1. Drosophila Stocks and Crosses
2.2. Behavior Tests
2.3. Genetic Analysis of Courtship Traits
3. Results
3.1. Variability of Courtship Elements in Courtship-Partner Pairs with Various Genotype Combinations
3.2. Selection of Optimal Candidate Models
- The Y chromosome carries a very small number of protein-coding genes, just over two dozen, which is significantly fewer than the set of genes present on the X chromosome and autosomes;
- In the case of these two species, only one direction of crosses is possible to obtain F1, namely a female D. americana and a male D. virilis. Consequently, it is not possible to obtain a complete combination of chromosomes from both species, including those with the Y-chromosome of D. americana. Offspring with the corresponding Y chromosome were only obtained from crossing F1 females with D. americana males. It is evident that D. americana males also possessed the Y chromosome of their species. The analysis of the role of sex chromosomes and autosomes included six male genotype combinations, with a single D. americana Y chromosome genotype in Group 1 and two genotypes in Group 2 (Table 1);
- Comparisons of test groups differing in the species origin of the Y chromosome are possible between the male genotypes YAm/X(Am,Vi); A(Am,Vi)/AAm, and YVi/X(Am,Vi); AAm/AVi, which have an equal ratio of X chromosomes of both species and a 25% advantage of D. virilis autosomes in the second case, when evaluating their traits in pairs with D. americana females. For all traits, comparisons using the Mann–Whitney U Test and Wald–Wolfowitz Runs Test did not reveal significant differences (Tables S25 and S26).
3.3. Indirect Influence of Chromosomes on Copulation Success
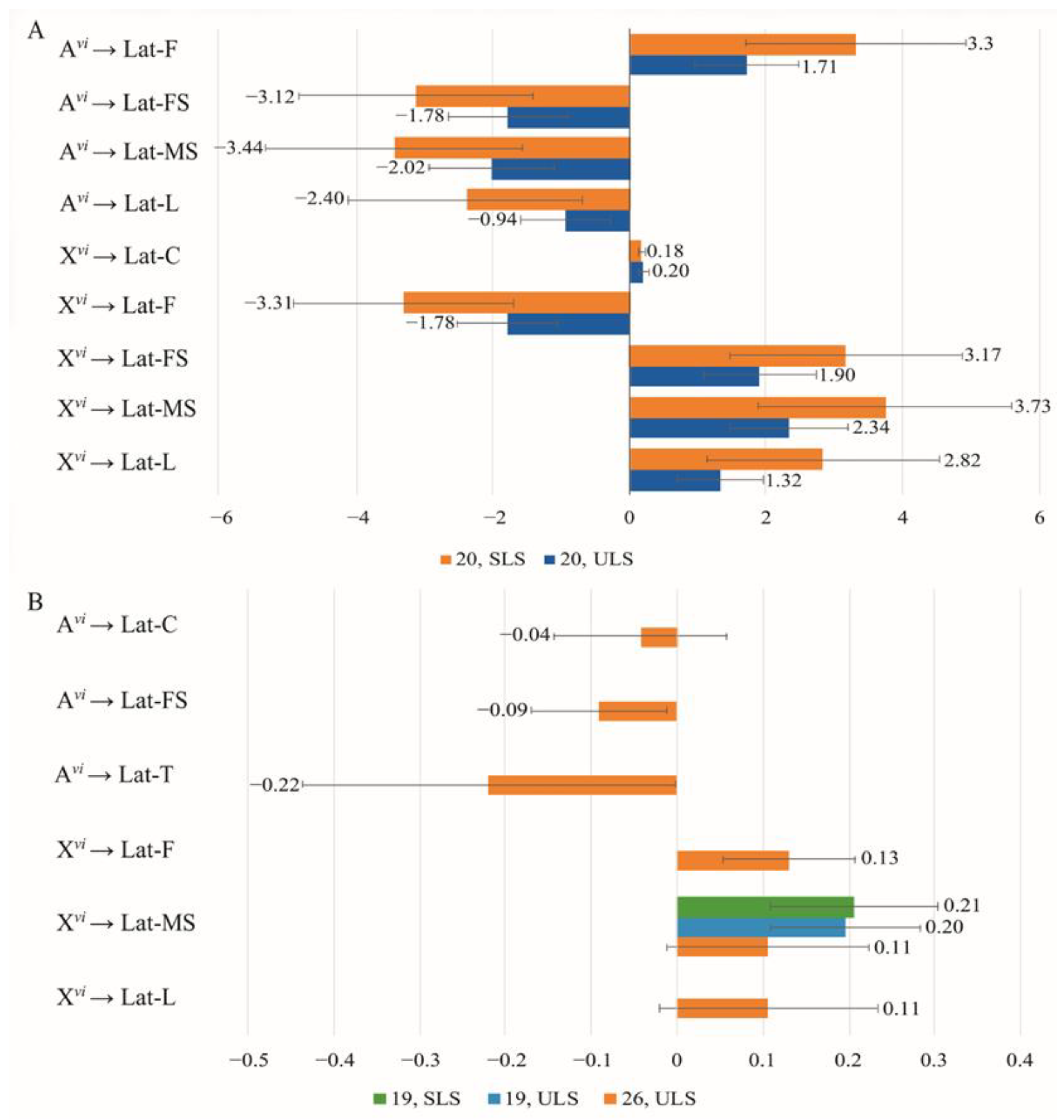
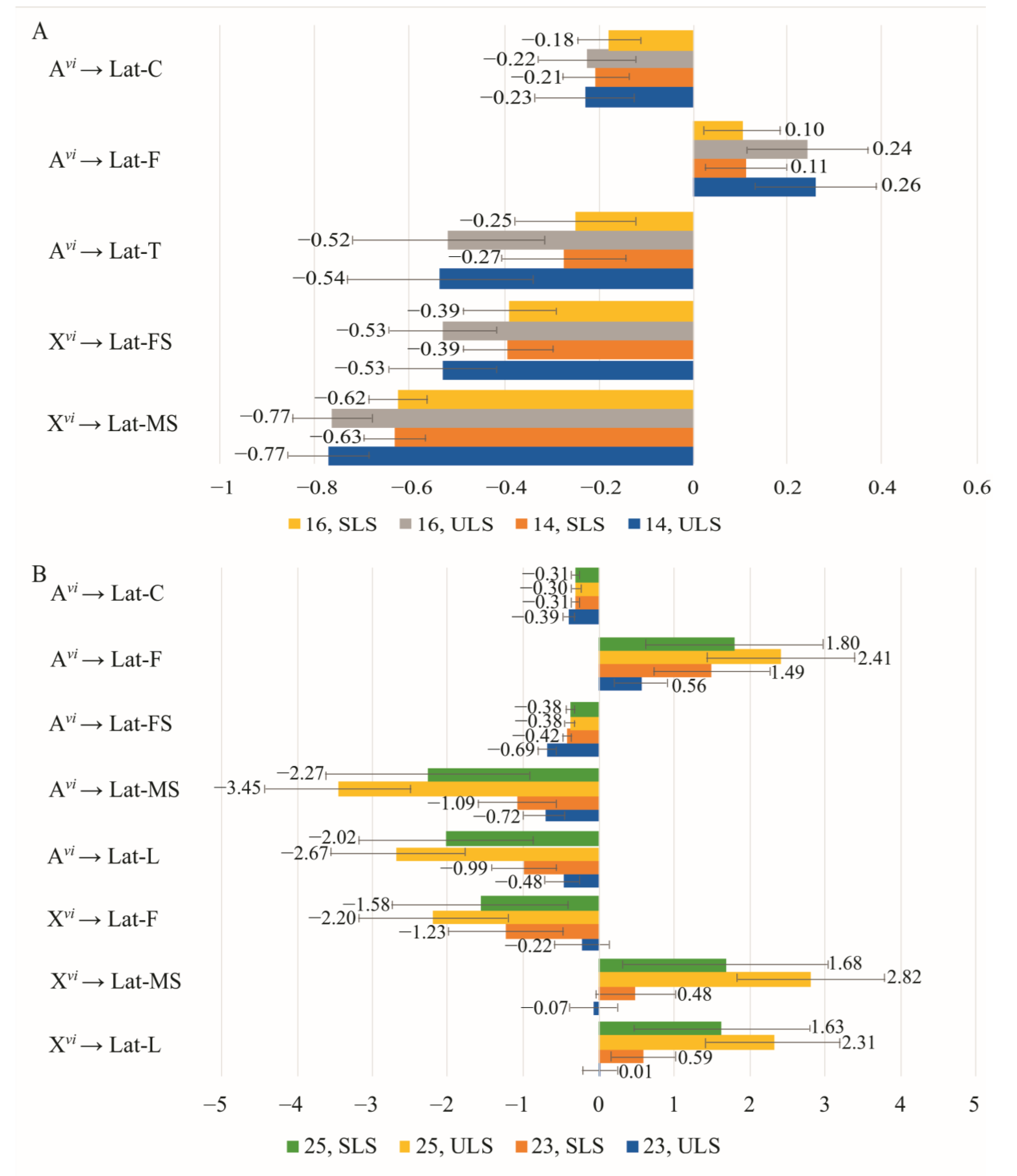
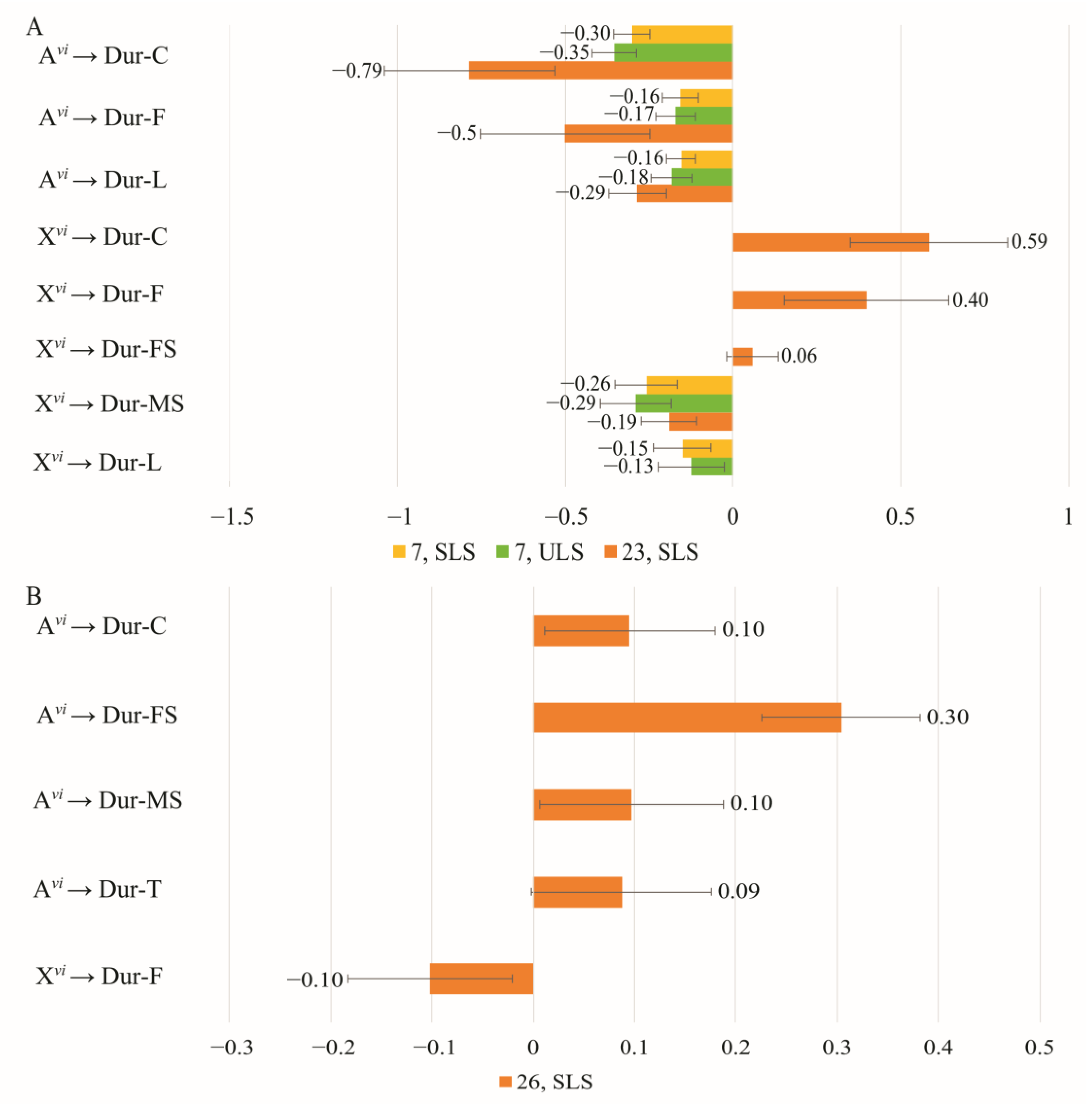
3.4. The Effect of Chromosomes on Courtship Elements
3.5. Effect of Courtship Elements on Copulation Efficiency
4. Discussion
4.1. The Effect of Sex Chromosomes and Autosomes on the Efficiency of Copulation
- (i)
- Recently emerged deleterious alleles that have low frequencies and lead to reduced fitness;
- (ii)
- Neutral and slightly deleterious alleles that are in equilibrium;
- (iii)
4.2. Variability of the Courtship Structure
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Spieth, H.T. Courtship behavior in Drosophila. Annu. Rev. Entomol. 1974, 19, 385–405. [Google Scholar] [CrossRef]
- Spieth, H.T. Mating behavior and sexual isolation in the Drosophila virilis species group. Behavior 1951, 3, 105–145. [Google Scholar] [CrossRef]
- Shorey, H.H. Nature of the sound produced by Drosophila melanogaster during courtship. Science 1962, 137, 677–678. [Google Scholar] [CrossRef]
- Jallon, J.M.; Hotta, Y. Genetic and behavioral studies of female sex appeal in Drosophila. Behav. Genet. 1979, 9, 257–275. [Google Scholar] [CrossRef]
- Ewing, A.W. Functional aspects of Drosophila courtship. Biol. Rev. 1983, 58, 275–292. [Google Scholar] [CrossRef]
- Markow, T.A.; O’Grady, P.M. Evolutionary genetics of reproductive behavior in Drosophila: Connecting the dots. Annu. Rev. Genet. 2005, 39, 263–291. [Google Scholar] [CrossRef] [PubMed]
- LaRue, K.M.; Clemens, J.; Berman, G.J.; Murthy, M. Acoustic duetting in Drosophila virilis relies on the integration of auditory and tactile signals. eLife 2015, 4, e07277. [Google Scholar] [CrossRef] [PubMed]
- Manning, A. The sexual behavior of two sibling Drosophila species. Behavior 1959, 15, 123–145. [Google Scholar] [CrossRef]
- Brown, R.G.B. Courtship behavior in the Drosophila obscura group. Pt II: Comparative studies. Behavior 1965, 25, 281–323. [Google Scholar] [CrossRef] [PubMed]
- Cobb, M.; Burnet, B.; Connolly, K. The structure of courtship in the Drosophila melanogaster species subgroup. Behavior 1985, 97, 182–212. [Google Scholar]
- Cobb, M.; Burnet, B.; Blizard, R.; Jallon, J.M. Courtship in Drosophila sechellia: Its structure, functional aspects, and relationship to those of other members of the Drosophila melanogaster species subgroup. J. Insect Behav. 1989, 2, 63–89. [Google Scholar] [CrossRef]
- Liimatainen, J.O.; Hoikkala, A. Interactions of the males and females of three sympatric Drosophila virilis- group species, D. montana, D. littoralis, and D. lummei (Diptera Drosophilidae) in intra-and interspecific courtships in the wild and in the laboratory. J. Insect Behav. 1998, 11, 399–417. [Google Scholar] [CrossRef]
- Hoikkala, A.; Crossley, S.A. Copulatory courtship in Drosophila: Behavior and songs of D. birchii and D. serrate. J. Insect Behav. 2000, 13, 71–86. [Google Scholar] [CrossRef]
- Saarikettu, M.; Liimatainen, J.O.; Hoikkala, A. The role of male courtship song in species recognition in Drosophila montana. Behav. Genet. 2005, 35, 257–263. [Google Scholar] [CrossRef]
- Dankert, H.; Wang, L.; Hoopfer, E.D.; Anderson, D.J.; Perona, P. Automated monitoring and analysis of social behavior in Drosophila. Nat. Methods 2009, 6, 297–303. [Google Scholar] [CrossRef] [PubMed]
- Nickel, D.; Civetta, A. An X chromosome effect responsible for asymmetric reproductive isolation between male Drosophila virilis and heterospecific females. Genome 2009, 52, 49–56. [Google Scholar] [CrossRef]
- Bollen, K.A.; Noble, M.D. Structural equation models and the quantification of behavior. Proc. Natl. Acad. Sci. USA 2011, 108, 15639–15646. [Google Scholar] [CrossRef] [PubMed]
- Butovskaya, M.L.; Lazebny, O.E.; Vasilyev, V.A.; Dronova, D.A.; Karelin, D.V.; Mabulla, A.Z.; Shibalev, D.V.; Shackelford, T.K.; Fink, B.; Ryskov, A.P. Androgen Receptor gene polymorphism, aggression, and reproduction in Tanzanian foragers and pastoralists. PLoS ONE 2015, 10, e0136208. [Google Scholar] [CrossRef]
- Park, A.Y.S. What advances information sharing for sustainability performance management? Empirical evidence from U.S. local governments. Urban Gov. 2021, 1, 72–80. [Google Scholar] [CrossRef]
- Snell, D.M.; Turner, J.M.A. Sex chromosome effects on male–female differences in Mammals. Curr. Biol. 2018, 28, 1313–1324. [Google Scholar] [CrossRef] [PubMed]
- Patterson, A.B.; Stone, W.S.; Griffin, R.K. Genetic and Cytological Analysis of the virilis Species Group; UT Press: Austin, TX, USA,, 1942; Volume 4228, pp. 162–200. [Google Scholar]
- Sweigart, A.L. The genetics of postmating, prezygotic reproductive isolation between Drosophila virilis and D. americana. Genetics 2010, 184, 401–410. [Google Scholar] [CrossRef] [PubMed]
- Mather, K.; Jinks, J.L. Biometrical Genetics: The Study of Continuous Variation; Springer Science and Business Media: Cham, Switzerland, 2013; p. 396. [Google Scholar]
- Heikkinen, E.; Lumme, J. Sterility of male and female hybrids of Drosophila virilis and Drosophila lummei. Heredity 1991, 67, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Falileeva, L.I.; Mitrofanov, V.G. Geneticheskiĭ analiz steril’nosti samtsov u gribridov D. virilis x D. lummei [Genetic analysis of sterile hybrid D. virilis × D. lummei males]. Dokl Akad Nauk. 1995, 341, 717–718. [Google Scholar]
- Falileeva, L.I.; Mitrofanov, V.G. Genomnaia nesovmestimost’ u gibridov Drosophila virilis X Drosophila lummei Hackman [Genomic incompatibility in Drosophila virilis Sturt. X Drosophila lummei Hackman hybrids]. Genetika 1997, 33, 458–463. [Google Scholar]
- Belkina, E.G.; Lazebny, O.E.; Vedenina, V.Y. The role of acoustic signals in courtship behavior of Drosophila virilis. Biol. Bull. 2016, 43, 561–566. [Google Scholar] [CrossRef]
- Belkina, E.G.; Vedenina, V.Y.; Sorokina, S.Y.; Lazebny, O.E. Courtship behavior analysis in three sibling species of the Drosophila virilis group. Entom Rev. 2018, 98, 1023–1037. [Google Scholar] [CrossRef]
- Belkina, E.G.; Lazebny, O.E.; Vedenina, V.Y. The importance of acoustic signals in multimodal courtship behavior in Drosophila virilis, D. lummei and D. littoralis. J. Insect Behav. 2021, 34, 280–295. [Google Scholar] [CrossRef]
- Arbuckle, J.L. Amos (Version 23.0) [Computer Program]; IBM SPSS: Chicago, IL, USA, 2014. [Google Scholar]
- Shah, R.; Goldstein, S.M. Use of structural equation modeling in operations management research: Looking back and forward. J. Oper. Manag. 2006, 24, 148–169. [Google Scholar] [CrossRef]
- Muller, H.J. Bearing of the Drosophila Work on Systematics. In The New Systematics; Huxley, J.S., Ed.; Clarendon Press: Oxford, UK, 1940; pp. 185–268. [Google Scholar]
- Throckmorton, L.H. The virilis Species Group. In The Genetics and Biology of Drosophila; Ashburnehr, M., Carson, L., Thompson, J.N., Eds.; Academic Press: London, UK, 1982; Volume 3b, pp. 227–296. [Google Scholar]
- Bone, J.R.; Kuroda, M.I. Dosage compensation regulatory proteins and the evolution of sex chromosomes in Drosophila. Genetics 1996, 144, 705–713. [Google Scholar] [CrossRef]
- Marin, I.; Franke, G.I.; Bradshaw, G.I.; Baker, B.S. The dosage compensation system of Drosophila is co-opted by newly evolved X chromosomes. Nature 1996, 383, 160–163. [Google Scholar] [CrossRef]
- Charlesworth, B.; Nordborg, M.; Charlesworth, D. The effects of local selection, balanced polymorphism and background selection on equilibrium patterns of genetic diversity in subdivided populations. Genet. Res. 1997, 70, 155–174. [Google Scholar] [CrossRef] [PubMed]
- Charlesworth, B.; Coyne, J.A.; Barton, N. The relative rates of evolution of sex chromosomes and autosomes. Am. Nat. 1987, 130, 113–146. [Google Scholar] [CrossRef]
- Kirkpatrick, M.; Hall, D.W. Male-biased mutation, sex linkage, and the rate of adaptive evolution. Evolution 2004, 57, 437–440. [Google Scholar]
- Wilson Sayres, M.A.; Makova, K.D. Genome analyses substantiate male mutation bias in many species. Bioessays 2011, 33, 938–945. [Google Scholar] [CrossRef] [PubMed]
- Johnson, N.A.; Lachance, J. The genetics of sex chromosomes: Evolution and implications for hybrid incompatibility. Ann. NY Acad. Sci. 2012, 1256, E1–E22. [Google Scholar] [CrossRef] [PubMed]
- Sturgill, D.; Zhang, Y.; Parisi, M.; Oliver, B. Demasculinization of X chromosomes in the Drosophila genus. Nature 2007, 450, 238–241. [Google Scholar] [CrossRef]
- Falconer, D.S.; Mackay, T.F.C. Introduction to Quantitative Genetics, 4th ed.; Pearson Education Ltd.: Essex, UK, 1996; p. 480. [Google Scholar]
- Frankham, R. Genetic Architecture of Reproductive Fitness and its Consequences. In Adaptation and Fitness in Animal Populations; van der Werf, J.H.J., Graser, H.-U., Frankham, R., Gondro, C., Eds.; Cambridge University Press: Cambridge, UK, 2009; Volume XII, p. 260. [Google Scholar]
- Houle, D.; Hughes, K.A.; Hoffmaster, D.K.; Ihara, J.; Assimacopoulos, S.; Canada, D.; Charlesworth, B. The effects of spontaneous mutation on quantitative traits. I. Variances and covariances of life history traits. Genetics 1994, 138, 773–785. [Google Scholar] [CrossRef]
- Houle, D.; Morikawa, B.; Lynch, M. Comparing Mutational Variabilities. Genetics 1996, 143, 1467–1483. [Google Scholar] [CrossRef]
- Charlesworth, B.; Hughes, K.A. The maintenance of genetic variation in life history traits. In Evolutionary Genetics from Molecules to Morphology; Singh, R.S., Krimbas, C.B., Eds.; Cambridge University Press: Cambridge, UK, 2000; pp. 369–391. [Google Scholar]
- Charlesworth, B.; Betancourt, A.J.; Kaiser, V.B.; Gordo, I. Genetic recombination and molecular evolution. Cold Spring Harb. Symp. Quant. Biol. 2009, 74, 177–186. [Google Scholar] [CrossRef]
- Kelly, J.K.; Willis, J.H. Deleterious mutations and genetic variation for flower size in Mimulus guttatus. Evolution 2001, 55, 937–942. [Google Scholar] [CrossRef] [PubMed]
- Kelly, J.K. Deleterious mutations and the genetic variance of male fitness components in Mimulus guttatus. Genetics 2003, 164, 1071–1085. [Google Scholar] [CrossRef] [PubMed]
- Kelly, J.K. An experimental method for evaluating the contribution of deleterious mutations to quantitative trait variation. Genet. Res. 1999, 73, 263–273. [Google Scholar] [CrossRef] [PubMed]
- Kulikov, A.M.; Sorokina, S.Y.; Melnikov, A.I.; Gornostaev, N.G.; Seleznev, D.G.; Lazebny, O.E. The effects of the sex chromosomes on the inheritance of species-specific traits of the copulatory organ shape in Drosophila virilis and Drosophila lummei. PLoS ONE 2020, 15, e0244339. [Google Scholar] [CrossRef]
- Vedenina, V.Y.; Ivanova, T.I.; Lazebny, O.E. Analysis of courtship behaviour in closely related species of Drosophila virilis group: A new approach arises new questions. J. Ins. Behav. 2012, 26, 402–415. [Google Scholar] [CrossRef]
- Saarikettu, M.; Liimatainen, J.O.; Hoikkala, A. Intraspecific variation in mating behavior does not cause sexual isolation between Drosophila virilis strains. Anim. Behav. 2005, 70, 417–426. [Google Scholar] [CrossRef]
- Stocker, R.F. The organization of the chemosensory system in Drosophila melanogaster: A review. Cell Tissue Res. 1994, 275, 3–26. [Google Scholar] [CrossRef]
- Hu, Y.; Han, Y.; Shao, Y.; Wang, X.; Ma, Y.; Ling, E.; Xue, L. Gr33a Modulates Drosophila male courtship preference. Sci. Rep. 2015, 5, 7777. [Google Scholar] [CrossRef]
- Spieth, H.T. Mating behavior within the genus Drosophila (Diptera). Bull. Am. Mus. Nat. Hist. 1952, 99, 395–474. [Google Scholar]
- Ahmed-Braimah, Y.H. Multiple genes cause postmating prezygotic reproductive isolation in the Drosophila virilis group. G3 2016, 6, 4067–4076. [Google Scholar] [CrossRef]


| Female | Male | Chromosome Composition * | XVi/AVi (%) | Sum Cop. | Fisher’s Exact Test, p |
|---|---|---|---|---|---|
| Group 1 (♀ D. virilis–♂ D. virilis → D. americana) | |||||
| ♀ D. virilis | ♂ D. virilis | YVi/XVi; AVi/AVi | 100/100 | 30 | |
| ♀ D. virilis | ♂ FB (♀♀ D. virilis × ♂♂ F1) | YVi/XVi; A(Am,Vi)/AVi | 100/75 | 29 | 0.5000 |
| ♀ D. virilis | ♂ FB (♀♀ F1 × ♂♂ D. virilis) | YVi/X(Am,Vi); A(Am,Vi)/AVi | 50/75 | 28 | 0.2458 |
| ♀ D. virilis | ♂ F2 | YVi/X(Am,Vi); AAm/AVi | 50/50 | 25 | 0.0261 |
| ♀ D. virilis | ♂ F1 | YVi/XAm; AAm/AVi | 0/50 | 23 | 0.0053 |
| ♀ D. virilis | ♂ D. americana | YAm/XAm; AAm/AAm | 0/0 | 11 | 0.0000 |
| Group 2 (♀ D. americana–♂ D. americana → D. virilis) | |||||
| ♀ D. americana | ♂ D. americana | YAm/XAm; AAm/AAm | 0/0 | 29 | |
| ♀ D. americana | ♂ FB (♀♀ D. americana × ♂♂ F1) | YVi/XAm; A(Am,Vi)/AAm | 0/25 | 28 | 0.5000 |
| ♀ D. americana | ♂ F1 | YVi/XAm; AAm/AVi | 0/50 | 27 | 0.3060 |
| ♀ D. americana | ♂ FB (♀♀ F1 × ♂♂ D. americana) | YAm/X(Am,Vi); A(Am,Vi)/AAm | 50/25 | 24 | 0.0514 |
| ♀ D. americana | ♂ F2 | YVi/X(Am,Vi); AAm/AVi | 50/50 | 27 | 0.3060 |
| ♀ D. americana | ♂ D. virilis | YVi/XVi; AVi/AVi | 100/100 | 1 | 0.0000 |
| Group 3 (♀ D. virilis → D. americana–♂ D. virilis) | |||||
| ♀ D. virilis | ♂ D. virilis | XVi/XVi; AVi/AVi | 100/100 | 30 | |
| ♀ FB (♀♀ F1 × ♂♂ D. virilis) | ♂ D. virilis | XAm/X3Vi; A(Am,Vi)/AVi | 75/75 | 29 | 0.5000 |
| ♀ FB (♀♀ D. virilis × ♂♂ F1 ) | ♂ D. virilis | XAm/XVi; A(Am,Vi/AVi | 50/75 | 28 | 0.2458 |
| ♀ F2 | ♂ D. virilis | XAm/X3Vi; AAmAVi | 75/50 | 28 | 0.2458 |
| ♀ F1 | ♂ D. virilis | XAm/XVi; AAmAVi | 50/50 | 29 | 0.5000 |
| ♀ D. americana | ♂ D. virilis | XAm/XAm; AAm/AAm | 0/0 | 1 | 0.0000 |
| Group 4 (♀ D. americana → D. virilis–♂ D. americana) | |||||
| ♀ D. americana | ♂ D. americana | XAm/XAm; AAm/AAm | 0/0 | 29 | |
| ♀ FB (♀♀ D. americana × ♂♂ F1 ) | ♂ D. americana | XAm/XAm; A(Am,Vi)/AAm | 0/25 | 24 | 0.0514 |
| ♀ FB (♀♀ F1 × ♂♂ D. americana) | ♂ D. americana | X3Am/XVi; A(Am,Vi)/AAm | 25/25 | 23 | 0.0262 |
| ♀ F1 | ♂ D. americana | XAm/XVi; AAm/AVi | 50/50 | 17 | 0.0002 |
| ♀ F2 | ♂ D. americana | XAm/X3Vi; AAm/AVi | 75/50 | 25 | 0.0973 |
| ♀ D. virilis | ♂ D. americana | XVi/XVi; AVi/AVi | 100/100 | 11 | 0.0000 |
| Traits | N | Mean | Median | Min. | Max. | Lower Quartile | Upper Quartile |
|---|---|---|---|---|---|---|---|
| Latency | |||||||
| Following | 600 | 1175.5 | 1801.0 | 0 | 1801 | 15.5 | 1801.0 |
| Tapping | 600 | 5.0 | 0.0 | 0 | 1801 | 0.0 | 0.0 |
| Licking | 600 | 77.0 | 2.0 | 0 | 1801 | 1.0 | 6.0 |
| Male singing | 600 | 145.5 | 4.5 | 0 | 1801 | 2.0 | 18.0 |
| Circling | 600 | 1119.1 | 1801.0 | 0 | 1801 | 33.0 | 1801.0 |
| Copulation attempt | 600 | 1375.6 | 1801.0 | 2 | 1801 | 1722.0 | 1801.0 |
| Copulation | 600 | 439.8 | 44.0 | 2 | 1801 | 9.0 | 477.0 |
| Female singing | 600 | 42.3 | 2.0 | 0 | 1801 | 0.0 | 7.0 |
| Duration | |||||||
| Following | 600 | 4.4 | 0.0 | 0 | 107 | 0.0 | 3.0 |
| Tapping | 600 | 45.4 | 16.0 | 0 | 896 | 7.0 | 42.0 |
| Licking | 600 | 33.6 | 10.0 | 0 | 665 | 4.0 | 27.0 |
| Male singing | 600 | 11.9 | 5.0 | 0 | 210 | 3.0 | 13.0 |
| Circling | 600 | 2.9 | 0.0 | 0 | 108 | 0.0 | 3.0 |
| Copulation attempt | 600 | 4.4 | 0.0 | 0 | 204 | 0.0 | 0.5 |
| Copulation | 600 | 125.2 | 142.0 | 0 | 380 | 97.0 | 170.0 |
| Female singing | 600 | 11.7 | 4.0 | 0 | 435 | 2.0 | 12.0 |
| Constant Mate Genotype | Copulation Parameters | Model | Parameters | df | C | C − df | AIC | BCC | BIC | C/df |
|---|---|---|---|---|---|---|---|---|---|---|
| ♀ D. virilis (Group 1) | latency | 14 | 23 | 32 | 11.72 | −20.28 | 57.72 | 60.74 | 131.16 | 0.37 |
| 16 | 25 | 30 | 9.12 | −20.88 | 59.12 | 62.39 | 138.94 | 0.3 | ||
| ♀ D. americana (Group 2) | 20 | 23 | 22 | 4.54 | −17.46 | 50.54 | 53.26 | 123.97 | 0.21 | |
| ♀ D. virilis (Group 1) | duration | 23 | 31 | 14 | 5.51 | −8.49 | 67.51 | 71.18 | 166.49 | 0.39 |
| ♀ D. americana (Group 2) | 7 | 28 | 17 | 61 | 44 | 117 | 120 | 206 | 4 | |
| 23 | 31 | 14 | 17 | 3 | 79 | 83 | 178 | 1 | ||
| ♂ D. virilis (Group 3) | latency | 23 | 31 | 14 | 20.21 | 6.21 | 82.21 | 85.87 | 181.19 | 1.44 |
| 25 | 33 | 12 | 16.81 | 4.81 | 82.81 | 86.71 | 188.18 | 1.4 | ||
| ♂ D. americana (Group 4) | 19 | 27 | 18 | 4.99 | −13.01 | 58.99 | 62.19 | 145.2 | 0.28 | |
| 26 | 34 | 11 | 2.54 | −8.46 | 70.54 | 74.56 | 179.1 | 0.23 | ||
| ♂ D. virilis (Group 3) | duration | 27 | 35 | 10 | 41.02 | 31.02 | 111.02 | 115.16 | 222.77 | 4.1 |
| 32 | 40 | 5 | 24.9 | 19.9 | 104.9 | 109.63 | 232.61 | 4.98 | ||
| Sat | 45 | 0 | 0 | 0 | 90 | 95.33 | 233.68 | |||
| ♂ D. americana (Group 4) | 26 | 35 | 10 | 2.55 | −7.45 | 72.55 | 76.69 | 184.3 | 0.25 |
| Copulation Parameters | Constant Partner Genotype | Changing Partner Genotype | Effect * | Method | Model | |||
|---|---|---|---|---|---|---|---|---|
| XVi | XVi s.e. | AuVi | AuVi s.e. | |||||
| Latency | ♀ D. virilis (Group 1) | ♂♂ vi → am | −0.427 | 0.07 | −0.06 | 0.03 | ULS | 16 |
| −0.433 | 0.071 | −0.06 | 0.03 | ULS | 14 | |||
| ♀ D. americana (Group 2) | ♂♂ am → vi | 1.775 | 0.627 | −1.233 | 0.759 | ULS | 20 | |
| Duration | ♀ D. virilis (Group 1) | ♂♂ vi → am | 0 | - | 0.531 | 0.068 | ULS | 23 |
| ♀ D. americana (Group 2) | ♂♂ am → vi | −0.725 | - | 0.022 | - | ULS | 23 | |
| Latency | ♂ D. virilis (Group 3) | ♀♀ vi → am | −0.03 | 0.259 | −0.65 | 0.230 | ULS | 23 |
| 0.54 | 0.590 | −1.12 | 0.578 | SLS | 23 | |||
| 2.16 | 0.93 | −2.69 | 0.93 | ULS | 25 | |||
| 1.55 | 1.518 | −2.12 | 1.513 | SLS | 25 | |||
| ♂ D. americana (Group 4) | ♀♀ am → vi | 0.049 | 0.206 | 0.302 | 0.23 | ULS | 26 | |
| Duration | ♂ D. virilis (Group 3) | ♀♀ vi → am | −0.455 | 0.144 | 1.183 | 0.158 | ULS | Sat* |
| ♂ D. americana (Group 4) | ♀♀ am → vi | −0.352 | - | −0.061 | - | ULS | 26 | |
| −0.369 | 0.098 | −0.043 | 0.126 | SLS | 26 | |||
| Fixed Mate Genotype; Model; Method | Latency | |||||
|---|---|---|---|---|---|---|
| Tapping | Licking | Male Singing | Circling | Following | Female Singing | |
| ♀ D. virilis (Group 1); 16; ULS | 0.56 | −0.248 | ||||
| s.e. | 0.057 | 0.061 | ||||
| ♀ D. virilis (Group 1); 16; SLS | 0.685 | −0.288 | ||||
| s.e. | 0.059 | 0.061 | ||||
| ♀ D. americana (Group 2); 20; ULS | −2.306 | 2.935 | −2.222 | −1.738 | ||
| s.e. | 1.409 | 1.872 | 1.79 | 1.07 | ||
| ♀ D. americana (Group 2); 20; SLS | −1.393 | 2.739 | −2.228 | −1.02 | ||
| s.e. | 1.28 | 2.289 | 2.387 | 1.115 | ||
| ♂ D. virilis (Group 3); 25; ULS | −0.618 | 1.08 | −0.246 | 0.041 | ||
| s.e. | 0.784 | 0.793 | 0.109 | 0.217 | ||
| ♂ D. virilis (Group 3); 25; SLS | −1.187 | 1.691 | −0.407 | −0.141 | ||
| s.e. | 1.166 | 1.351 | 0.296 | 0.592 | ||
| ♂ D. virilis (Group 3); 23; ULS | −0.522 | 0.957 | −0.234 | 0.111 | ||
| s.e. | 0.673 | 0.689 | 0.091 | 0.172 | ||
| ♂ D. virilis (Group 3); 23; SLS | −0.949 | 1.375 | −0.357 | 0.066 | ||
| s.e. | 1.114 | 1.213 | 0.146 | 0.3 | ||
| ♂ D. virilis (Group 3); 15; ULS | 0.432 | −0.233 | 0.18 | |||
| s.e. | 0.069 | 0.061 | 0.069 | |||
| ♂ D. virilis (Group 3); 15; SLS | 0.477 | −0.272 | 0.264 | |||
| s.e. | 0.067 | 0.06 | 0.058 | |||
| ♂ D. americana (Group 4); 26; ULS | −1.344 | 1.544 | −0.448 | −0.181 | −0.751 | |
| s.e. | 1.376 | 1.993 | 1.484 | 0.119 | 0.574 | |
| ♂ D. americana (Group 4); 19; ULS | −0.055 | 0.461 | −0.141 | −0.194 | ||
| s.e. | 0.37 | 0.238 | 0.076 | 0.229 | ||
| ♂ D. americana (Group 4); 19; SLS | −0.127 | 0.591 | −0.131 | −0.132 | ||
| s.e. | 1.182 | 0.733 | 0.343 | 1.139 | ||
| Fixed Mate Genotype; Model; Method | Duration | |||||
|---|---|---|---|---|---|---|
| Tapping | Licking | Male Singing | Circling | Following | Female Singing | |
| ♀ D. virilis (Group 1); 23; ULS | −2.43 | 2.248 | ||||
| s.e. | 0.744 | 0.748 | ||||
| ♀ D. virilis (Group 1); 23; SLS | −2.588 | 2.404 | ||||
| s.e. | 0.833 | 0.837 | ||||
| ♀ D. americana (Group 2); 23; SLS | −8.402 | 8.591 | ||||
| s.e. | 7.863 | 7.888 | ||||
| ♀ D. americana (Group 2); 7; ULS | −6.754 | 5.688 | 2.921 | 0.811 | ||
| s.e. | 3.643 | 3.079 | 1.568 | 0.576 | ||
| ♀ D. americana (Group 2); 7; SLS | −11.405 | 9.432 | 4.975 | 1.674 | ||
| s.e. | 7.534 | 6.343 | 3.143 | 1.243 | ||
| ♂ D. virilis (Group 3); base; ULS | −4.124 | 4.79 | −1.471 | 0.395 | 0.626 | −0.026 |
| s.e. | 1.07 | 1.954 | 1.235 | 0.247 | 0.201 | 0.137 |
| ♂ D. virilis (Group 3); base; SLS | −4.739 | 5.992 | −2.118 | 0.366 | 0.655 | 0.001 |
| s.e. | 2.255 | 4.434 | 2.848 | 0.432 | 0.351 | 0.256 |
| ♂ D. virilis (Group 3); 32; ULS | −3.019 | 2.544 | 0.416 | −0.081 | 0.293 | |
| s.e. | 0.537 | 0.522 | 0.117 | 0.099 | 0.186 | |
| ♂ D. virilis (Group 3); 32; SLS | −3.164 | 2.718 | 0.386 | −0.074 | 0.27 | |
| s.e. | 0.595 | 0.576 | 0.115 | 0.099 | 0.187 | |
| ♂ D. virilis (Group 3); 27; ULS | −1.786 | 1.37 | 0.325 | −0.154 | 0.388 | |
| s.e. | 0.195 | 0.245 | 0.127 | 0.099 | 0.163 | |
| ♂ D. virilis (Group 3); 27; SLS | −1.876 | 1.425 | 0.325 | −0.147 | 0.429 | |
| s.e. | 0.22 | 0.27 | 0.14 | 0.099 | 0.173 | |
| ♂ D. americana (Group 4); 29; SLS | −10.179 | 10.194 | −2.691 | 0.992 | 0.667 | 1.498 |
| s.e. | 4.635 | 4.887 | 1.849 | 0.62 | 0.545 | 0.999 |
| ♂ D. americana (Group 4); 26; SLS | −6.982 | 6.759 | −1.616 | 0.623 | 0.423 | 1.101 |
| s.e. | 5.752 | 5.541 | 1.757 | 0.845 | 0.616 | 1.191 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Belkina, E.G.; Seleznev, D.G.; Sorokina, S.Y.; Kulikov, A.M.; Lazebny, O.E. The Effect of Chromosomes on Courtship Behavior in Sibling Species of the Drosophila virilis Group. Insects 2023, 14, 609. https://doi.org/10.3390/insects14070609
Belkina EG, Seleznev DG, Sorokina SY, Kulikov AM, Lazebny OE. The Effect of Chromosomes on Courtship Behavior in Sibling Species of the Drosophila virilis Group. Insects. 2023; 14(7):609. https://doi.org/10.3390/insects14070609
Chicago/Turabian StyleBelkina, Elena G., Dmitry G. Seleznev, Svetlana Yu. Sorokina, Alex M. Kulikov, and Oleg E. Lazebny. 2023. "The Effect of Chromosomes on Courtship Behavior in Sibling Species of the Drosophila virilis Group" Insects 14, no. 7: 609. https://doi.org/10.3390/insects14070609
APA StyleBelkina, E. G., Seleznev, D. G., Sorokina, S. Y., Kulikov, A. M., & Lazebny, O. E. (2023). The Effect of Chromosomes on Courtship Behavior in Sibling Species of the Drosophila virilis Group. Insects, 14(7), 609. https://doi.org/10.3390/insects14070609





