The Study of Cell-Penetrating Peptides to Deliver dsRNA and siRNA by Feeding in the Desert Locust, Schistocerca gregaria
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Rearing of the Desert Locust, S. gregaria
2.2. CPPs
2.2.1. CPP Synthesis and Purification
2.2.2. CPP Quantification
2.3. Synthesis of Long dsRNA
2.4. Synthesis of Fluorescent dsRNA
2.5. Synthesis of (Fluorescent) siRNAs
2.6. Formation of CPP:dsRNA and CPP:siRNA Complexes
2.7. Assessment of Complex Stability in an S. gregaria Ex Vivo Gut Environment
2.7.1. Collection of Midgut Enzyme Solution
2.7.2. Ex Vivo Degradation Assay
2.8. Visualization of EB1:dsRNA and EB1:siRNA Complexes
2.9. RNAi Assays
2.9.1. Feeding Assays
2.9.2. Injection Assays
2.10. Tissue Collection
2.11. RNA Extraction and cDNA Synthesis
2.12. Quantitative Real-Time PCR (qRT-PCR)
3. Results
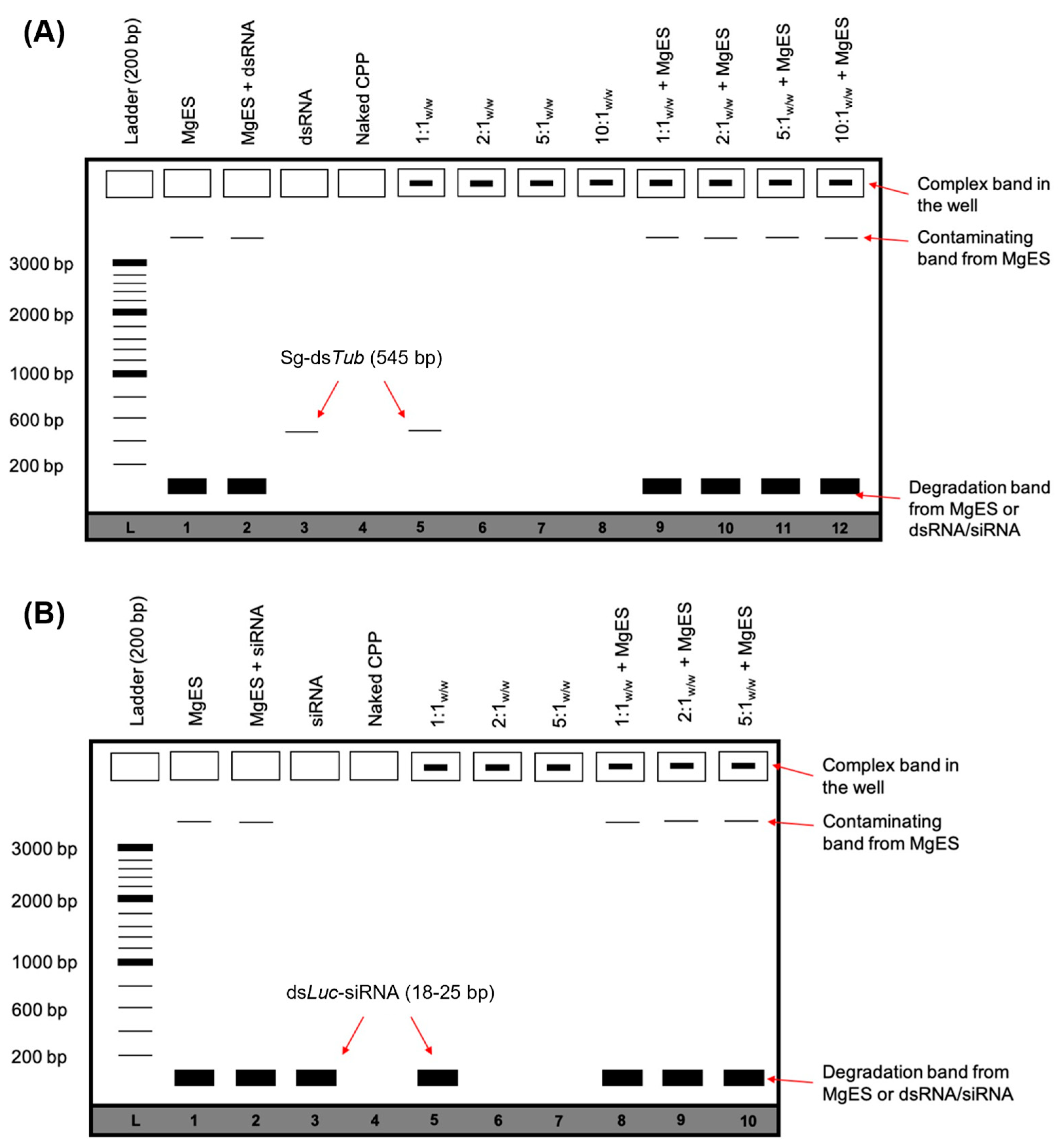
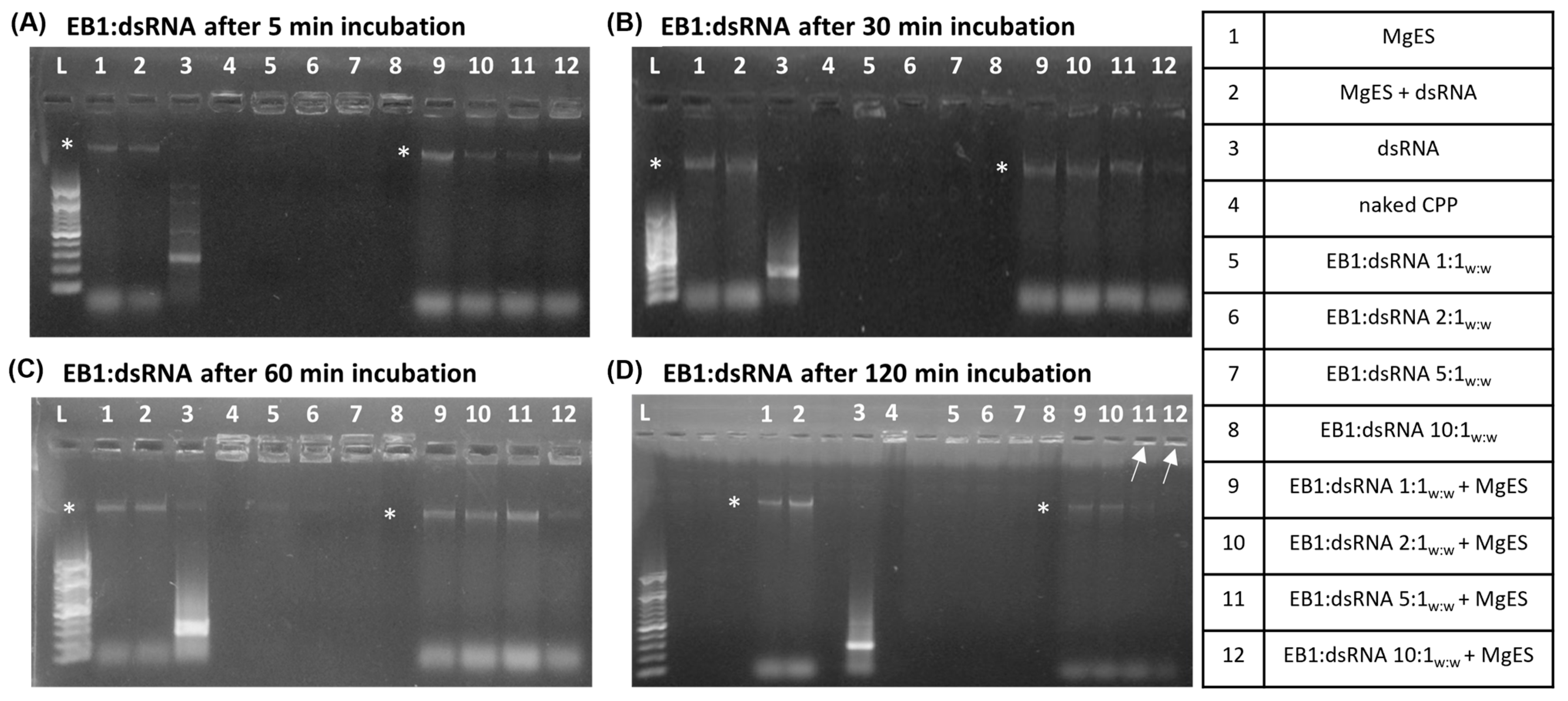
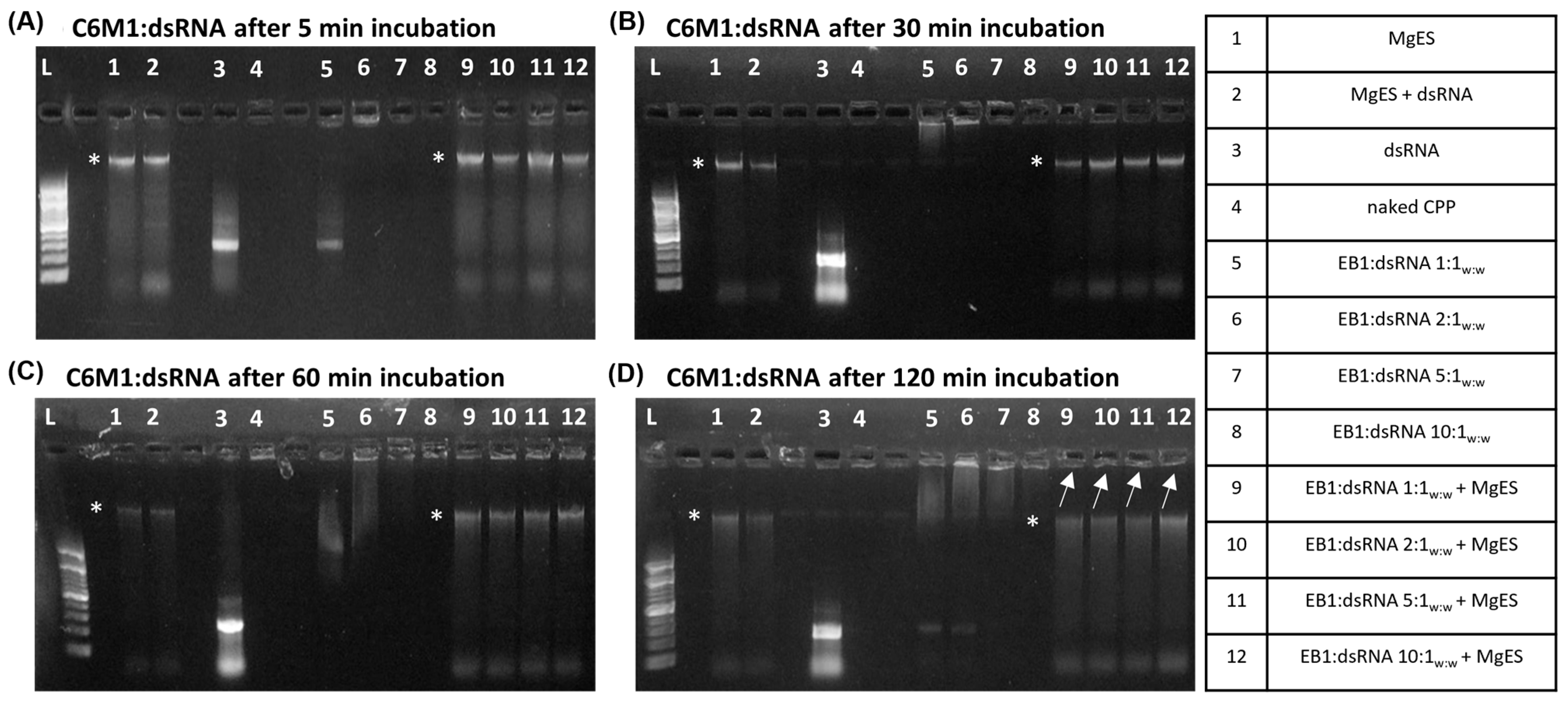
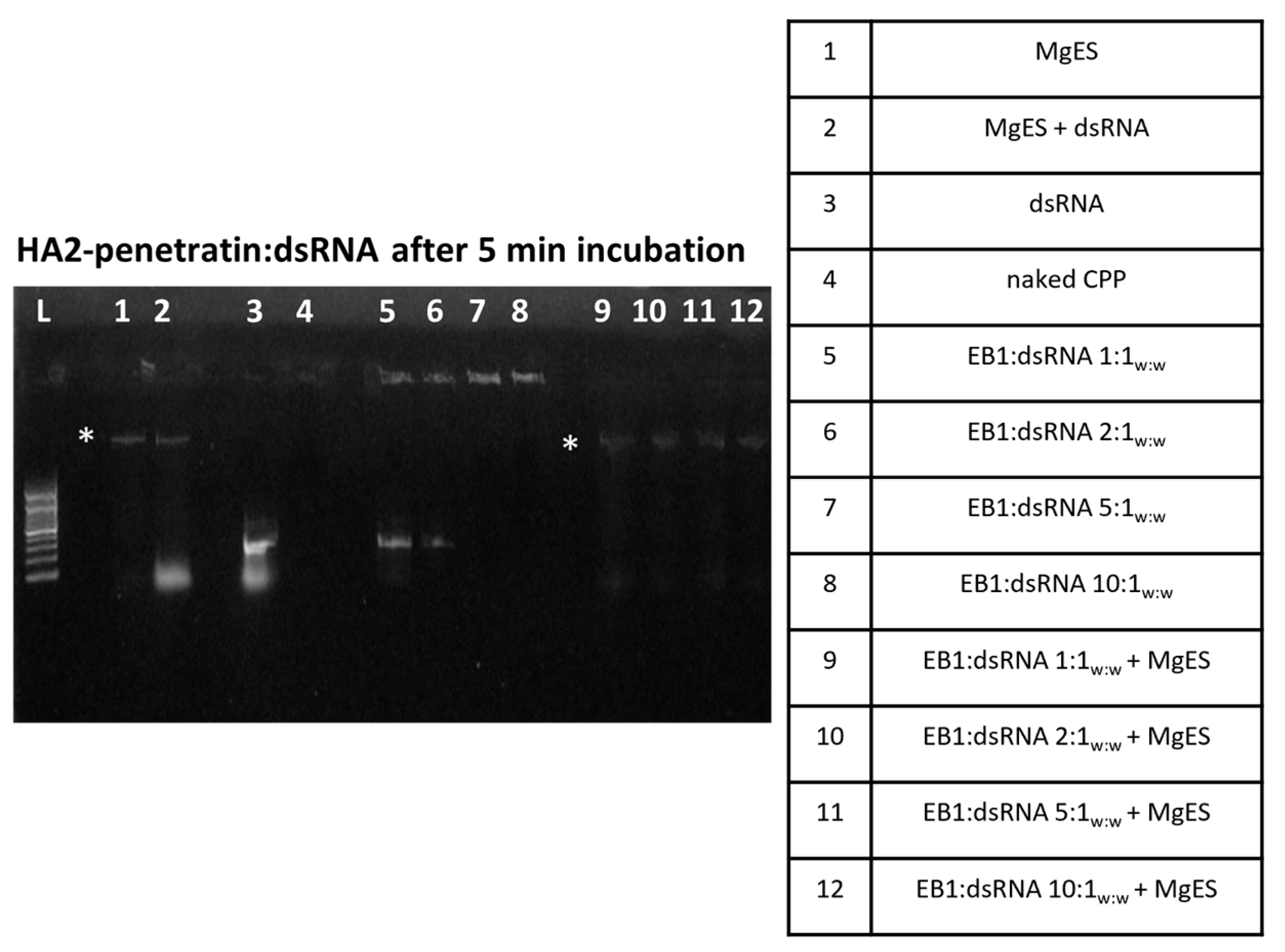
3.1. CPP:dsRNA Complexation and Stability in Ex Vivo Locust Midgut Environment
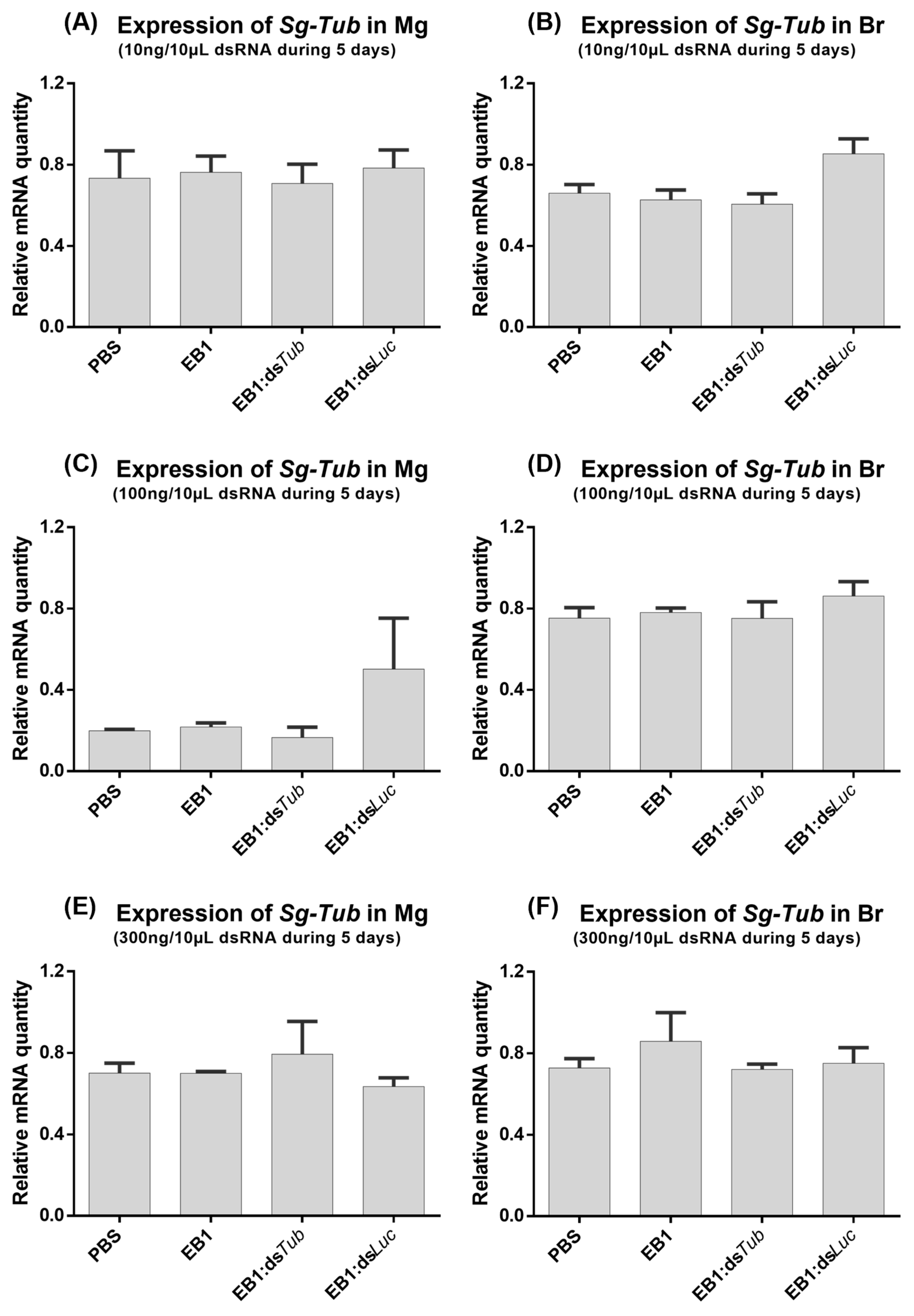
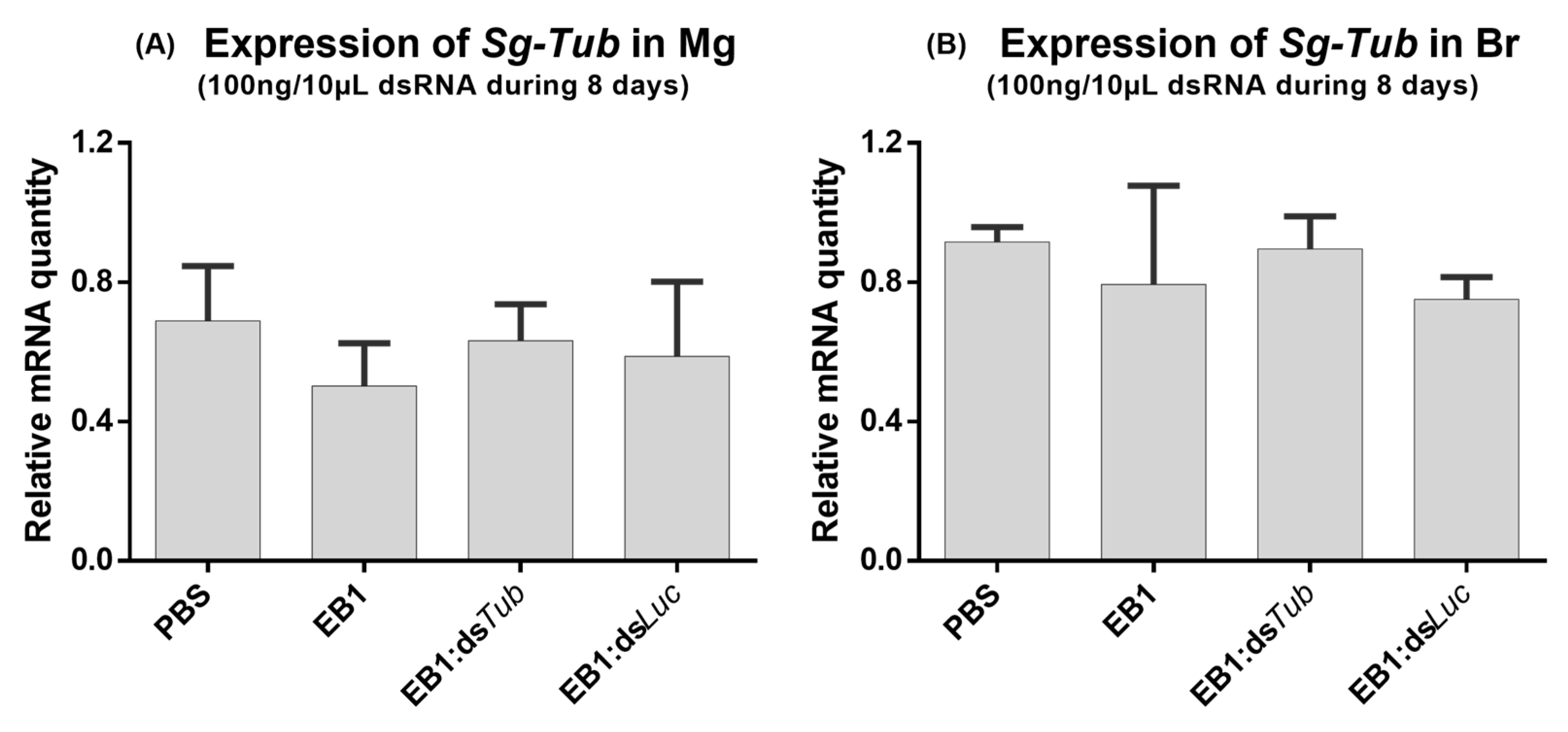
3.2. Feeding EB1:dsRNA
3.3. Visualization of EB1:dsRNA and EB1:siRNA Complexes
3.4. EB1:siRNA Complexation and Stability in Ex Vivo Locust Midgut Environment
3.5. Injecting Naked siRNA
3.6. Injecting EB1:siRNA
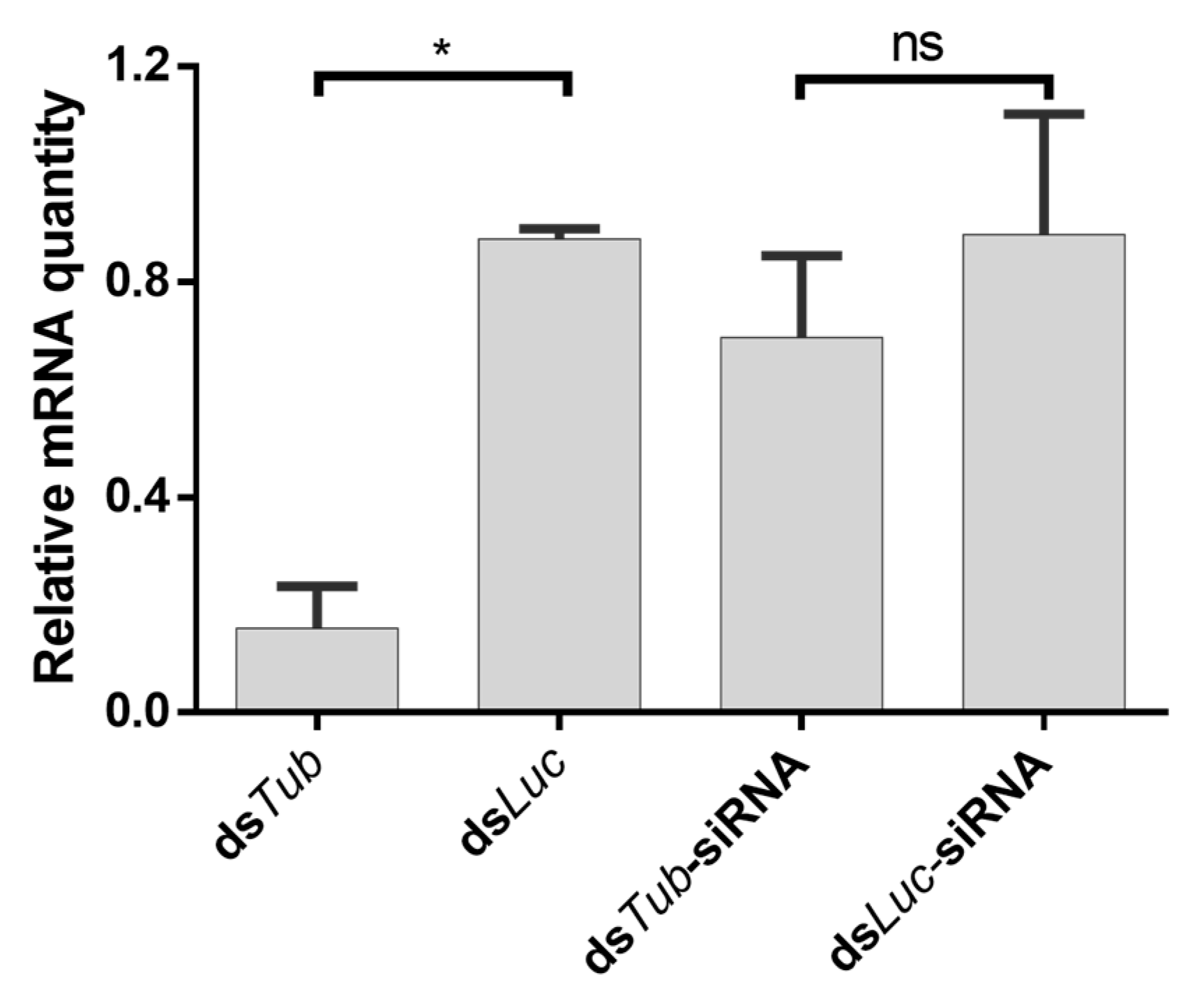
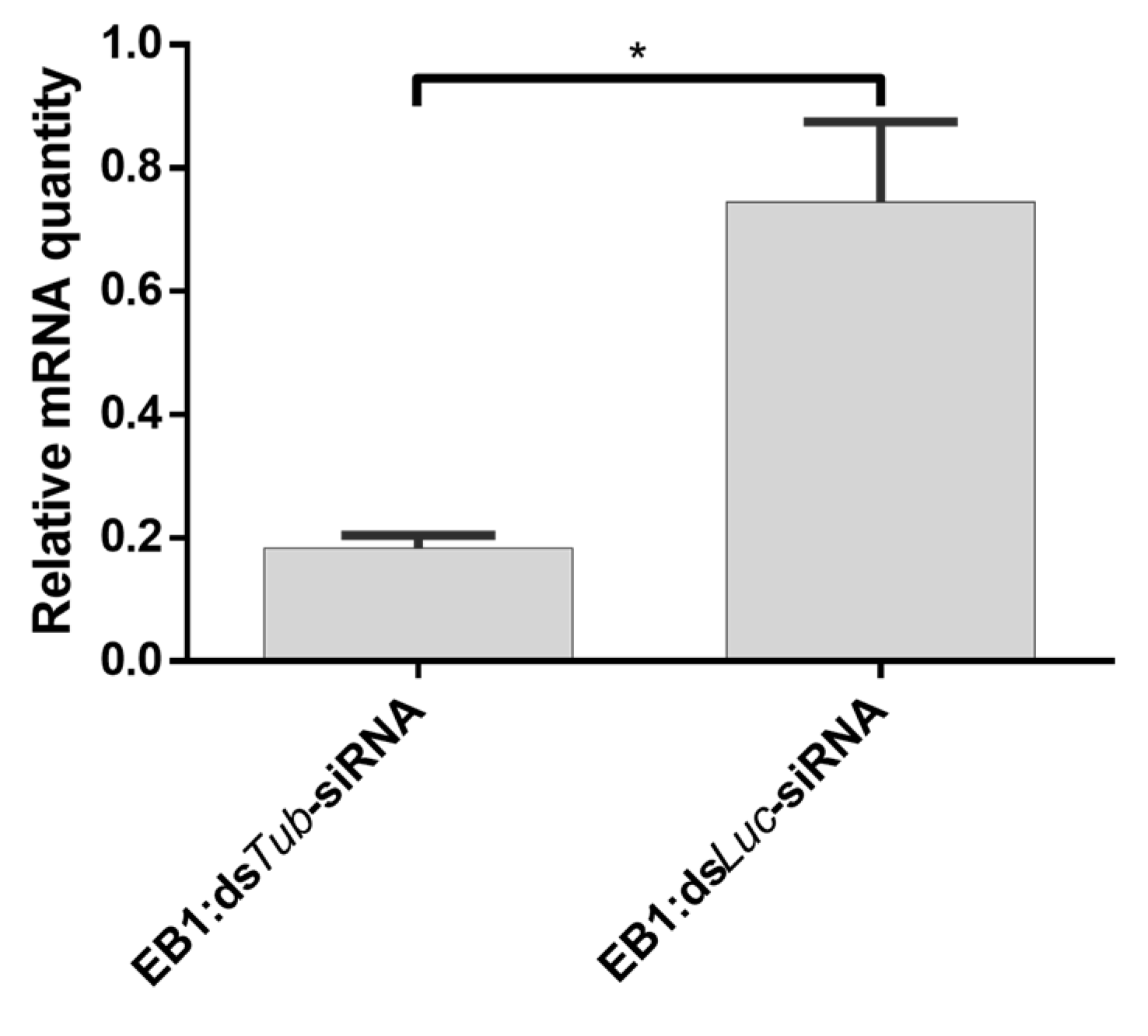
3.7. Feeding EB1:siRNA
4. Discussion
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Vogel, E.; Santos, D.; Mingels, L.; Verdonckt, T.W.; Broeck, J. vanden RNA Interference in Insects: Protecting Beneficials and Controlling Pests. Front. Physiol. 2019, 10, 1912. [Google Scholar] [CrossRef] [PubMed]
- Huvenne, H.; Smagghe, G. Mechanisms of DsRNA Uptake in Insects and Potential of RNAi for Pest Control: A Review. J. Insect Physiol. 2010, 56, 227–235. [Google Scholar] [CrossRef] [PubMed]
- Santos, D.; vanden Broeck, J.; Wynant, N. Systemic RNA Interference in Locusts: Reverse Genetics and Possibilities for Locust Pest Control. Curr. Opin. Insect Sci. 2014, 6, 9–14. [Google Scholar] [CrossRef]
- Wynant, N.; Verlinden, H.; Breugelmans, B.; Simonet, G.; Vanden Broeck, J. Tissue-Dependence and Sensitivity of the Systemic RNA Interference Response in the Desert Locust, Schistocerca gregaria. Insect Biochem. Mol. Biol. 2012, 42, 911–917. [Google Scholar] [CrossRef] [PubMed]
- Wynant, N.; Santos, D.; Verdonck, R.; Spit, J.; van Wielendaele, P.; vanden Broeck, J. Identification, Functional Characterization and Phylogenetic Analysis of Double Stranded RNA Degrading Enzymes Present in the Gut of the Desert Locust, Schistocerca gregaria. Insect Biochem. Mol. Biol. 2014, 46, 1–8. [Google Scholar] [CrossRef]
- Tian, H.; Peng, H.; Yao, Q.; Chen, H.; Xie, Q.; Tang, B.; Zhang, W. Developmental Control of a Lepidopteran Pest Spodoptera Exigua by Ingestion of Bacteria Expressing DsRNA of a Non-Midgut Gene. PLoS ONE 2009, 4, e6225. [Google Scholar] [CrossRef]
- Vatanparast, M.; Kim, Y. Optimization of Recombinant Bacteria Expressing DsRNA to Enhance Insecticidal Activity against a Lepidopteran Insect, Spodoptera Exigua. PLoS ONE 2017, 12, e0183054. [Google Scholar] [CrossRef]
- Kim, E.; Park, Y.; Kim, Y. A Transformed Bacterium Expressing Double-Stranded RNA Specific to Integrin Β1 Enhances Bt Toxin Efficacy against a Polyphagous Insect Pest, Spodoptera Exigua. PLoS ONE 2015, 10, e0132631. [Google Scholar] [CrossRef]
- Yang, J.; Han, Z. Efficiency of Different Methods for DsRNA Delivery in Cotton Bollworm (Helicoverpa Armigera). J. Integr. Agric. 2014, 13, 115–123. [Google Scholar] [CrossRef]
- Kontogiannatos, D.; Swevers, L.; Maenaka, K.; Park, E.Y.; Iatrou, K.; Kourti, A. Functional Characterization of a Juvenile Hormone Esterase Related Gene in the Moth Sesamia Nonagrioides through RNA Interference. PLoS ONE 2013, 8, e73834. [Google Scholar] [CrossRef]
- Caccia, S.; Astarita, F.; Barra, E.; di Lelio, I.; Varricchio, P.; Pennacchio, F. Enhancement of Bacillus Thuringiensis Toxicity by Feeding Spodoptera Littoralis Larvae with Bacteria Expressing Immune Suppressive DsRNA. J. Pest Sci. 2020, 93, 303–314. [Google Scholar] [CrossRef]
- Bento, F.M.M.; Marques, R.N.; Campana, F.B.; Demétrio, C.G.B.; Leandro, R.A.; Parra, J.R.P.; Figueira, A. Gene Silencing by RNAi via Oral Delivery of DsRNA by Bacteria in the South American Tomato Pinworm, Tuta Absoluta. Pest Manag. Sci. 2020, 76, 287–295. [Google Scholar] [CrossRef] [PubMed]
- Zhu, F.; Xu, J.; Palli, R.; Ferguson, J.; Palli, S.R. Ingested RNA Interference for Managing the Populations of the Colorado Potato Beetle, Leptinotarsa Decemlineata. Pest Manag. Sci. 2011, 67, 175–182. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Xu, L.; Li, S.; Zhang, J. Bacteria-Mediated RNA Interference for Management of Plagiodera Versicolora (Coleoptera: Chrysomelidae). Insects 2019, 10, 415. [Google Scholar] [CrossRef] [PubMed]
- Murphy, K.A.; Tabuloc, C.A.; Cervantes, K.R.; Chiu, J.C. Ingestion of Genetically Modified Yeast Symbiont Reduces Fitness of an Insect Pest via RNA Interference. Sci. Rep. 2016, 6, 22587. [Google Scholar] [CrossRef]
- Van Ekert, E.; Powell, C.A.; Shatters, R.G.; Borovsky, D. Control of Larval and Egg Development in Aedes Aegypti with RNA Interference against Juvenile Hormone Acid Methyl Transferase. J. Insect Physiol. 2014, 70, 143–150. [Google Scholar] [CrossRef]
- Whitten, M.M.A.; Facey, P.D.; del Sol, R.; Fernández-Martínez, L.T.; Evans, M.C.; Mitchell, J.J.; Bodger, O.G.; Dyson, P.J. Symbiont-Mediated RNA Interference in Insects. Proc. Biol. Sci. 2016, 283, 20160042. [Google Scholar] [CrossRef]
- He, B.; Chu, Y.; Yin, M.; Müllen, K.; An, C.; Shen, J. Fluorescent Nanoparticle Delivered DsRNA Toward Genetic Control of Insect Pests. Adv. Mater. 2013, 25, 4580–4584. [Google Scholar] [CrossRef]
- Christiaens, O.; Tardajos, M.G.; Reyna, Z.L.M.; Dash, M.; Dubruel, P.; Smagghe, G. Increased RNAi Efficacy in Spodoptera Exigua via the Formulation of DsRNA with Guanylated Polymers. Front. Physiol. 2018, 9, 316. [Google Scholar] [CrossRef]
- Mysore, K.; Flannery, E.M.; Tomchaney, M.; Severson, D.W.; Duman-Scheel, M. Disruption of Aedes Aegypti Olfactory System Development through Chitosan/SiRNA Nanoparticle Targeting of Semaphorin-1a. PLoS Negl. Trop. Dis. 2013, 7, e2215. [Google Scholar] [CrossRef]
- Mysore, K.; Andrews, E.; Li, P.; Duman-Scheel, M. Chitosan/SiRNA Nanoparticle Targeting Demonstrates a Requirement for Single-Minded during Larval and Pupal Olfactory System Development of the Vector Mosquito Aedes Aegypti. BMC Dev. Biol. 2014, 14, 9. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Zhang, J.; Zhu, K.Y. Chitosan/Double-Stranded RNA Nanoparticle-Mediated RNA Interference to Silence Chitin Synthase Genes through Larval Feeding in the African Malaria Mosquito (Anopheles Gambiae). Insect Mol. Biol. 2010, 19, 683–693. [Google Scholar] [CrossRef] [PubMed]
- Whyard, S.; Singh, A.D.; Wong, S. Ingested Double-Stranded RNAs Can Act as Species-Specific Insecticides. Insect Biochem. Mol. Biol. 2009, 39, 824–832. [Google Scholar] [CrossRef] [PubMed]
- Taning, C.N.T.; Christiaens, O.; Berkvens, N.; Casteels, H.; Maes, M.; Smagghe, G. Oral RNAi to Control Drosophila Suzukii: Laboratory Testing against Larval and Adult Stages. J. Pest Sci. 2016, 89, 803–814. [Google Scholar] [CrossRef]
- Bedoya-Pérez, L.P.; Cancino-Rodezno, A.; Flores-Escobar, B.; Soberón, M.; Bravo, A. Role of UPR Pathway in Defense Response of Aedes Aegypti against Cry11Aa Toxin from Bacillus Thuringiensis. Int. J. Mol. Sci. 2013, 14, 8467. [Google Scholar] [CrossRef]
- Cancino-Rodezno, A.; Alexander, C.; Villaseñor, R.; Pacheco, S.; Porta, H.; Pauchet, Y.; Soberón, M.; Gill, S.S.; Bravo, A. The Mitogen-Activated Protein Kinase P38 Is Involved in Insect Defense against Cry Toxins from Bacillus Thuringiensis. Insect Biochem. Mol. Biol. 2010, 40, 58–63. [Google Scholar] [CrossRef]
- Derakhshankhah, H.; Jafari, S. Cell Penetrating Peptides: A Concise Review with Emphasis on Biomedical Applications. Biomed. Pharmacother. 2018, 108, 1090–1096. [Google Scholar] [CrossRef]
- Durzyńska, J.; Przysiecka, Ł.; Nawrot, R.; Barylski, J.; Nowicki, G.; Warowicka, A.; Musidlak, O.; Goździcka-Józefiak, A. Viral and Other Cell-Penetrating Peptides as Vectors of Therapeutic Agents in Medicine. J. Pharmacol. Exp. Ther. 2015, 354, 32–42. [Google Scholar] [CrossRef]
- Agrawal, P.; Bhalla, S.; Usmani, S.S.; Singh, S.; Chaudhary, K.; Raghava, G.P.S.; Gautam, A. CPPsite 2.0: A Repository of Experimentally Validated Cell-Penetrating Peptides. Nucleic Acids Res. 2016, 44, D1098. [Google Scholar] [CrossRef]
- Cermenati, G.; Terracciano, I.; Castelli, I.; Giordana, B.; Rao, R.; Pennacchio, F.; Casartelli, M. The CPP Tat Enhances EGFP Cell Internalization and Transepithelial Transport by the Larval Midgut of Bombyx Mori (Lepidoptera, Bombycidae). J. Insect Physiol. 2011, 57, 1689–1697. [Google Scholar] [CrossRef]
- Chen, Y.J.; Liu, B.R.; Dai, Y.H.; Lee, C.Y.; Chan, M.H.; Chen, H.H.; Chiang, H.J.; Lee, H.J. A Gene Delivery System for Insect Cells Mediated by Arginine-Rich Cell-Penetrating Peptides. Gene 2012, 493, 201–210. [Google Scholar] [CrossRef]
- Zhou, Z.; Li, Y.; Yuan, C.; Zhang, Y.; Qu, L.; Bendena, B. Oral Administration of TAT-PTD–Diapause Hormone Fusion Protein Interferes With Helicoverpa armigera (Lepidoptera: Noctuidae) Development. J. Insect Sci. 2015, 15, 123. [Google Scholar] [CrossRef] [PubMed]
- Gillet, F.X.; Garcia, R.A.; Macedo, L.L.P.; Albuquerque, E.V.S.; Silva, M.C.M.; Grossi-de-Sa, M.F. Investigating Engineered Ribonucleoprotein Particles to Improve Oral RNAi Delivery in Crop Insect Pests. Front. Physiol. 2017, 8, 256. [Google Scholar] [CrossRef] [PubMed]
- Lundberg, P.; El-Andaloussi, S.; Sütlü, T.; Johansson, H.; Langel, Ü. Delivery of Short Interfering RNA Using Endosomolytic Cell-Penetrating Peptides. FASEB J. 2007, 21, 2664–2671. [Google Scholar] [CrossRef]
- Deshayes, S.; Heitz, A.; Morris, M.C.; Charnet, P.; Divita, G.; Heitz, F. Insight into the Mechanism of Internalization of the Cell-Penetrating Carrier Peptide Pep-1 through Conformational Analysis†. Biochemistry 2004, 43, 1449–1457. [Google Scholar] [CrossRef] [PubMed]
- Raja, S.M.; Rawat, S.S.; Chattopadhyay, A.; Lala, A.K. Localization and Environment of Tryptophans in Soluble and Membrane-Bound States of a Pore-Forming Toxin from Staphylococcus Aureus. Biophys. J. 1999, 76, 1469–1479. [Google Scholar] [CrossRef] [PubMed]
- Tilley, S.J.; Saibil, H.R. The Mechanism of Pore Formation by Bacterial Toxins. Curr. Opin. Struct. Biol. 2006, 16, 230–236. [Google Scholar] [CrossRef] [PubMed]
- Futaki, S.; Suzuki, T.; Ohashi, W.; Yagami, T.; Tanaka, S.; Ueda, K.; Sugiura, Y. Arginine-Rich Peptides. J. Biol. Chem. 2001, 276, 5836–5840. [Google Scholar] [CrossRef]
- Wender, P.A.; Mitchell, D.J.; Pattabiraman, K.; Pelkey, E.T.; Steinman, L.; Rothbard, J.B. The Design, Synthesis, and Evaluation of Molecules That Enable or Enhance Cellular Uptake: Peptoid Molecular Transporters. Proc. Natl. Acad. Sci. USA 2000, 97, 13003–13008. [Google Scholar] [CrossRef]
- Herce, H.D.; Garcia, A.E. Molecular Dynamics Simulations Suggest a Mechanism for Translocation of the HIV-1 TAT Peptide across Lipid Membranes. Proc. Natl. Acad. Sci. USA 2007, 104, 20805–20810. [Google Scholar] [CrossRef]
- Rothbard, J.B.; Jessop, T.C.; Wender, P.A. Adaptive Translocation: The Role of Hydrogen Bonding and Membrane Potential in the Uptake of Guanidinium-Rich Transporters into Cells. Adv. Drug Deliv. Rev. 2005, 57, 495–504. [Google Scholar] [CrossRef]
- Plank, C.; Oberhauser, B.; Mechtler, K.; Koch, C.; Wagner, E. The Influence of Endosome-Disruptive Peptides on Gene Transfer Using Synthetic Virus-like Gene Transfer Systems. J. Biol. Chem. 1994, 269, 12918–12924. [Google Scholar] [CrossRef] [PubMed]
- Luneberg, J.; Martin, I.; Nussler, F.; Ruysschaert, J.M.; Herrmann, A. Structure and Topology of the Influenza Virus Fusion Peptide in Lipid Bilayers. J. Biol. Chem. 1995, 270, 27606–27614. [Google Scholar] [CrossRef] [PubMed]
- Xu, W.; Jafari, M.; Yuan, F.; Pan, R.; Chen, B.; Ding, Y.; Sheinin, T.; Chu, D.; Lu, S.; Yuan, Y.; et al. In Vitro and in Vivo Therapeutic SiRNA Delivery Induced by a Tryptophan-Rich Endosomolytic Peptide. J. Mater. Chem. B 2014, 2, 6010–6019. [Google Scholar] [CrossRef]
- Jafari, M.; Karunaratne, D.N.; Sweeting, C.M.; Chen, P. Modification of a Designed Amphipathic Cell-Penetrating Peptide and Its Effect on Solubility, Secondary Structure, and Uptake Efficiency. Biochemistry 2013, 52, 3428–3435. [Google Scholar] [CrossRef]
- Jafari, M.; Xu, W.; Pan, R.; Sweeting, C.M.; Karunaratne, D.N.; Chen, P. Serum Stability and Physicochemical Characterization of a Novel Amphipathic Peptide C6M1 for SiRNA Delivery. PLoS ONE 2014, 9, e97797. [Google Scholar] [CrossRef]
- Wadia, J.S.; Stan, R.V.; Dowdy, S.F. Transducible TAT-HA Fusogenic Peptide Enhances Escape of TAT-Fusion Proteins after Lipid Raft Macropinocytosis. Nat. Med. 2004, 10, 310–315. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.H.; Seo, H.H.; Han, S.J.; Yoon, E.K.; Yang, M.S.; Lee, W.S. Phytohormone Abscisic Acid Control RNA-Dependent RNA Polymerase 6 Gene Expression and Post-Transcriptional Gene Silencing in Rice Cells. Nucleic Acids Res. 2008, 36, 1220. [Google Scholar] [CrossRef] [PubMed]
- Unnamalai, N.; Kang, B.G.; Lee, W.S. Cationic Oligopeptide-Mediated Delivery of DsRNA for Post-Transcriptional Gene Silencing in Plant Cells. FEBS Lett. 2004, 566, 307–310. [Google Scholar] [CrossRef]
- Van Hiel, M.B.; van Wielendaele, P.; Temmerman, L.; van Soest, S.; Vuerinckx, K.; Huybrechts, R.; vanden Broeck, J.; Simonet, G. Identification and Validation of Housekeeping Genes in Brains of the Desert Locust Schistocerca gregaria under Different Developmental Conditions. BMC Mol. Biol. 2009, 10, 56. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2−ΔΔCT Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Huang, J.H.; Jing, X.; Douglas, A.E. The Multi-Tasking Gut Epithelium of Insects. Insect Biochem. Mol. Biol. 2015, 67, 15. [Google Scholar] [CrossRef]
- Holtof, M.; Lenaerts, C.; Cullen, D.; vanden Broeck, J. Extracellular Nutrient Digestion and Absorption in the Insect Gut. Cell Tissue Res. 2019, 377, 397–414. [Google Scholar] [CrossRef] [PubMed]
- Chapman, R.F. The Head, Ingestion, Utilization and Distribution Of Food. In The Insects: Structure and Function; Cambridge University Press: Cambridge, UK, 2013; pp. 3–11. [Google Scholar]
- Saleh, M.C.; van Rij, R.P.; Hekele, A.; Gillis, A.; Foley, E.; O’Farrell, P.H.; Andino, R. The Endocytic Pathway Mediates Cell Entry of DsRNA to Induce RNAi Silencing. Nat. Cell Biol. 2006, 8, 793. [Google Scholar] [CrossRef] [PubMed]
- Miller, S.C.; Miyata, K.; Brown, S.J.; Tomoyasu, Y. Dissecting Systemic RNA Interference in the Red Flour Beetle Tribolium Castaneum: Parameters Affecting the Efficiency of RNAi. PLoS ONE 2012, 7, e47431. [Google Scholar] [CrossRef]
- He, W.; Xu, W.; Xu, L.; Fu, K.; Guo, W.; Bock, R.; Zhang, J. Length-Dependent Accumulation of Double-Stranded RNAs in Plastids Affects RNA Interference Efficiency in the Colorado Potato Beetle. J. Exp. Bot. 2020, 71, 2670–2677. [Google Scholar] [CrossRef] [PubMed]
- Kumar, M.; Gupta, G.P.; Rajam, M.V. Silencing of Acetylcholinesterase Gene of Helicoverpa armigera by SiRNA Affects Larval Growth and Its Life Cycle. J. Insect Physiol. 2009, 55, 273–278. [Google Scholar] [CrossRef]
- Yamaguchi, J.; Mizoguchi, T.; Fujiwara, H. SiRNAs Induce Efficient RNAi Response in Bombyx Mori Embryos. PLoS ONE 2011, 6, e25469. [Google Scholar] [CrossRef]
- Zhang, Y.; Li, Z.; Wang, Z.L.; Zhang, L.Z.; Zeng, Z.J. A Comparison of RNA Interference via Injection and Feeding in Honey Bees. Insects 2022, 13, 928. [Google Scholar] [CrossRef]
- Chen, M.; Li, B. The Effect of Molecular Weights on the Survivability of Casein-Derived Antioxidant Peptides after the Simulated Gastrointestinal Digestion. Innov. Food Sci. Emerg. Technol. 2012, 16, 341–348. [Google Scholar] [CrossRef]
- Copolovici, D.M.; Langel, K.; Eriste, E.; Langel, Ü. Cell-Penetrating Peptides: Design, Synthesis, and Applications. ACS Nano 2014, 8, 1972–1994. [Google Scholar] [CrossRef]
- Margus, H.; Padari, K.; Pooga, M. Cell-Penetrating Peptides as Versatile Vehicles for Oligonucleotide Delivery. Mol. Ther. 2012, 20, 525–533. [Google Scholar] [CrossRef]
- Allen, M.L.; Walker, W.B. Saliva of Lygus Lineolaris Digests Double Stranded Ribonucleic Acids. J. Insect Physiol. 2012, 58, 391–396. [Google Scholar] [CrossRef] [PubMed]
- Sharma, R.; Taning, C.N.T.; Smagghe, G.; Christiaens, O. Silencing of Double-Stranded Ribonuclease Improves Oral RNAi Efficacy in Southern Green Stinkbug Nezaraviridula. Insects 2021, 12, 115. [Google Scholar] [CrossRef] [PubMed]
- Spit, J.; Philips, A.; Wynant, N.; Santos, D.; Plaetinck, G.; vanden Broeck, J. Knockdown of Nuclease Activity in the Gut Enhances RNAi Efficiency in the Colorado Potato Beetle, Leptinotarsa Decemlineata, but Not in the Desert Locust, Schistocerca gregaria. Insect Biochem. Mol. Biol. 2017, 81, 103–116. [Google Scholar] [CrossRef]
- Vivès, E.; Josse, E.; Konate, K.; Deshayes, S.; Boisguérin, P. Cell-Penetrating Peptides for Nucleic Acid Delivery—An Update. Cell Penetrat. Pept. 2023, 262, 237–262. [Google Scholar] [CrossRef]
- Verdonckt, T.W.; Vanden Broeck, J. Methods for the Cost-Effective Production of Bacteria-Derived Double-Stranded RNA for in Vitro Knockdown Studies. Front. Physiol. 2022, 13, 559. [Google Scholar] [CrossRef]
- Wu, M.; Dong, Y.; Zhang, Q.; Li, S.; Chang, L.; Vanessa Loiacono, F.; Ruf, S.; Zhang, J.; Bock, R. Efficient Control of Western Flower Thrips by Plastid-Mediated RNA Interference. Proc. Natl. Acad. Sci. USA 2022, 119, e2120081119. [Google Scholar] [CrossRef]





| Name | Sequence | Properties | Origin | Charge (pH 7) | Mr (kDa) | Reference |
|---|---|---|---|---|---|---|
| EB1 | LIRLWSHLIHIWFQNRRLK WKKK-amide | Endosomolytic, amphipatic. | Penetratin derivative. | +8 | 3.66 | [34] |
| C6M1 | RLWRLLWRLWRRLWRLLR | Endosomolytic, amphipatic. | Synthetic—based on C6. | +7 | 2.67 | [44,45,46] |
| HA2-penetratin | GLFGAIAGFIENGWEGMI DGRQIKIWFQNRRMKW KK-amide | Fusogenic, amphipatic. | Chimeric—fusion of influenza virus hemagglutinin (HA) subunit and penetratin, derived from the third α-helix of the Drosophila Antennapedia homeodomain. | +5 | 4.28 | [34] |
| HA2-TAT | GLFGAIAGFIENGWEGLIEG WYGGRKKRRQRRR | Fusogenic, amphipatic. | Chimeric—fusion of influenza virus hemagglutinin (HA) subunit and HIV-1 Trans-Activator of Transcription (TAT) protein. | +5 | 3.84 | [47] |
| POA | RRRRRRRRRRRR | Cationic. | Synthetic—based on TAT peptide. | +12 | 1.89 | [48,49] |
| Forward Primer | Reverse Primer | |
|---|---|---|
| dsTub | taatacgactcactataggg attttttagcgaaactggtgctggg | taatacgactcactataggg tggtgtaagtcgggcgttcaatgt |
| CPP | Weight Ratio (CPP:dsRNA) | Molar Ratio (CPP:dsRNA) | Weight Ratio (CPP:siRNA) | Molar Ratio (CPP:siRNA) |
|---|---|---|---|---|
| EB1 | 1:1 | 93:1 | 1:1 | 4:1 |
| 2:1 | 187:1 | 2:1 | 7:1 | |
| 5:1 | 466:1 | 5:1 | 18:1 | |
| 10:1 | 933:1 | |||
| C6M1 | 1:1 | 128:1 | ||
| 2:1 | 256:1 | |||
| 5:1 | 639:1 | |||
| 10:1 | 1278:1 | |||
| HA2-penetratin | 1:1 | 80:1 | ||
| 2:1 | 159:1 | |||
| 5:1 | 399:1 | |||
| 10:1 | 797:1 | |||
| HA2-TAT | 1:1 | 89:1 | ||
| 2:1 | 178:1 | |||
| 5:1 | 444:1 | |||
| 10:1 | 889:1 | |||
| POA | 1:1 | 181:1 | ||
| 2:1 | 361:1 | |||
| 5:1 | 903:1 | |||
| 10:1 | 1806:1 |
| Fw Primer | Rv Primer | |
|---|---|---|
| Tub | Tgacaatgaggccatctatg | cgcaaagatgctgtgattga |
| GAPDH | Gtctgatgacaacagtgcat | gtccatcacgccacacttc |
| EF1a | Gatgctccaggccacagaga | tgcacagtcggcctgtgat |
| Ubi | Gactttgaggtgtggcgtag | ggatcacaaacacagaacga |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Vogel, E.; Santos, D.; Huygens, C.; Peeters, P.; Van den Brande, S.; Wynant, N.; Vanden Broeck, J. The Study of Cell-Penetrating Peptides to Deliver dsRNA and siRNA by Feeding in the Desert Locust, Schistocerca gregaria. Insects 2023, 14, 597. https://doi.org/10.3390/insects14070597
Vogel E, Santos D, Huygens C, Peeters P, Van den Brande S, Wynant N, Vanden Broeck J. The Study of Cell-Penetrating Peptides to Deliver dsRNA and siRNA by Feeding in the Desert Locust, Schistocerca gregaria. Insects. 2023; 14(7):597. https://doi.org/10.3390/insects14070597
Chicago/Turabian StyleVogel, Elise, Dulce Santos, Cissy Huygens, Paulien Peeters, Stijn Van den Brande, Niels Wynant, and Jozef Vanden Broeck. 2023. "The Study of Cell-Penetrating Peptides to Deliver dsRNA and siRNA by Feeding in the Desert Locust, Schistocerca gregaria" Insects 14, no. 7: 597. https://doi.org/10.3390/insects14070597
APA StyleVogel, E., Santos, D., Huygens, C., Peeters, P., Van den Brande, S., Wynant, N., & Vanden Broeck, J. (2023). The Study of Cell-Penetrating Peptides to Deliver dsRNA and siRNA by Feeding in the Desert Locust, Schistocerca gregaria. Insects, 14(7), 597. https://doi.org/10.3390/insects14070597







