Genetic Differentiation of Aedes aegypti (Diptera: Culicidae) in Areas with High Rates of Infestation in Mid-North Region of Brazil
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Area and Sample Collection
2.2. Extraction and Amplification of the DNA, and Genotyping
2.3. Data Analysis
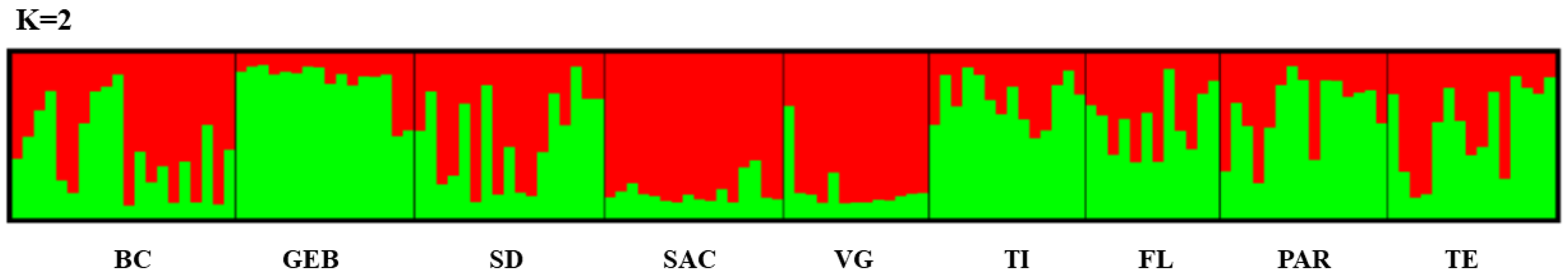
3. Results
4. Discussion
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Consoli, R.A.G.B.; Oliveira, R.L. Principais Mosquitos de Importância Sanitária No Brasil; Editora Fiocruz: Rio de Janeiro, Brazil, 1994; ISBN 8585676035. [Google Scholar]
- Bryant, J.E.; Holmes, E.C.; Barrett, A.D.T. Out of Africa: A Molecular Perspective on the Introduction of Yellow Fever Virus into the Americas. PLoS Pathog. 2007, 3, 0668–0673. [Google Scholar] [CrossRef]
- De Oliveira, N.C.; de Faria Souza, D.T.; Martins, M.; de Souza, E.A.; Medeiros, M.O. Dinâmica Populacional de Aedes (Stegomyia) aegypti (Linnaeus, 1762) (DIPTERA: CULICIDAE); Campus Universitário de Rondonópolis: Rondonópolis, Brazil, 2017; Volume 16. [Google Scholar]
- Forattini, O.P. Culicidologia Médica: Identificaçäo, Biologia e Epidemiologia: V.2; Universidade de São Paulo: São Paulo, Brazil, 2002. [Google Scholar]
- Lima-Camara, T.N. Emerging Arboviruses and Public Health Challenges in Brazil. Rev. Saude Publica 2016, 50, 36. [Google Scholar] [CrossRef]
- Soares-da-Silva, J.; Ibiapina, S.S.; Bezerra, J.M.T.; Tadei, W.P.; Pinheiro, V.C.S. Variation in Aedes Aegypti (Linnaeus) (Diptera, Culicidae) Infestation in Artificial Containers in Caxias, State of Maranhão, Brazil. Rev. Soc. Bras. Med. Trop. 2012, 45, 174–179. [Google Scholar] [CrossRef]
- Bezerra, J.; Silva Santana, I.; Miranda, J.; Tadei, W.; Soares Pinheiro, V. Breeding Sites of Aedes Aegypti (Linnaeus) (Diptera, Culicidae): Study About the Containers in Dry and Rainy Seasons in Dengue-Endemic City. Rev. Pesq. Saude 2017, 18, 102–107. [Google Scholar]
- Vargas, L.D.L.; Ferreira, S.M.B.; Souza, M.D.; da Silva, C.A.L.; Shimoya-Bittencourt, W. Resistência Das Populações de Aedes (Stegomyia) aegypti (Linnaeus, 1762) (Insecta, Diptera, Culicidae) Aos Inseticidas Utilizados Para o Controle. Rev. Ciências Médicas E Biológicas 2022, 21, 98–116. [Google Scholar] [CrossRef]
- Scarpassa, V.M.; Cardoza, T.B.; Cardoso, R.P. Population Genetics and Phylogeography of Aedes Aegypti (Diptera: Culicidae) from Brazil. Am. J. Trop. Med. Hyg. 2008, 78, 895–903. [Google Scholar] [CrossRef]
- Brasil Boletim Epidemiológico Vol. 53 N° 24—Português (Brasil). Available online: https://www.gov.br/saude/pt-br/centrais-de-conteudo/publicacoes/boletins/boletins-epidemiologicos/edicoes/2022/boletim-epidemiologico-vol-53-no24/view (accessed on 20 July 2022).
- Kasprzykowski, J.I.; Fukutani, K.F.; Fabio, H.; Fukutani, E.R.; Costa, L.C.; Andrade, B.B.; Queiroz, A.T.L. A Recursive Sub-Typing Screening Surveillance System Detects the Appearance of the ZIKV African Lineage in Brazil: Is There a Risk of a New Epidemic? Int. J. Infect. Dis. 2020, 96, 579–581. [Google Scholar] [CrossRef]
- Li, Y.C.; Korol, A.B.; Fahima, T.; Nevo, E. Microsatellites within Genes: Structure, Function, and Evolution. Mol. Biol. Evol. 2004, 21, 991–1007. [Google Scholar] [CrossRef]
- Paupy, C.; Brengues, C.; Ndiath, O.; Toty, C.; Hervé, J.P.; Simard, F. Morphological and Genetic Variability within Aedes aegypti in Niakhar, Senegal. Infect. Genet. Evol. 2010, 10, 473–480. [Google Scholar] [CrossRef]
- Paupy, C.; le Goff, G.; Brengues, C.; Guerra, M.; Revollo, J.; Barja Simon, Z.; Hervé, J.P.; Fontenille, D. Genetic Structure and Phylogeography of Aedes aegypti, the Dengue and Yellow-Fever Mosquito Vector in Bolivia. Infect. Genet. Evol. 2012, 12, 1260–1269. [Google Scholar] [CrossRef]
- Fonzi, E.; Higa, Y.; Bertuso, A.G.; Futami, K.; Minakawa, N. Human-mediated marine dispersal influences the population structure of Aedes aegypti in the Philippine archipelago. PLoS Negl. Trop. 2015, 9, e0003829. [Google Scholar] [CrossRef]
- Maitra, A.; Cunha-Machado, A.S.; de Souza Leandro, A.; da Costa, F.M.; Scarpassa, V.M. Exploring Deeper Genetic Structures: Aedes aegypti in Brazil. Acta Trop. 2019, 195, 68–77. [Google Scholar] [CrossRef]
- Kumar, M.S.; Kalimuthu, M.; Selvam, A.; Mathivanan, A.; Paramasivan, R.; Kumar, A.; Gupta, B. Genetic structure and connectivity among Aedes aegypti populations within Madurai city in Southern India. Infect. Genet. Evol. 2021, 95, 105031. [Google Scholar] [CrossRef]
- Brasil Boletim Epidemiológico Vol. 51 N° 5—Português (Brasil). Available online: https://www.gov.br/saude/pt-br/centrais-de-conteudo/publicacoes/boletins/boletins-epidemiologicos/edicoes/2020/boletim_epidemiologico_svs_51.pdf/view (accessed on 21 July 2022).
- Brasil Boletim Epidemiológico Vol. 52-N° 48—Português (Brasil). Available online: https://www.gov.br/saude/pt-br/centrais-de-conteudo/publicacoes/boletins/boletins-epidemiologicos/edicoes/2021/boletim-epidemiologico-vol-52-no-48.pdf/view (accessed on 21 July 2022).
- Aragão, C.F.; Cruz, A.C.R.; Neto, J.P.N.; de Oliveira Monteiro, H.A.; da Silva, E.V.P.; da Silva, S.P.; dos Santos Andrade, A.T.; Tadei, W.P.; Pinheiro, V.C.S. Circulation of Chikungunya Virus in Aedes aegypti in Maranhão, Northeast Brazil. Acta Trop. 2018, 186, 1–4. [Google Scholar] [CrossRef]
- Aragão, C.F.; Pinheiro, V.C.S.; Neto, J.P.N.; da Silva, E.V.P.; Pereira, G.J.G.; do Nascimento, B.L.S.; Castro, K.D.S.; Maia, A.M.; Catete, C.P.; Martins, L.C.; et al. Natural Infection of Aedes aegypti by Chikungunya and Dengue Type 2 Virus in a Transition Area of North-Northeast Brazil. Viruses 2019, 11, 1126. [Google Scholar] [CrossRef]
- De Carvalho, A.C.P.; Couto Portela, F.; Azevedo Feitosa Ferro, T.; Quaresma Bomfim, M.R. Epidemiologia do vírus do dengue em São Luís, Maranhão, no período de 2002 a 2012. Rev. Patol. Trop. 2016, 45, 243. [Google Scholar] [CrossRef]
- Fraga, E.C.; Oliveira, D.R.S.; Aragão, D.G.; Sampaio, H.S.; Barros, M.C. Genetic Variability and Evidence of Two Distinct Lineages of Aedes aegypti (Diptera, Culicidae) on São Luís Island in Maranhão, Brazil. Open Trop. Med. J. 2013, 6, 11–18. [Google Scholar] [CrossRef]
- De Sousa, A.A.; Fraga, E.; Sampaio, I.; Schneider, H.; Barros, M.C. Genetic Differentiation in Populations of Aedes aegypti (Diptera, Culicidae) Dengue Vector from the Brazilian State of Maranhão. Rev. Bras. Entomol. 2017, 61, 51–59. [Google Scholar] [CrossRef]
- Rebouças, A.d.C. Água Na Região Nordeste: Desperdício e Escassez. Estud. Avançados 1997, 11, 127–154. [Google Scholar] [CrossRef]
- Brasil Liraa-1-2019.Pdf—Português (Brasil). Available online: https://www.gov.br/saude/pt-br/centrais-de-conteudo/arquivos/liraa-1-2019-pdf/view (accessed on 21 July 2022).
- Fay, R.W.; Perry, A.S. Laboratory Studies of Oviposilional Preferences of Aedes aegypti. Mosq. News 1965, 25, 276–281. [Google Scholar]
- Dos Santos, J.M.M.; Contel, E.P.B.; Kerr, W.E. Biologia de anofelinos amazônicos. 1—Ciclo biológico, postura e estádios larvais de Anopheles darlingi Root 1926 (Diptera: Culicidae) da Rodovia Manaus—Boa Vista. Acta Amaz. 1981, 11, 789–797. [Google Scholar] [CrossRef]
- Huber, K.; Mousson, L.; Rodhain, F.; Failloux, A.-B. Isolation and Variability of Polymorphic Microsatellite Loci in Aedes aegypti, the Vector of Dengue Viruses. Mol. Ecol. Notes 2001, 1, 219–222. [Google Scholar] [CrossRef]
- Chambers, E.W.; Meece, J.K.; McGowan, J.A.; Lovin, D.D.; Hemme, R.R.; Chadee, D.D.; McAbee, K.; Brown, S.E.; Knudson, D.L.; Severson, D.W. Microsatellite Isolation and Linkage Group Identification in the Yellow Fever Mosquito Aedes aegypti. J. Hered. 2007, 98, 202–210. [Google Scholar] [CrossRef]
- Hulce, D.; Li, X.; Snyder-Leiby, T.; Liu, C.J. GeneMarker® genotyping software: Tools to increase the statistical power of DNA fragment analysis. J. Biomol. Tech. JBT 2011, 22, S35. [Google Scholar]
- Coombs, J.A.; Letcher, B.H.; Nislow, K.H. Create: A Software to Create Input Files from Diploid Genotypic Data for 52 Genetic Software Programs. Mol. Ecol. Resour. 2008, 8, 578–580. [Google Scholar] [CrossRef] [PubMed]
- van Oosterhout, C.; Hutchinson, W.F.; Wills, D.P.M.; Shipley, P. MICRO-CHECKER: Software for Identifying and Correcting Genotyping Errors in Microsatellite Data. Mol. Ecol. Notes 2004, 4, 535–538. [Google Scholar] [CrossRef]
- Peakall, R.O.D.; Smouse, P.E. GENALEX 6: Genetic analysis in Excel. Population genetic software for teaching and research. Mol. Ecol. Notes 2006, 6, 288–295. [Google Scholar] [CrossRef]
- Rousset, F. GENEPOP’007: A Complete Re-Implementation of the GENEPOP Software for Windows and Linux. Mol. Ecol. Resour. 2008, 8, 103–106. [Google Scholar] [CrossRef]
- GOUDET, J. FSTAT (Version 1.2): A Computer Program to Calculate F-Statistics. J. Hered. 1995, 86, 485–486. [Google Scholar] [CrossRef]
- Weir, B.S. Genetic Data Analysis II. Biometrics 1997, 53, 392. [Google Scholar] [CrossRef]
- Excoffier, L.; Lischer, H.E.L. Arlequin Suite Ver 3.5: A New Series of Programs to Perform Population Genetics Analyses under Linux and Windows. Mol. Ecol. Resour. 2010, 10, 564–567. [Google Scholar] [CrossRef] [PubMed]
- Novembre, J. Pritchard, Stephens, and Donnelly on Population Structure. Genetics 2016, 204, 391–393. [Google Scholar] [CrossRef] [PubMed]
- Evanno, G.; Regnaut, S.; Goudet, J. Detecting the Number of Clusters of Individuals Using the Software STRUCTURE: A Simulation Study. Mol. Ecol. 2005, 14, 2611–2620. [Google Scholar] [CrossRef] [PubMed]
- Earl, D.A.; vonHoldt, B.M. STRUCTURE HARVESTER: A Website and Program for Visualizing STRUCTURE Output and Implementing the Evanno Method. Conserv. Genet. Resour. 2012, 4, 359–361. [Google Scholar] [CrossRef]
- Jakobsson, M.; Rosenberg, N.A. CLUMPP: A cluster matching and permutation program for dealing with label switching and multimodality in analysis of population structure. Bioinformatics 2007, 23, 1801–1806. [Google Scholar] [CrossRef]
- Li, Y.L.; Liu, J.X. StructureSelector: A Web-Based Software to Select and Visualize the Optimal Number of Clusters Using Multiple Methods. Mol. Ecol. Resour. 2018, 18, 176–177. [Google Scholar] [CrossRef]
- Do, C.; Waples, R.S.; Peel, D.; Macbeth, G.M.; Tillett, B.J.; Ovenden, J.R. NeEstimator v2: Re-Implementation of Software for the Estimation of Contemporary Effective Population Size (Ne) from Genetic Data. Mol. Ecol. Resour. 2014, 14, 209–214. [Google Scholar] [CrossRef] [PubMed]
- Piry, S.; Luikart, G.; Cornuet, J.M. Computer note. BOTTLENECK: A computer program for detecting recent reductions in the effective size using allele frequency data. J. Hered. 1999, 90, 502–503. [Google Scholar] [CrossRef]
- Louise, C.; Vidal, P.O.; Suesdek, L. Microevolution of Aedes aegypti. PLoS ONE 2015, 10, e0137851. [Google Scholar] [CrossRef]
- Mendonça, B.A.A.; de Sousa, A.C.B.; de Souza, A.P.; Scarpassa, V.M. Temporal Genetic Structure of Major Dengue Vector Aedes aegypti from Manaus, Amazonas, Brazil. Acta Trop. 2014, 134, 80–88. [Google Scholar] [CrossRef]
- Paupy, C.; Brengues, C.; Kamgang, B.; Hervé, J.P.; Fontenille, D.; Simard, F. Gene flow between domestic and sylvan populations of Aedes aegypti (Diptera: Culicidae) in North Cameroon. J. Med. Entomol. 2014, 45, 391–400. [Google Scholar] [CrossRef]
- Honório, N.A.; Silva, W.C.; Leite, P.J.; Gonçalves, J.M.; Lounibos, L.P.; Lourenço-De-Oliveira, R. Dispersal of Aedes aegypti and Aedes albopictus (Diptera: Culicidae) in an urban endemic dengue area in the State of Rio de Janeiro, Brazil. Mem. Inst. Oswaldo Cruz 2003, 98, 191–198. [Google Scholar] [CrossRef] [PubMed]
- Harrington, L.C.; Edman, J.D.; Kittayapong, P.; Coleman, R.C.; Clark, G.G.; Sithiprasasna, R.; Kitthawee, S.; Lerdthusnee, K.; Scott, T.W.; Costero, A.; et al. Dispersal of the dengue vector aedes aegypti within and BETWEEN rural communities. Am. J. Trop. Med. Hyg. 2005, 72, 209–220. [Google Scholar] [CrossRef]
- Bosio, C.F.; Harrington, L.C.; Norris, D.; Scott, T.W.; Jones, J.W.; Sithiprasasna, R. Genetic structure of aedes aegypti populations in thailand using mitochondrial dna. Am. J. Trop. Med. Hyg. 2005, 72, 434–442. [Google Scholar] [CrossRef]
- Da Silva, A.G.; Cunha, I.C.L.; Santos, W.S.; Luz, S.L.B.; Ribolla, P.E.M.; Abad-Franch, F. Gene flow networks among American Aedes aegypti populations. Evol. Appl. 2012, 5, 664–676. [Google Scholar] [CrossRef]
- Kotsakiozi, P.; Gloria-Soria, A.; Caccone, A.; Evans, B.; Schama, R.; Martins, A.J.; Powell, J.R. Tracking the return of Aedes aegypti to Brazil, the major vector of the dengue, chikungunya and Zika viruses. PLOS Negl. Trop. Dis. 2017, 11, e0005653. [Google Scholar] [CrossRef] [PubMed]
- Silva, F.B.; Rafael, J.; Santos, N.; Emerson, F.; Silva, C.; Dirceu, I.; Silva, C.; Lucyanne, M.; de Araújo, S.; Guterres, C.E.; et al. Evidências de Mudanças Climáticas Na Região de Transição Amazônia-Cerrado No Estado Do Maranhão Evidence of Climate Change in the Amazon-Savanna Transition Region in Maranhão State. Rev. Bras. Meteorol. 2016, 31, 330–336. [Google Scholar] [CrossRef]
- Carvajal, T.M.; Ogishi, K.; Yaegeshi, S.; Hernandez, L.F.T.; Viacrusis, K.M.; Ho, H.T.; Amalin, D.M.; Watanabe, K. Fine-scale population genetic structure of dengue mosquito vector, Aedes aegypti, in Metropolitan Manila, Philippines. PLOS Negl. Trop. Dis. 2020, 14, e0008279. [Google Scholar] [CrossRef]
- Olanratmanee, P.; Kittayapong, P.; Chansang, C.; Hoffmann, A.A.; Weeks, A.R.; Endersby, N.M. Population Genetic Structure of Aedes (Stegomyia) aegypti (L.) at a Micro-Spatial Scale in Thailand: Implications for a Dengue Suppression Strategy. PLOS Neglected Trop. Dis. 2013, 7, e1913. [Google Scholar] [CrossRef]
- Saarman, N.P.; Gloria-Soria, A.; Anderson, E.C.; Evans, B.R.; Pless, E.; Cosme, L.V.; Gonzalez-Acosta, C.; Kamgang, B.; Wesson, D.M.; Powell, J.R. Effective Population Sizes of a Major Vector of Human Diseases, Aedes aegypti. Evol. Appl. 2017, 10, 1031–1039. [Google Scholar] [CrossRef]
- Pless, E.; Gloria-Soria, A.; Evans, B.R.; Kramer, V.; Bolling, B.G.; Tabachnick, W.J.; Powell, J.R. Multiple introductions of the dengue vector, Aedes aegypti, into California. PLOS Neglected Trop. Dis. 2017, 11, e0005718. [Google Scholar] [CrossRef]
- Zanluca, C.; de Melo, V.C.A.; Mosimann, A.L.P.; Santos, G.I.V.; dos Santos, C.N.D.; dos Luz, K. First Report of Autochthonous Transmission of Zika Virus in Brazil. Mem. Inst. Oswaldo Cruz 2015, 110, 569–572. [Google Scholar] [CrossRef]
- Wilke, A.B.B.; Wilk-Da-Silva, R.; Marrelli, M.T. Microgeographic population structuring of Aedes aegypti (Diptera: Culicidae). PLoS ONE 2017, 12, e0185150. [Google Scholar] [CrossRef]

| State | Population | Abbreviation | Coordinates (Lat./Long.) | Sample Size |
|---|---|---|---|---|
| Maranhão | Barra do Corda | BC | 5°30′21″ S | 20 |
| 45°14′6″ W | ||||
| Governador Eugênio Barros | GEB * | 5°18′39″ S | 16 | |
| 44°14′51″ W | ||||
| São Domingos | SD | 5°34′46″ S | 17 | |
| 44°22′59″ W | ||||
| Senador Alexandre Costa | SAC | 5°15′21″ S | 16 | |
| 44°03′21″ W | ||||
| Vargem Grande | VG | 3°32′34″ S | 13 | |
| 43°54′57″ W | ||||
| Timon | TI * | 5°05′38″ S | 14 | |
| 42°50′13″ W | ||||
| Piauí | Floriano | FL | 6°46′2″ S | 12 |
| 43°1′33″ W | ||||
| Parnaíba | PAR | 2°54′14″ S | 15 | |
| 41°46′35″ W | ||||
| Teresina | TE | 5°05′20″ S | 15 | |
| 42°48′07″ W |
| Locus | Type of Repeat | F | pb | r (Median) | References |
|---|---|---|---|---|---|
| 3472 | GAAA(GA)6 CAGACAGGAAA | FAM | 82–117 | −0.0071 | [26] |
| B19 | CAT7 | VIC | 101–205 | 0.0970 | [27] |
| B07 | GA15 | NED | 95–195 | 0.1040 | [27] |
| M201 | ATA36 | PET | 92–140 | −0.3894 | [27] |
| H08 | TCG7 | VIC | 101–218 | −0.0097 | [27] |
| T3A7 | (TTTA)7(T)14 | PET | 107–263 | 0.0045 | [26] |
| Locus | BC | GEB | SD | SAC | VG | PI | |
|---|---|---|---|---|---|---|---|
| 3472 | N | 20 | 16 | 17 | 16 | 13 | 56 |
| Na | 4 | 1 | 3 | 4 | 2 | 6 | |
| Ne(allele) | 2.694 | 1.000 | 2.181 | 3.413 | 1.166 | 2.899 | |
| Ar | 3.952 | 1.000 | 3.000 | 4.000 | 2.000 | 4.669 | |
| Ap | - | - | - | - | - | 2 | |
| r | 0.244 | - | - | - | 0.238 | - | |
| B19 | Na | 4 | 3 | 5 | 3 | 2 | 3 |
| Ne(allele) | 3.606 | 2.522 | 2.249 | 2.667 | 1.988 | 2.154 | |
| Ar | 4.000 | 3.000 | 4.665 | 3.000 | 2.000 | 2.858 | |
| Ap | - | - | 1 | - | - | - | |
| r | 0.300 | 0.392 | - | 0.195 | 0.035 | 0.017 | |
| B07 | Na | 5 | 3 | 5 | 3 | 5 | 9 |
| Ne(allele) | 3.980 | 2.024 | 2.833 | 1.679 | 3.449 | 3.670 | |
| Ar | 4.952 | 3.000 | 4.705 | 2.970 | 5.000 | 5.368 | |
| Ap | - | - | - | - | 1 | 2 | |
| r | 0.193 | 0.039 | 0.189 | 0,075 | 0.176 | 0.126 | |
| M201 | Na | 2 | 2 | 2 | 3 | 2 | 2 |
| Ne(allele) | 1.471 | 2.000 | 1.940 | 1.962 | 1.257 | 1.849 | |
| Ar | 2.000 | 2.000 | 2.000 | 2.813 | 2.000 | 2.000 | |
| Ap | - | - | - | 1 | - | - | |
| r | 0.029 | - | - | - | - | - | |
| H08 | Na | 3 | 2 | 3 | 1 | 2 | 3 |
| Ne(allele) | 2.986 | 1.800 | 2.261 | 1.000 | 1.257 | 2.974 | |
| Ar | 3.000 | 2.000 | 2.986 | 1.000 | 2.000 | 3.000 | |
| Ap | - | - | - | - | - | - | |
| r | - | - | 0,281 | - | - | 0.122 | |
| T3A7 | Na | 4 | 2 | 2 | 3 | 2 | 4 |
| Ne(allele) | 1.867 | 1.882 | 1.841 | 2.977 | 1.649 | 2.544 | |
| Ar | 3.651 | 2.000 | 2.000 | 3.000 | 2.000 | 3.411 | |
| Ap | 1 | - | - | - | - | 1 | |
| r | 0.064 | 0.402 | - | - | 0.011 | 0.003 | |
| Median | Na | 3.666 | 2.166 | 3.333 | 2.833 | 2.500 | 4.500 |
| Ne(allele) | 2.747 | 1.871 | 2.218 | 2.334 | 1.794 | 2.682 | |
| Ar | 3.593 | 2.167 | 3.226 | 2.797 | 2.500 | 3.551 |
| Locus | BC | GEB | SD | SAC | VG | PI | |
|---|---|---|---|---|---|---|---|
| 3472 | Ho | 0.300 | 0.000 | 0.647 | 0.750 | 0.000 | 0.673 |
| He | 0.644 | 0.000 | 0.542 | 0.707 | 0.142 | 0.655 | |
| P-HWE | 0.001 | - | 0.744 | 0.720 | 0.040 | 0.022 | |
| FIS | 0.541 | NA | −0.166 | −0.029 | 1.000 | −0.018 | |
| B19 | Ho | 0.250 | 0.063 | 0.529 | 0.357 | 0.462 | 0.509 |
| He | 0.713 | 0.604 | 0.555 | 0.630 | 0.497 | 0.536 | |
| P-HWE | 0.000 | 0.000 | 0.000 | 0.006 | 0.560 | 0.085 | |
| FIS | 0.668 | 0.903 | 0.077 | 0.463 | 0.111 | 0.059 | |
| B07 | Ho | 0.450 | 0.438 | 0.412 | 0.438 | 0.462 | 0.537 |
| He | 0.749 | 0.506 | 0.647 | 0.486 | 0.710 | 0.728 | |
| P-HWE | 0.000 | 0.027 | 0.079 | 0.370 | 0.021 | 0.008 | |
| FIS | 0.420 | 0.167 | 0.390 | 0.132 | 0.385 | 0.271 | |
| M201 | Ho | 0.320 | 1.000 | 0.824 | 0.750 | 0.231 | 0.607 |
| He | 0.328 | 0.500 | 0.484 | 0.490 | 0.204 | 0.459 | |
| P-HWE | 0.578 | 1.000 | 1.000 | 1.000 | 1.000 | 0.997 | |
| FIS | 0.088 | −1.000 | −0.684 | −0.506 | −0.091 | −0.314 | |
| H08 | Ho | 1.000 | 0.667 | 0.200 | 0.000 | 0.231 | 0.500 |
| He | 0.665 | 0.444 | 0.558 | 0.000 | 0.204 | 0.664 | |
| P-HWE | 1.000 | 1.000 | 0.001 | - | 1.000 | 0.007 | |
| FIS | −0.482 | −0.474 | −0.684 | NA | −0.091 | 0.256 | |
| T3A7 | Ho | 0.389 | 0.000 | 0.706 | 0.750 | 0.385 | 0.500 |
| He | 0.465 | 0.469 | 0.457 | 0.664 | 0.393 | 0.607 | |
| P-HWE | 0.183 | 0.000 | 1.000 | 0.591 | 0.667 | 0.001 | |
| FIS | 0.190 | 1.000 | −0.524 | −0.098 | 0.063 | 0.185 | |
| Median | Ho | 0.448 | 0.361 | 0.553 | 0.507 | 0.295 | 0.519 |
| He | 0.590 | 0.420 | 0.540 | 0.496 | 0.358 | 0.566 | |
| FIS | 0.237 | 0.099 | −0.041 | −0.006 | 0.000 | 0.073 |
| Groups of Samples | Source of Variation | Variation (%) | Fixation Index |
|---|---|---|---|
| All: BC, GEB, SD, SAC, VG, TI, FL, PAR, and TE | Among population | 14.34 | FST: 0.14340 *** |
| Within populations | 85.66 | ||
| Two Groups: Maranhão: BC, SD, SAC and VG Piauí: GEB, TI, FL, PAR, and TE | Among groups | 4.36 | FST: 0.15971 *** |
| Among populations within groups | 13.47 | FSC: 0.12144 *** | |
| Within populations | 85.01 | FCT: 0.04357 |
| POP | BC | GEB | SD | SAC | VG | TI | FL | PAR | TE |
|---|---|---|---|---|---|---|---|---|---|
| BC | 2.406 | 5.505 | 2.215 | 4.020 | 5.909 | 5.591 | 4.990 | 12.239 | |
| GEB | 0.172 (130.6) | 4.331 | 1.163 | 0.819 | 4.079 | 2.382 | 5.419 | 3.168 | |
| SD | 0.083 (132.9) | 0.103 (68.5) | 2.369 | 1.463 | 4.372 | 4.339 | 3.737 | 7.277 | |
| SAC | 0.184 (163) | 0.300 (38.8) | 0.174 (100.9) | 1.353 | 1.705 | 1.842 | 2.440 | 2.362 | |
| VG | 0.110 (322) | 0.378 (241.6) | 0.254 (265) | 0.269 (280.3) | 1.357 | 1.655 | 1.366 | 2.277 | |
| TI | 0.078 (331.9) | 0.109 (207.8) | 0.102 (245.4) | 0.226 (169.7) | 0.269 (305.7) | 12.317 | 15.054 | 35.328 | |
| FL | 0.0820 (388.9) | 0.173 (324.5) | 0.103 (256.6) | 0.213 (330.3) | 0.231 (555.2) | 0.039 (249.7) | 6.050 | 17.486 | |
| PAR | 0.091 (641.4) | 0.084 (517.3) | 0.117 (554.9) | 0.170 (512.3) | 0.267 (318.2) | 0.032 (343.5) | 0.076 (572.6) | 8.141 | |
| TE | 0.039 (342.8) | 0.136 (212.3) | 0.064 (256.3) | 0.174 (180.6) | 0.180 (316.6) | 0.013 (13.8) | 0.027 (255.5) | 0.057 (335.4) |
| Model | Maranhão | Piauí | |
|---|---|---|---|
| LD | 9.6 | 45.5 | |
| 95% CI | 4.1–16.3 | 19.6–193.4 | |
| IAM | He < Heq | 0 | 0 |
| He > Heq | 6 | 6 | |
| P (He > Heq) | 0.0078 | 0.0078 | |
| TPM | He < Heq | 0 | 1 |
| He > Heq | 6 | 5 | |
| P (He > Heq) | 0.0078 | 0.0234 | |
| SMM | He < Heq | 2 | 2 |
| He > Heq | 4 | 4 | |
| P (He > Heq) | 0.2812 | 0.5781 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rodrigues, L.F.; Sousa, A.A.d.; Mendes Júnior, W.P.; Cardoso e Silva, A.C.; Nascimento, M.H.S.d.; Barros, M.C.; Sampaio, I.; Fraga, E.d.C. Genetic Differentiation of Aedes aegypti (Diptera: Culicidae) in Areas with High Rates of Infestation in Mid-North Region of Brazil. Insects 2023, 14, 530. https://doi.org/10.3390/insects14060530
Rodrigues LF, Sousa AAd, Mendes Júnior WP, Cardoso e Silva AC, Nascimento MHSd, Barros MC, Sampaio I, Fraga EdC. Genetic Differentiation of Aedes aegypti (Diptera: Culicidae) in Areas with High Rates of Infestation in Mid-North Region of Brazil. Insects. 2023; 14(6):530. https://doi.org/10.3390/insects14060530
Chicago/Turabian StyleRodrigues, Luzianny Farias, Andrelina Alves de Sousa, Walter Pinheiro Mendes Júnior, Amanda Caroline Cardoso e Silva, Maria Histelle Sousa do Nascimento, Maria Claudene Barros, Iracilda Sampaio, and Elmary da Costa Fraga. 2023. "Genetic Differentiation of Aedes aegypti (Diptera: Culicidae) in Areas with High Rates of Infestation in Mid-North Region of Brazil" Insects 14, no. 6: 530. https://doi.org/10.3390/insects14060530
APA StyleRodrigues, L. F., Sousa, A. A. d., Mendes Júnior, W. P., Cardoso e Silva, A. C., Nascimento, M. H. S. d., Barros, M. C., Sampaio, I., & Fraga, E. d. C. (2023). Genetic Differentiation of Aedes aegypti (Diptera: Culicidae) in Areas with High Rates of Infestation in Mid-North Region of Brazil. Insects, 14(6), 530. https://doi.org/10.3390/insects14060530






