Gene Co-Expression Network Analysis Reveals Key Regulatory Genes in Metisa plana Hormone Pathways
Abstract
Simple Summary
Abstract
1. Introduction
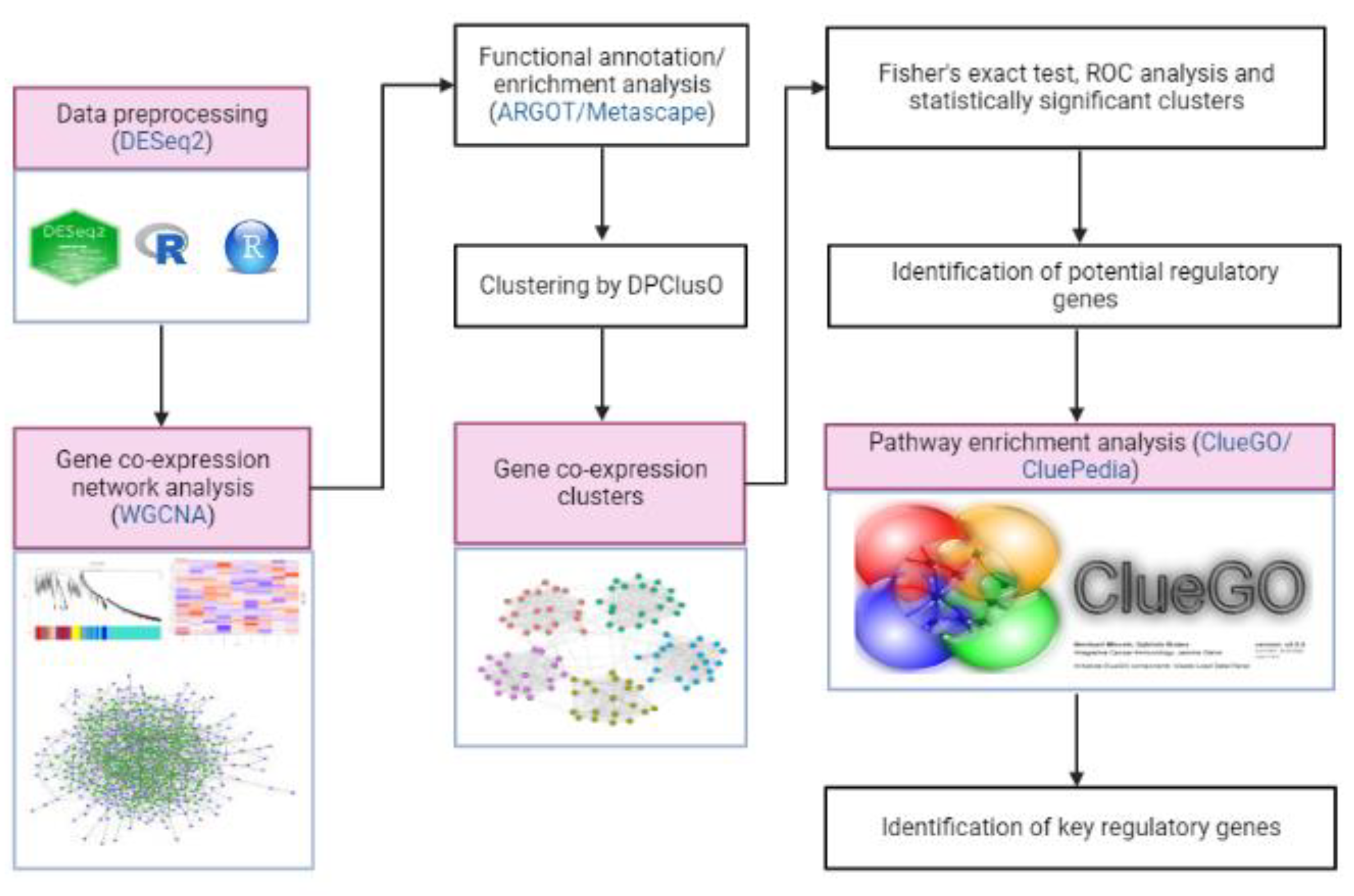
2. Materials and Methods
2.1. Data Pre-Processing
2.2. Construction of Weighted Gene Co-Expression Network Analysis (WGCNA)
2.3. Key Modules Selection and Hub Genes Identification
2.4. Functional Annotation of Unannotated Genes
2.5. Functional Enrichment Analysis of the Key Modules
2.6. Network Clustering Analysis
2.7. Fisher’s Exact Test Analysis
2.8. SScore and ROC Statistical Analysis
2.9. Pathway Enrichment Analysis
3. Results
3.1. Data Processing
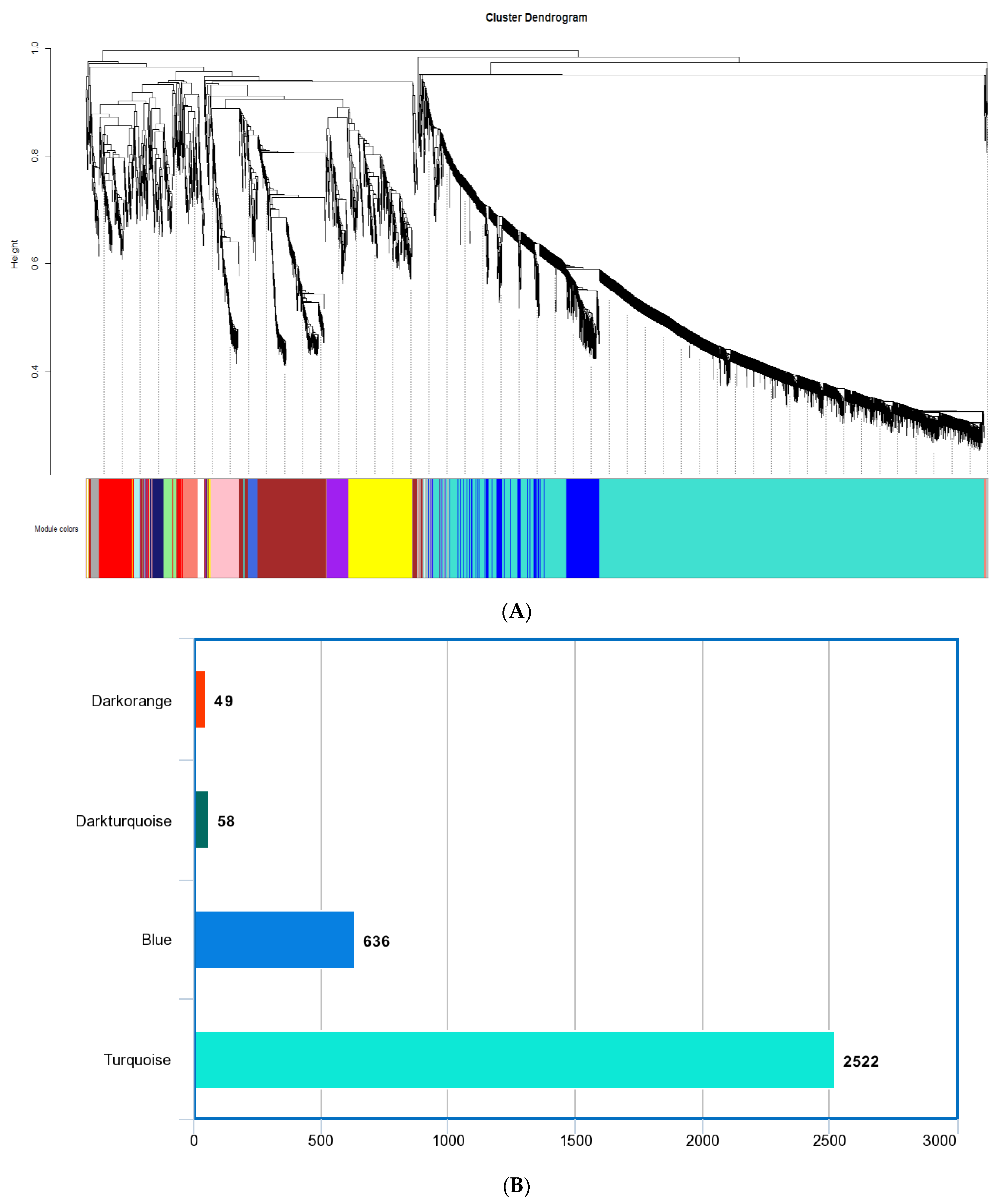
3.2. Weighted Gene Co-Expression Network Construction and Key Modules Selection
3.3. Functional Annotation
3.4. Functional Enrichment Analysis of Key Modules
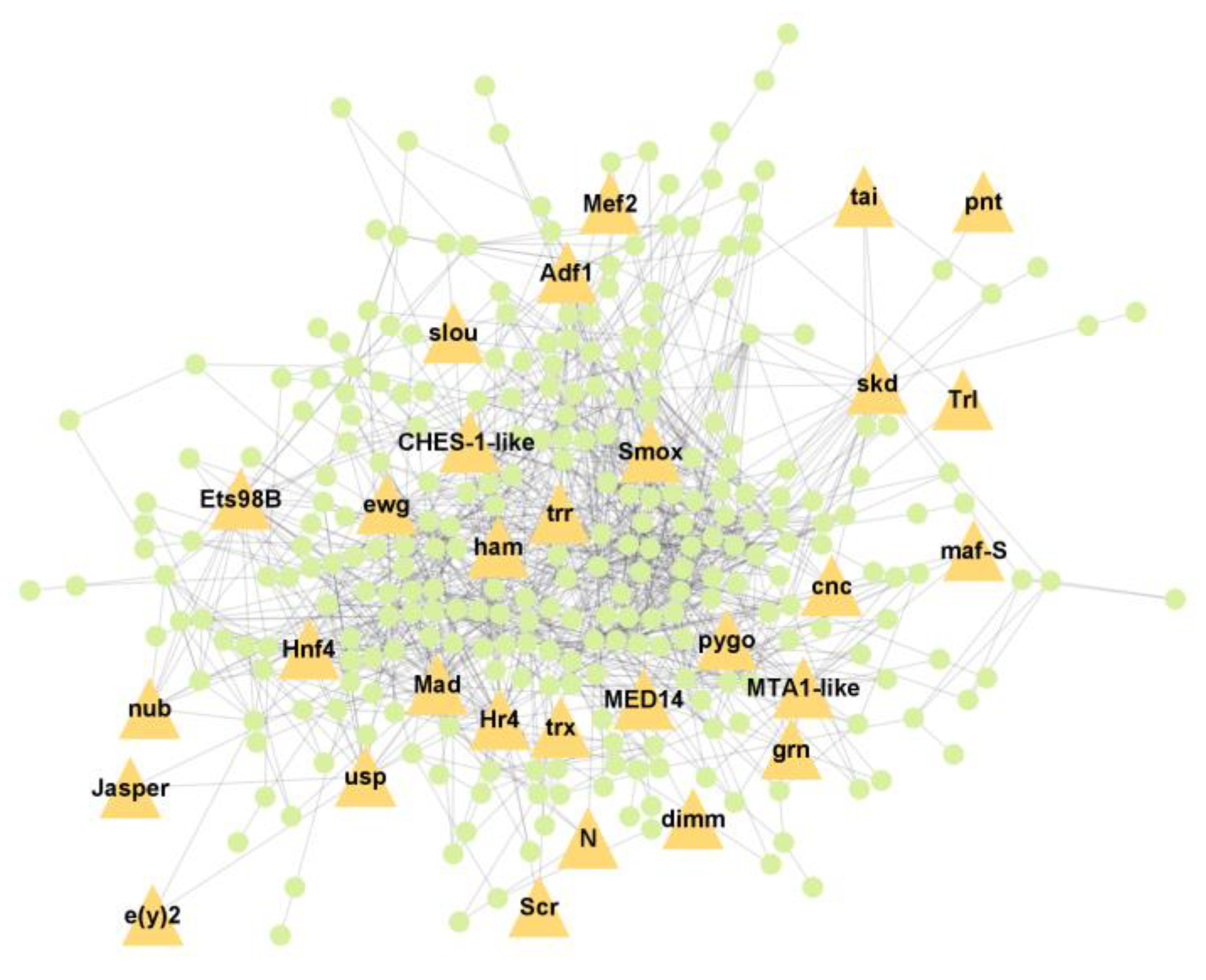
3.5. Protein–Protein Interaction Network Analysis
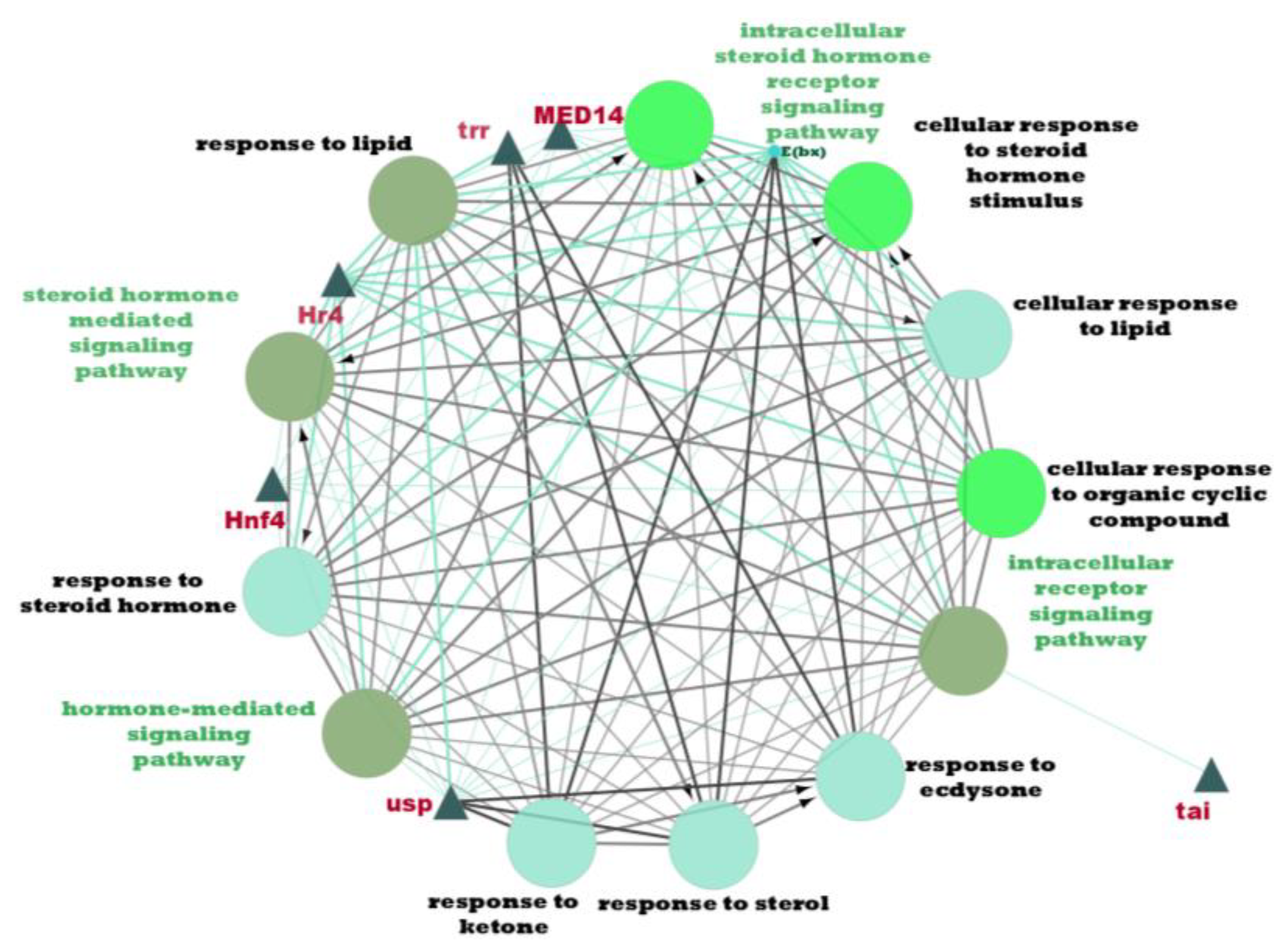
3.6. Network Clustering Analysis and Pathway Enrichment Analysis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Murphy, D.J.; Goggin, K.; Paterson, R.R.M. Oil palm in the 2020s and beyond: Challenges and solutions. CABI Agric. Biosci. 2021, 2, 39. [Google Scholar] [CrossRef] [PubMed]
- Carter, C.; Finley, W.; Fry, J.; Jackson, D.; Willis, L. Palm oil markets and future supply. Eur. J. Lipid Sci. Technol. 2007, 109, 307–314. [Google Scholar] [CrossRef]
- Shevade, V.S.; Loboda, T.V. Oil palm plantations in Peninsular Malaysia: Determinants and constraints on expansion. PLoS ONE 2019, 14, e0210628. [Google Scholar] [CrossRef] [PubMed]
- Wicke, B.; Sikkema, R.; Dornburg, V.; Faaij, A. Exploring land use changes and the role of palm oil production in Indonesia and Malaysia. Land Use Policy 2011, 28, 193–206. [Google Scholar] [CrossRef]
- Nambiappan, B. Malaysia: 100 years of resilient palm oil economic performance. J. Oil Palm Res. 2018, 30, 13–25. [Google Scholar] [CrossRef]
- Koh, L.P.; Wilcove, D.S. Cashing in palm oil for conservation. Nature 2007, 448, 993–994. [Google Scholar] [CrossRef]
- Foster, W.A.; Snaddon, J.L.; Turner, E.; Fayle, T.; Cockerill, T.D.; Ellwood, M.D.F.; Broad, G.R.; Chung, A.Y.C.; Eggleton, P.; Khen, C.V.; et al. Establishing the evidence base for maintaining biodiversity and ecosystem function in the oil palm landscapes of South East Asia. Philos. Trans. R. Soc. B Biol. Sci. 2011, 366, 3277–3291. [Google Scholar] [CrossRef]
- Jamian, S.; Norhisham, A.; Ghazali, A.; Zakaria, A.; Azhar, B. Impacts of 2 species of predatory Reduviidae on bagworms in oil palm plantations. Insect Sci. 2016, 24, 285–294. [Google Scholar] [CrossRef]
- Thaer, S.; Kassim, F.A.; Hasbullah, N.A.; Al-obaidi, J.R. Evaluation of Bagworm, Metisa plana (Lepidoptera: Psychidae) Infestation and Beneficial Parasitoid in an Oil Palm Plantation, Perak, Malaysia. J. Sci. Math. Lett. 2021, 9, 19–35. [Google Scholar] [CrossRef]
- Halim, M.; Muhaimin, A.M.D.; Zulaikha, S.A.S.; Atikah, A.R.N.; Masri, M.M.M.; Yaakop, S. Evaluation of infestation in parasitoids on Metisa plana Walker (Lepidoptera: Psychidae) in three oil palm plantations in peninsular Malaysia. Serangga 2017, 22, 135–149. [Google Scholar]
- Production of Crude Palm Oil in Malaysia 2012–2021. 2022. Available online: https://www.statista.com/statistics/489441/palm-oil-consumption-malaysia/ (accessed on 28 December 2022).
- Tey, C.-C.; Loong, C.-Y. Understanding Pest Biology and Behaviour for Effective Control of Oil Palm Bagworms. Plant. Kuala Lumpur. 2012, 88, 699–715. [Google Scholar]
- Pradana, M.G.; Priwiratama, H.; Rozziansha, T.A.P.; Prasetyo, A.E.; Susanto, A. Field evaluation of Bacillus thuringiensis product to control Metisa plana bagworm in oil palm plantation. IOP Conf. Ser. Earth Environ. Sci. 2022, 974, 012025. [Google Scholar] [CrossRef]
- Kamarudin, N.; Ali, S.R.A.; Masri, M.M.M.; Ahmad, M.N.; Manan, C.A.H.C.; Kamarudin, N. Controlling Metisa plana Walker (Lepidoptera: Psychidae) outbreak using bacillus thuringiensis at an oil palm plantation in slim river, perak, malaysia. J. Oil Palm Res. 2017, 29, 47–54. [Google Scholar] [CrossRef]
- Kok, C.C.; Eng, O.K.; Razak, A.R.; Arshad, A.M. Microstructure and life cycle of Metisa plana Walker (Lepidoptera: Psychidae). J. Sustain. Sci. Manag. 2011, 6, 51–59. [Google Scholar]
- Salim, H.; Rawi, C.S.; Ahmad, A.H.; Al-Shami, S.A. Efficacy of Insecticide and Bioinsecticide Ground Sprays to Control Metisa plana Walker (Lepidoptera: Psychidae) in Oil Palm Plantations, Malaysia. Trop. Life Sci. Res. 2015, 26, 73–83. [Google Scholar] [PubMed]
- Mirth, C.K.; Tang, H.Y.; Makohon-Moore, S.C.; Salhadar, S.; Gokhale, R.H.; Warner, R.D.; Koyama, T.; Riddiford, L.M.; Shingleton, A.W. Juvenile hormone regulates body size and perturbs insulin signaling in Drosophila. Proc. Natl. Acad. Sci. USA 2014, 111, 7018–7023. [Google Scholar] [CrossRef] [PubMed]
- Semsey, S.; Adhya, S. Regulatory Genes. In Brenner’s Encyclopedia of Genetics, 2nd ed.; Elsevier Inc.: Amsterdam, The Netherlands, 2013; pp. 119–122. [Google Scholar] [CrossRef]
- Uryu, O.; Ou, Q.; Komura-Kawa, T.; Kamiyama, T.; Iga, M.; Syrzycka, M.; Hirota, K.; Kataoka, H.; Honda, B.M.; King-Jones, K.; et al. Cooperative Control of Ecdysone Biosynthesis in Drosophila by Transcription Factors Séance, Ouija Board, and Molting Defective. Genetics 2018, 208, 605–622. [Google Scholar] [CrossRef]
- Guo, Z.; Qin, J.; Zhou, X.; Zhang, Y. Insect Transcription Factors: A Landscape of Their Structures and Biological Functions in Drosophila and beyond. Int. J. Mol. Sci. 2018, 19, 3691. [Google Scholar] [CrossRef]
- Britannica, “Gene,” Encyclopedia Britannica. 2023. Available online: https://www.britannica.com/science/gene (accessed on 3 January 2023).
- Zeng, B.; Huang, Y.; Xu, J.; Shiotsuki, T.; Bai, H.; Palli, S.R.; Huang, Y.; Tan, A. The FOXO transcription factor controls insect growth and development by regulating juvenile hormone degradation in the silkworm, Bombyx mori. J. Biol. Chem. 2017, 292, 11659–11669. [Google Scholar] [CrossRef]
- Rahmat, N.L.; Zifruddin, A.N.; Abidin, C.M.R.Z.; Muhammad, N.-A.N.; Hassan, M. The Developmental Transcriptome of Bagworm, Metisa plana (Lepidoptera: Psychidae) and Insights into Chitin Biosynthesis Genes. Genes 2020, 12, 7. [Google Scholar] [CrossRef]
- Zainuddin, N.; Maidin, M.S.T.; Kamarudin, N.; Napiah, N.R.A.M.A.; Keni, M.F.; Masri, M.M.M. De novo transcriptome analysis of bagworm Metisa plana from highly infested oil palm estate in Perak revealed detoxification genes and potential insecticide targets. J. Asia-Pac. Èntomol. 2023, 26, 102039. [Google Scholar] [CrossRef]
- Langfelder, P.; Horvath, S. WGCNA: An R package for weighted correlation network analysis. BMC Bioinform. 2008, 9, 559. [Google Scholar] [CrossRef] [PubMed]
- Bakhtiarizadeh, M.R.; Hosseinpour, B.; Shahhoseini, M.; Korte, A.; Gifani, P. Weighted Gene Co-expression Network Analysis of Endometriosis and Identification of Functional Modules Associated With Its Main Hallmarks. Front. Genet. 2018, 9, 453. [Google Scholar] [CrossRef] [PubMed]
- Bakhtiarizadeh, M.R.; Mirzaei, S.; Norouzi, M.; Sheybani, N.; Sadi, M.S.V. Identification of Gene Modules and Hub Genes Involved in Mastitis Development Using a Systems Biology Approach. Front. Genet. 2020, 11, 722. [Google Scholar] [CrossRef] [PubMed]
- Fan, X.-B.; Pang, R.; Li, W.-X.; Ojha, A.; Li, D.; Zhang, W.-Q. An Overview of Embryogenesis: External Morphology and Transcriptome Profiling in the Hemipteran Insect Nilaparvata lugens. Front. Physiol. 2020, 11, 106. [Google Scholar] [CrossRef]
- Ding, W.-F.; Ling, X.-F.; Lu, Q.; Wang, W.-W.; Zhang, X.; Feng, Y.; Chen, X.-M.; Chen, H. Identification of the Key Pathways and Genes Involved in the Wax Biosynthesis of the Chinese White Wax Scale Insect (Ericerus pela Chavannes) by Integrated Weighted Gene Coexpression Network Analysis. Genes 2022, 13, 1364. [Google Scholar] [CrossRef] [PubMed]
- Katoch, R.; Sethi, A.; Thakur, N.; Murdock, L.L. RNAi for Insect Control: Current Perspective and Future Challenges. Appl. Biochem. Biotechnol. 2013, 171, 847–873. [Google Scholar] [CrossRef]
- Mehlhorn, S.; Hunnekuhl, V.S.; Geibel, S.; Nauen, R.; Bucher, G. Establishing RNAi for basic research and pest control and identification of the most efficient target genes for pest control: A brief guide. Front. Zool. 2021, 18, 60. [Google Scholar] [CrossRef]
- Whyard, S.; Singh, A.D.; Wong, S. Ingested double-stranded RNAs can act as species-specific insecticides. Insect Biochem. Mol. Biol. 2009, 39, 824–832. [Google Scholar] [CrossRef]
- Christiaens, O.; Whyard, S.; Vélez, A.M.; Smagghe, G. Double-Stranded RNA Technology to Control Insect Pests: Current Status and Challenges. Front. Plant Sci. 2020, 11, 451. [Google Scholar] [CrossRef]
- Zhu, F.; Xu, J.; Palli, R.; Ferguson, J.; Palli, S.R. Ingested RNA interference for managing the populations of the Colorado potato beetle, Leptinotarsa decemlineata. Pest Manag. Sci. 2010, 67, 175–182. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Ma, Y.; Ma, X.; Hu, H.; Wang, D.; Song, X.; Ren, X.; Ma, Y. Combined Transcriptomic Analysis and RNA Interference Reveal the Effects of Methoxyfenozide on Ecdysone Signaling Pathway of Spodoptera exigua. Int. J. Mol. Sci. 2021, 22, 9080. [Google Scholar] [CrossRef] [PubMed]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Falda, M.; Toppo, S.; Pescarolo, A.; Lavezzo, E.; Di Camillo, B.; Facchinetti, A.; Cilia, E.; Velasco, R.; Fontana, P. Argot2: A large scale function prediction tool relying on semantic similarity of weighted Gene Ontology terms. BMC Bioinform. 2012, 13, S14. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Zhou, B.; Pache, L.; Chang, M.; Khodabakhshi, A.H.; Tanaseichuk, O.; Benner, C.; Chanda, S.K. Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat. Commun. 2019, 10, 1523. [Google Scholar] [CrossRef] [PubMed]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software environment for integrated models of Biomolecular Interaction Networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef]
- Karim, M.B.; Wakamatsu, N.; Altaf-Ul-Amin, M. DPClusOST: A Software Tool for General Purpose Graph Clustering. J. Comput. Aided Chem. 2017, 18, 76–93. [Google Scholar] [CrossRef]
- Harun, S.; Afiqah-Aleng, N.; Karim, M.B.; Amin, A.U.; Kanaya, S.; Mohamed-Hussein, Z.-A. Potential Arabidopsis thaliana glucosinolate genes identified from the co-expression modules using graph clustering approach. PeerJ 2021, 9, e11876. [Google Scholar] [CrossRef]
- Karim, M.B.B.; Huang, M.; Ono, N.; Kanaya, S.; Amin, A.-U. BiClusO: A Novel Biclustering Approach and Its Application to Species-VOC Relational Data. IEEE/ACM Trans. Comput. Biol. Bioinform. 2019, 17, 1955–1965. [Google Scholar] [CrossRef]
- Eguchi, R.; Karim, M.B.; Hu, P.; Sato, T.; Ono, N.; Kanaya, S.; Amin, A.U. An integrative network-based approach to identify novel disease genes and pathways: A case study in the context of inflammatory bowel disease. BMC Bioinform. 2018, 19, 264. [Google Scholar] [CrossRef]
- Fisher, R.A. Statistical Methods for Research Workers; Springer: New York, NY, USA, 1992; pp. 66–70. [Google Scholar] [CrossRef]
- Fisher, R.A. On the Interpretation of χ2 from Contingency Tables, and the Calculation of P. J. R. Stat. Soc. 1922, 85, 87. [Google Scholar] [CrossRef]
- Metz, C.E. Basic principles of ROC analysis. Semin Nucl. Med. 1978, 8, 283–298. [Google Scholar] [CrossRef]
- Sing, T.; Sander, O.; Beerenwinkel, N.; Lengauer, T. ROCR: Visualizing classifier performance in R. Bioinformatics 2005, 21, 3940–3941. [Google Scholar] [CrossRef] [PubMed]
- Bindea, G.; Mlecnik, B.; Hackl, H.; Charoentong, P.; Tosolini, M.; Kirilovsky, A.; Fridman, W.-H.; Pagès, F.; Trajanoski, Z.; Galon, J. ClueGO: A Cytoscape plug-in to decipher functionally grouped gene ontology and pathway annotation networks. Bioinformatics 2009, 25, 1091–1093. [Google Scholar] [CrossRef] [PubMed]
- Purnomo, H.; Okarda, B.; Dermawan, A.; Ilham, Q.P.; Pacheco, P.; Nurfatriani, F.; Suhendang, E. Reconciling oil palm economic development and environmental conservation in Indonesia: A value chain dynamic approach. For. Policy Econ. 2020, 111, 102089. [Google Scholar] [CrossRef]
- Jackson, T.A.; Crawford, J.W.; Traeholt, C.; Sanders, T.A.B. Learning to love the world’s most hated crop—Review Articles. J. Oil Palm Res. 2019, 31, 331–347. [Google Scholar] [CrossRef]
- Ting, G.A.; Jie, A.C.; Abidin, M.R.Z.; Hamid, N.H.; Salim, H. 16S rRNA Amplicon Sequencing of Bagworm Metisa plana Walker (Lepidoptera: Psychidae). BioRxiv 2021. [Google Scholar] [CrossRef]
- Kok, C.C.; Eng, O.K.; Razak, A.R.; Arshad, A.M.; Marcon, P.G. Susceptibility of Bagworm Metisa plana (Lepidoptera: Psychidae) to chlorantraniliprole. Pertanika J. Trop Agric. Sci. 2012, 35, 149–163. [Google Scholar]
- Riddiford, L.M. Molting. In Encyclopedia of Insects; Elsevier Inc.: Amsterdam, The Netherlands, 2009; pp. 649–654. [Google Scholar] [CrossRef]
- Kaleka, A.S.; Kaur, N.; Bali, G.K. Larval Development and Molting. In Edible Insects; IntechOpen: London, UK, 2019. [Google Scholar] [CrossRef]
- Tettamanti, G.; Casartelli, M. Cell death during complete metamorphosis. Philos. Trans. R. Soc. B Biol. Sci. 2019, 374, 20190065. [Google Scholar] [CrossRef]
- Yamanaka, N.; Rewitz, K.F.; O’Connor, M.B. Ecdysone Control of Developmental Transitions: Lessons from Drosophila Research. Annu. Rev. Èntomol. 2013, 58, 497–516. [Google Scholar] [CrossRef] [PubMed]
- Jindra, M.; Palli, S.R.; Riddiford, L.M. The Juvenile Hormone Signaling Pathway in Insect Development. Annu. Rev. Èntomol. 2013, 58, 181–204. [Google Scholar] [CrossRef] [PubMed]
- Afiqah-Aleng, N.; Altaf-Ul-Amin, M.; Kanaya, S.; Mohamed-Hussein, Z.-A. Graph cluster approach in identifying novel proteins and significant pathways involved in polycystic ovary syndrome. Reprod. Biomed. Online 2019, 40, 319–330. [Google Scholar] [CrossRef] [PubMed]
- Harun, S.; Abdullah-Zawawi, M.-R.; Goh, H.-H.; Mohamed-Hussein, Z.-A. A Comprehensive Gene Inventory for Glucosinolate Biosynthetic Pathway in Arabidopsis thaliana. J. Agric. Food Chem. 2020, 68, 7281–7297. [Google Scholar] [CrossRef] [PubMed]
- Harun, S.; Rohani, E.R.; Ohme-Takagi, M.; Goh, H.-H.; Mohamed-Hussein, Z.-A. ADAP is a possible negative regulator of glucosinolate biosynthesis in Arabidopsis thaliana based on clustering and gene expression analyses. J. Plant Res. 2021, 134, 327–339. [Google Scholar] [CrossRef] [PubMed]
- Harun, S.; Abdullah-Zawawi, M.-R.; A-Rahman, M.R.A.; Muhammad, N.A.N.; Mohamed-Hussein, Z.-A. SuCComBase: A manually curated repository of plant sulfur-containing compounds. Database 2019, 2019, baz021. [Google Scholar] [CrossRef]
- Amin, A.U.; Wada, M.; Kanaya, S. Partitioning a PPI Network into Overlapping Modules Constrained by High-Density and Periphery Tracking. ISRN Biomath. 2012, 2012, 11. [Google Scholar] [CrossRef]
- Jones, G.; Teal, P.; Henrich, V.C.; Krzywonos, A.; Sapa, A.; Wozniak, M.; Smolka, J.; Jones, D. Ligand binding pocket function of Drosophila USP is necessary for metamorphosis. Gen. Comp. Endocrinol. 2013, 182, 73–82. [Google Scholar] [CrossRef]
- King-Jones, K.; Charles, J.-P.; Lam, G.; Thummel, C.S. The Ecdysone-Induced DHR4 Orphan Nuclear Receptor Coordinates Growth and Maturation in Drosophila. Cell 2005, 121, 773–784. [Google Scholar] [CrossRef]
- Ou, Q.; Magico, A.; King-Jones, K. Nuclear Receptor DHR4 Controls the Timing of Steroid Hormone Pulses During Drosophila Development. PLoS Biol. 2011, 9, e1001160. [Google Scholar] [CrossRef]
- Barry, W.E.; Thummel, C.S. The Drosophila HNF4 nuclear receptor promotes glucose-stimulated insulin secretion and mitochondrial function in adults. eLife 2016, 5, e11183. [Google Scholar] [CrossRef]
- Xu, Q.-Y.; Du, J.-L.; Mu, L.-L.; Guo, W.-C.; Li, G.-Q. Importance of Taiman in Larval-Pupal Transition in Leptinotarsa decemlineata. Front. Physiol. 2019, 10, 724. [Google Scholar] [CrossRef] [PubMed]
- Johnston, D.M.; Sedkov, Y.; Petruk, S.; Riley, K.M.; Fujioka, M.; Jaynes, J.B.; Mazo, A. Ecdysone- and NO-Mediated Gene Regulation by Competing EcR/Usp and E75A Nuclear Receptors during Drosophila Development. Mol. Cell 2011, 44, 51–61. [Google Scholar] [CrossRef] [PubMed]
- Yu, R.; Xu, X.; Liang, Y.; Tian, H.; Pan, Z.; Jin, S.; Wang, N.; Zhang, W. The Insect Ecdysone Receptor is a Good Potential Target for RNAi-based Pest Control. Int. J. Biol. Sci. 2014, 10, 1171–1180. [Google Scholar] [CrossRef] [PubMed]
- Masterson, M.; Bittar, R.; Chu, H.; Yamanaka, N.; Haga-Yamanaka, S. Rapid Assessment of Insect Steroid Hormone Entry Into Cultured Cells. Front. Physiol. 2022, 12, 2544. [Google Scholar] [CrossRef]
- Yan, T.; Chen, H.; Sun, Y.; Yu, X.; Xia, L. RNA Interference of the Ecdysone Receptor Genes EcR and USP in Grain Aphid (Sitobion avenae F.) Affects Its Survival and Fecundity upon Feeding on Wheat Plants. Int. J. Mol. Sci. 2016, 17, 2098. [Google Scholar] [CrossRef]
- Xu, Y.; Li, Y.; Wang, Q.; Zheng, C.; Zhao, D.; Shi, F.; Liu, X.; Tao, J.; Zong, S. Identification of key genes associated with overwintering in Anoplophora glabripennis larva using gene co-expression network analysis. Pest Manag. Sci. 2020, 77, 805–816. [Google Scholar] [CrossRef]

| Regulatory Genes | Non-Regulatory Genes | |
|---|---|---|
| In cluster | a | b |
| Not in cluster | c | d |
| Total | a + c | b + d |
| Modules | Number of Co-Expressed Genes | Number of Genes with MM ≥ 0.80 |
|---|---|---|
| Blue | 636 | 576 |
| Dark-orange | 49 | 25 |
| Dark-turquoise | 58 | 71 |
| Turquoise | 2522 | 1918 |
| Modules | Number of Hub Genes | Number of Genes without Annotation | Number of Genes Annotated by ARGOT | Genes without Annotation by ARGOT |
|---|---|---|---|---|
| Blue | 576 | 216 | 149 | 67 |
| Dark-orange | 25 | 5 | 5 | 0 |
| Dark-turquoise | 71 | 14 | 9 | 5 |
| Turquoise | 1918 | 857 | 624 | 233 |
| Total | 2590 | 1092 | 787 | 305 |
| GO | Category | Description | Count | Log10(q) |
|---|---|---|---|---|
| GO:0035295 | GO BP | tube development | 67 | −8.89 |
| GO:0042692 | GO BP | muscle cell differentiation | 19 | −5.68 |
| GO:0060541 | GO BP | respiratory system development | 30 | −5.68 |
| GO:0003002 | GO BP | regionalisation | 45 | −5.38 |
| GO:0032989 | GO BP | cellular component morphogenesis | 47 | −5.35 |
| GO:0002376 | GO BP | immune system process | 32 | −4.93 |
| GO:0097305 | GO BP | response to alcohol | 18 | −4.22 |
| GO:0048565 | GO BP | digestive tract development | 17 | −3.97 |
| GO:0040008 | GO BP | regulation of growth | 30 | −3.89 |
| GO:0042592 | GO BP | homeostatic process | 38 | −3.89 |
| GO:0007166 | GO BP | cell surface receptor signalling pathway | 29 | −3.76 |
| GO:0007447 | GO BP | imaginal disc pattern formation | 16 | −3.48 |
| GO:0002164 | GO BP | larval development | 21 | −3.48 |
| GO:0044281 | GO BP | small molecule metabolic process | 52 | −3.45 |
| GO:0022408 | GO BP | negative regulation of cell-cell adhesion | 6 | −3.23 |
| R-DME−9006931 | Reactome Gene Sets | signalling by nuclear receptors | 15 | −3.16 |
| GO:0051604 | GO BP | protein maturation | 16 | −3.16 |
| GO:0012501 | GO BP | programmed cell death | 18 | −2.95 |
| GO:0008037 | GO BP | cell recognition | 15 | −2.86 |
| GO:0010876 | GO BP | lipid localisation | 14 | −2.81 |
| Density | Number of Clusters | Maximum Size of Clusters |
|---|---|---|
| 0.5 | 193 | 1071 |
| 0.6 | 227 | 991 |
| 0.7 | 275 | 1136 |
| 0.8 | 291 | 1074 |
| 0.9 | 307 | 913 |
| Density | Area under the Curve (AUC) |
|---|---|
| 0.5 | 0.913 |
| 0.6 | 0.950 |
| 0.7 | 0.824 |
| 0.8 | 0.947 |
| 0.9 | 0.934 |
| Cluster Number | Cluster Size | Regulatory Genes | Cluster | p-Value |
|---|---|---|---|---|
| Cluster 3 | 15 | MTA1-like, Nub, Grn, Usp, Hr4, Mad, Smox, Tai, Mef2 | 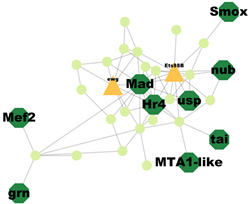 | 3.52 × 10−7 |
| Cluster 7 | 12 | Trx, Trr, Usp, Hr4 |  | 1.52 × 10−3 |
| Cluster 8 | 12 | Mad, Smox, Ches-1-like, Usp, Hr4, Tai, Mef2 | 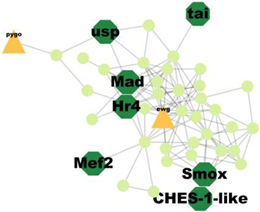 | 1.25 × 10−5 |
| Cluster 9 | 12 | Skd, MED14, Usp, Hr4, Mad, Smox, Tai, Mef2 | 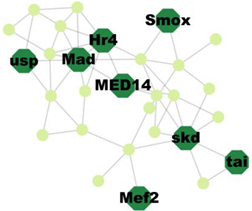 | 5.83 × 10−7 |
| Cluster 11 | 12 | Cnc, Mad, Smox, Usp, Hr4, Tai, Mef2 | 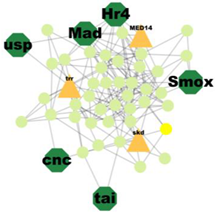 | 1.25 × 10−5 |
| Cluster 12 | 11 | Usp, Hr4, Mad, Smox, Tai, Mef2, Grn |  | 5.36 × 10−6 |
| Cluster 14 | 11 | Pnt, Mef2, Scr, Mad, Smox, Usp, Hr4, Tai | 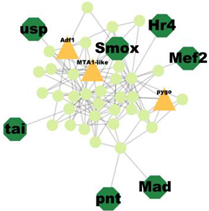 | 2.07 × 10−7 |
| Cluster 24 | 8 | Pygo, N, Trr | 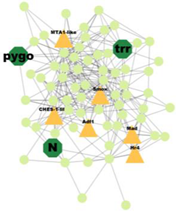 | 2.613 × 10−2 |
| Cluster 47 | 5 | Hr4, Hnf4, Usp | 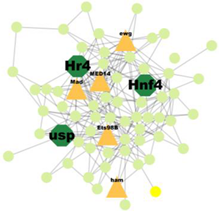 | 5.652 × 10−3 |
| Cluster 53 | 5 | Smox, Hnf4, Mad | 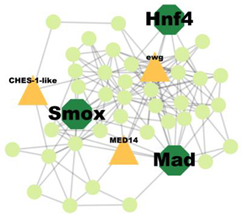 | 5.625 × 10−3 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Vengatharajuloo, V.; Goh, H.-H.; Hassan, M.; Govender, N.; Sulaiman, S.; Afiqah-Aleng, N.; Harun, S.; Mohamed-Hussein, Z.-A. Gene Co-Expression Network Analysis Reveals Key Regulatory Genes in Metisa plana Hormone Pathways. Insects 2023, 14, 503. https://doi.org/10.3390/insects14060503
Vengatharajuloo V, Goh H-H, Hassan M, Govender N, Sulaiman S, Afiqah-Aleng N, Harun S, Mohamed-Hussein Z-A. Gene Co-Expression Network Analysis Reveals Key Regulatory Genes in Metisa plana Hormone Pathways. Insects. 2023; 14(6):503. https://doi.org/10.3390/insects14060503
Chicago/Turabian StyleVengatharajuloo, Vinothienii, Hoe-Han Goh, Maizom Hassan, Nisha Govender, Suhaila Sulaiman, Nor Afiqah-Aleng, Sarahani Harun, and Zeti-Azura Mohamed-Hussein. 2023. "Gene Co-Expression Network Analysis Reveals Key Regulatory Genes in Metisa plana Hormone Pathways" Insects 14, no. 6: 503. https://doi.org/10.3390/insects14060503
APA StyleVengatharajuloo, V., Goh, H.-H., Hassan, M., Govender, N., Sulaiman, S., Afiqah-Aleng, N., Harun, S., & Mohamed-Hussein, Z.-A. (2023). Gene Co-Expression Network Analysis Reveals Key Regulatory Genes in Metisa plana Hormone Pathways. Insects, 14(6), 503. https://doi.org/10.3390/insects14060503






