Characterization and Functional Analysis of Toll Receptor Genes during Antibacterial Immunity in the Green Peach Aphid Myzus persicae (Sulzer)
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Insect Rearing and Collection
2.2. Characterization and Sequencing of M. persicae Toll Receptor Genes
2.3. Analysis of Developmental and Tissue Specificity Expression Patterns
2.4. Bacterial Challenge
2.5. Functional Analysis Based on RNAi
2.6. Statistical Analysis
3. Results
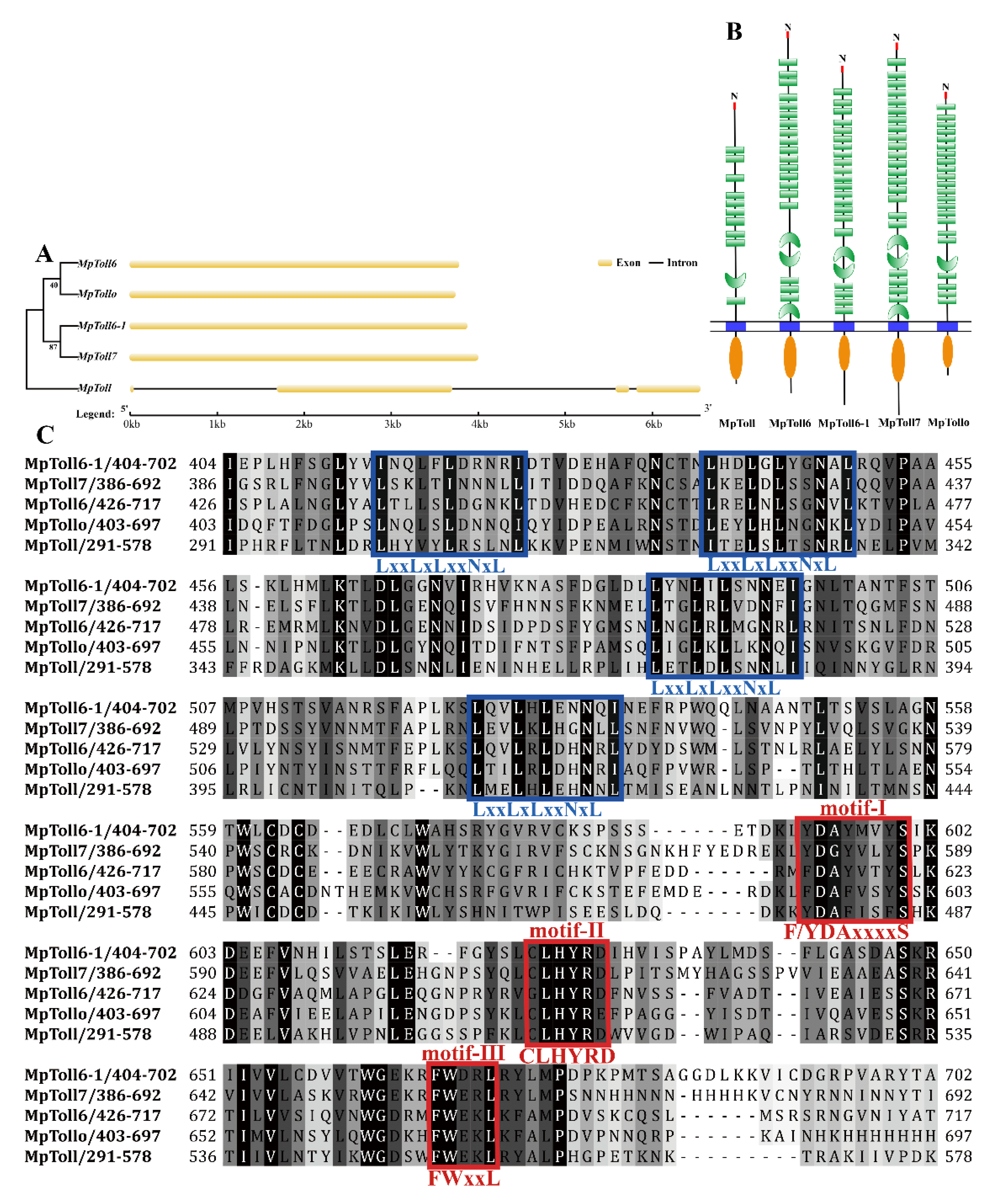
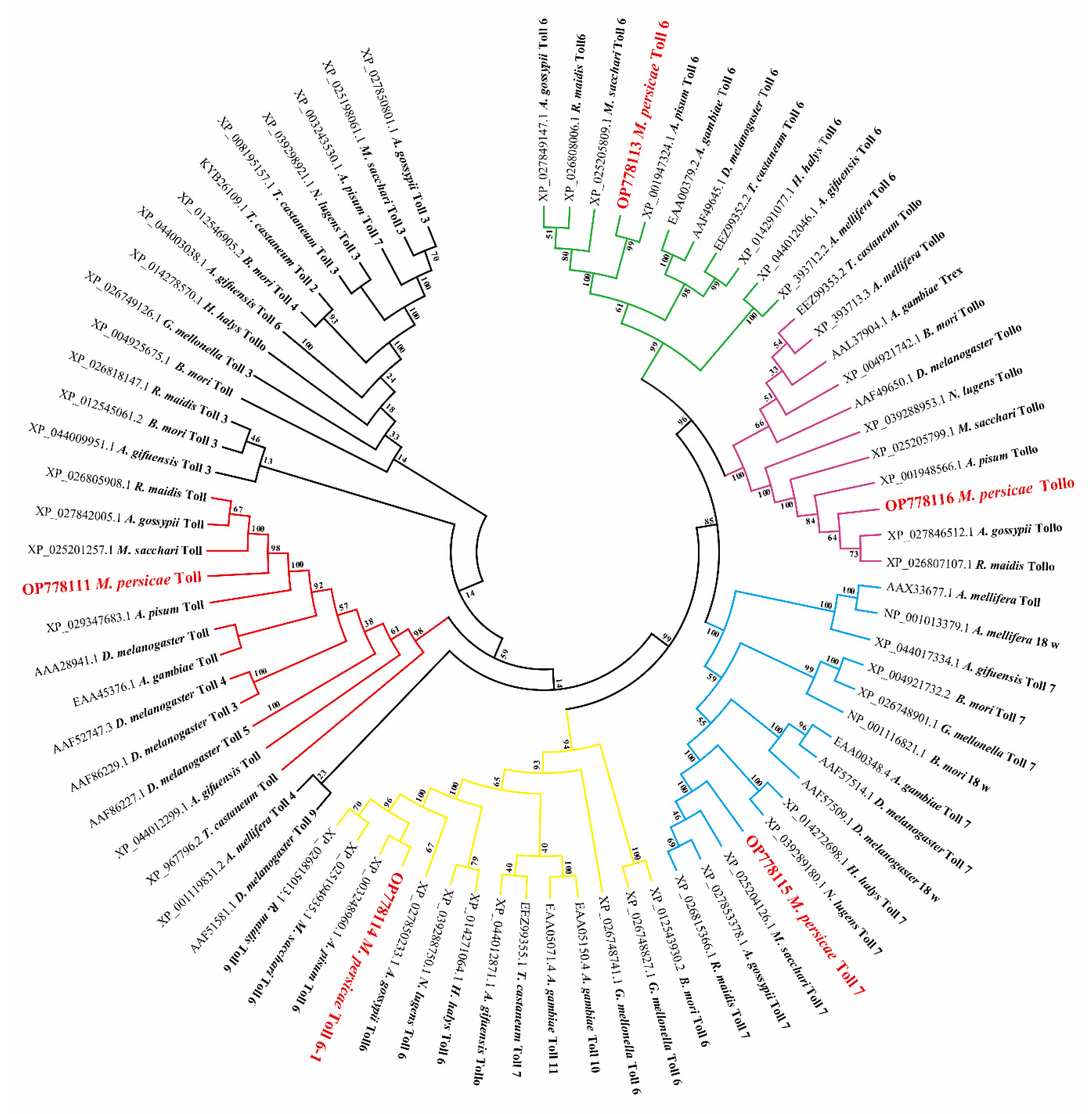
3.1. Bioinformatics and Phylogenetic Analysis of MpToll Genes
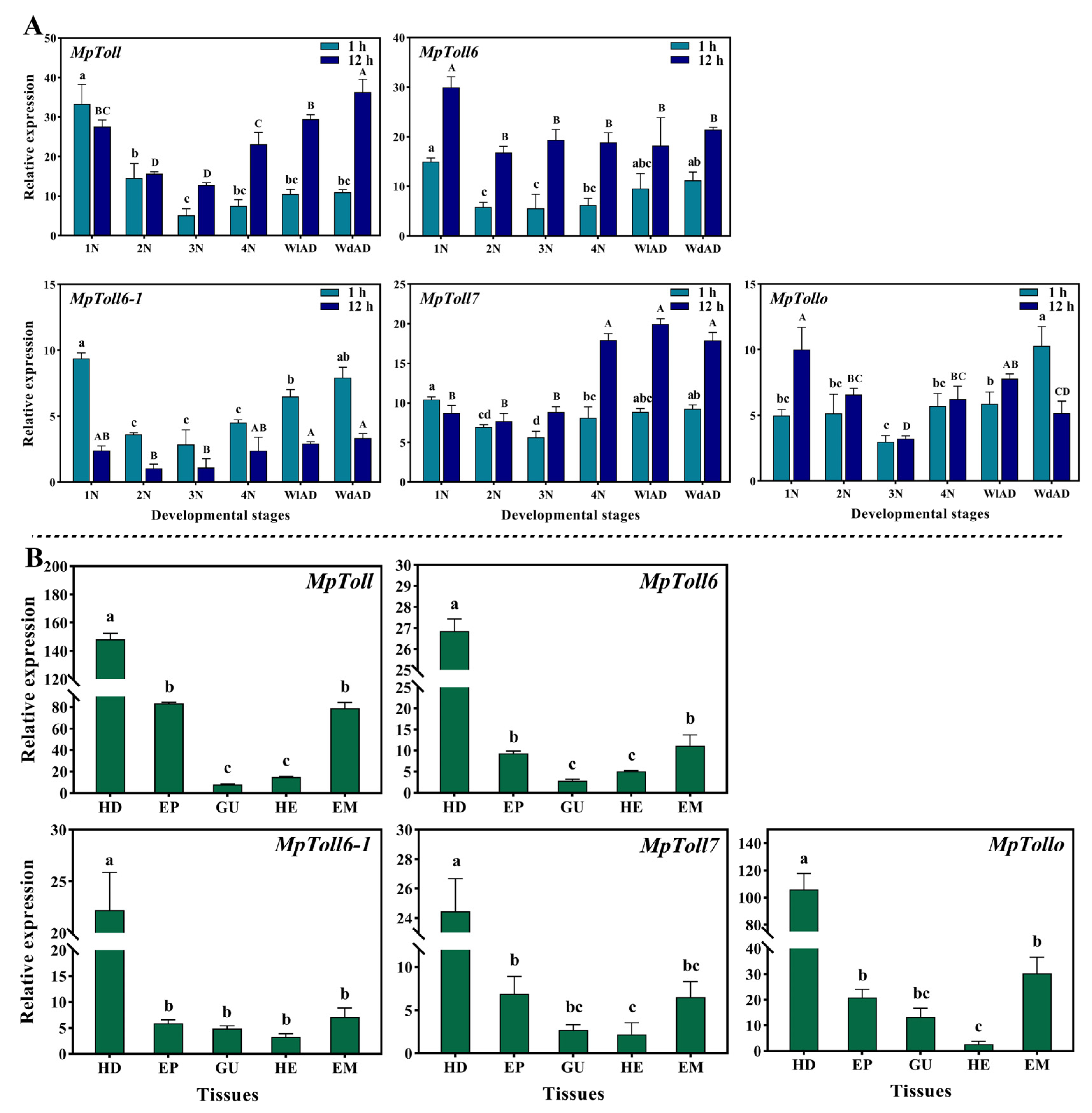
3.2. Developmental and Tissue Expression Profiles of MpToll Genes
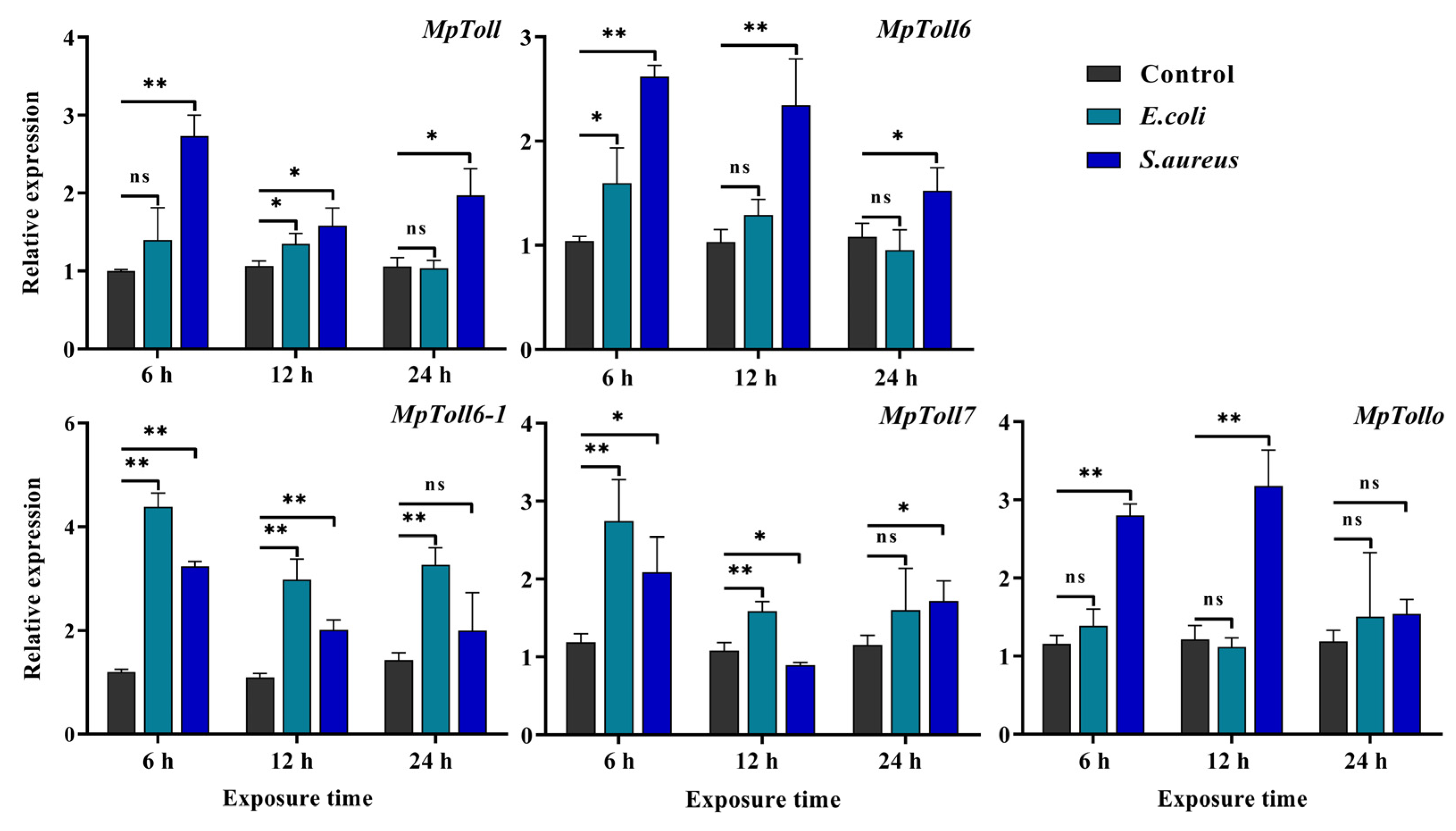
3.3. Expression Induced by Bacterial Challenge
3.4. Effect of Knockdown of MpToll Genes Expression on Susceptibility to Bacterial Infection
4. Discussion
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ali Mohammadie Kojour, M.; Han, Y.S.; Jo, Y.H. An overview of insect innate immunity. Entomol. Res. 2020, 50, 282–291. [Google Scholar] [CrossRef]
- Hoffmann, J.A. The immune response of Drosophila. Nature 2003, 426, 33–38. [Google Scholar] [CrossRef]
- Lemaitre, B.; Hoffmann, J. The host defense of Drosophila melanogaster. Annu. Rev. Immunol. 2007, 25, 697–743. [Google Scholar] [CrossRef]
- Nie, L.; Cai, S.Y.; Shao, J.Z.; Chen, J. Toll-like receptors, associated biological roles, and signaling networks in non-Mammals. Front. Immunol. 2018, 9, 1523. [Google Scholar] [CrossRef]
- Nüsslein–Volhard, C.; Lohs–Schardin, M.; Sander, K.; Cremer, C. A dorso–ventral shift of embryonic primordia in a new maternal–effect mutant of Drosophila. Nature 1980, 283, 474–476. [Google Scholar] [CrossRef]
- Lemaitre, B.; Nicolas, E.; Michaut, L.; Reichhart, J.M.; Hoffmann, J.A. The dorsoventral regulatory gene cassette spätzle/Toll/cactus controls the potent antifungal response in Drosophila adults. Cell 1996, 86, 973–983. [Google Scholar] [CrossRef]
- Leulier, F.; Parquet, C.; Pili-Floury, S.; Ryu, J.H.; Caroff, M.; Lee, W.J.; Mengin–Lecreulx, D.; Lemaitre, B. The Drosophila immune system detects bacteria through specific peptidoglycan recognition. Nat. Immunol. 2003, 4, 478–484. [Google Scholar] [CrossRef]
- Ye, C.; Wang, Z.W.; Sheng, Y.L.; Wang, Z.G.; Smagghe, G.; Christiaens, O.; Niu, J.Z.; Wang, J.J. GNBP1 as a potential RNAi target to enhance the virulence of Beauveria bassiana for aphid control. J. Pest Sci. 2022, 95, 87–100. [Google Scholar] [CrossRef]
- Stöven, S.; Ando, I.; Kadalayil, L.; Engström, Y.; Hultmark, D. Activation of the Drosophila NF-κB factor Relish by rapid endoproteolytic cleavage. EMBO Rep. 2000, 1, 347–352. [Google Scholar] [CrossRef]
- Weber, A.N.R.; Tauszig-Delamasure, S.; Hoffmann, J.A.; Lelièvre, E.; Gascan, H.; Ray, K.P.; Morse, M.A.; Imler, J.L.; Gay, N.J. Binding of the Drosophila cytokine Spätzle to Toll is direct and establishes signaling. Nat. Immunol. 2003, 4, 794–800. [Google Scholar] [CrossRef]
- Yagi, Y.; Nishida, Y.; Ip, Y.T. Functional analysis of Toll–related genes in Drosophila. Dev. Growth Differ. 2010, 52, 771–783. [Google Scholar] [CrossRef] [PubMed]
- Zhang, R.; Li, X.; Zhang, J.; Li, Y.; Wang, Y.; Song, Y.; Ren, F.; Yi, H.; Deng, X.; Zhong, Y.; et al. Toll9 from Bombyx mori functions as a pattern recognition receptor that shares features with toll-like receptor 4 from mammals. Proc. Natl. Acad. Sci. USA 2021, 118, e2103021118. [Google Scholar] [CrossRef]
- Hashimoto, C.; Hudson, K.L.; Anderson, K.V. The Toll gene of Drosophila, required for Dorsal-Ventral embryonic polarity, appears to encode a transmembrane protein. Cell 1988, 52, 269–279. [Google Scholar] [CrossRef] [PubMed]
- Imler, J.L.; Zheng, L. Biology of Toll receptors: Lessons from insects and mammals. J. Leukoc. Biol. 2004, 75, 18–26. [Google Scholar] [CrossRef]
- Xu, Y.; Tao, X.; Shen, B.; Horng, T.; Medzhitov, R.; Manley, J.L.; Tong, L. Structural basis for signal transduction by the Toll/interleukin–1 receptor domains. Nature 2000, 408, 111–115. [Google Scholar] [CrossRef]
- Christophides, G.K.; Zdobnov, E.; Barillas–Mury, C.; Birney, E.; Blandin, S.; Blass, C.; Brey, P.T.; Collins, F.H.; Danielli, A.; Dimopoulos, G.; et al. Immunity–related genes and gene families in Anopheles gambiae. Science 2002, 298, 159–165. [Google Scholar] [CrossRef]
- Evans, J.D.; Aronstein, K.; Chen, Y.P.; Hetru, C.; Imler, J.L.; Jiang, H.; Kanost, M.; Thompson, G.J.; Zou, Z.; Hultmark, D. Immune pathways and defence mechanisms in honey bees Apis mellifera. Insect Mol. Biol. 2006, 15, 645–656. [Google Scholar] [CrossRef]
- Altincicek, B.; Knorr, E.; Vilcinskas, A. Beetle immunity: Identification of immune–inducible genes from the model insect Tribolium castaneum. Dev. Comp. Immunol. 2008, 32, 585–595. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, H.; Ishibashi, J.; Fujita, K.; Nakajima, Y.; Sagisaka, A.; Tomimoto, K.; Suzuki, N.; Yoshiyama, M.; Kaneko, Y.; Iwasaki, T.; et al. A genome-wide analysis of genes and gene families involved in innate immunity of Bombyx mori. Insect Biochem. Mol. Biol. 2008, 38, 1087–1110. [Google Scholar] [CrossRef]
- Xia, X.; Yu, L.; Xue, M.; Yu, X.; Vasseur, L.; Gurr, G.M.; Baxter, S.W.; Lin, H.; Lin, J.; You, M. Genome–wide characterization and expression profiling of immune genes in the diamondback moth, Plutella xylostella (L.). Sci. Rep. 2015, 5, 9877. [Google Scholar] [CrossRef]
- Beutler, B. Inferences, questions and possibilities in Toll–like receptor signalling. Nature 2004, 430, 257–263. [Google Scholar] [CrossRef] [PubMed]
- Solbakken, M.H.; Torresen, O.K.; Nederbragt, A.J.; Seppola, M.; Gregers, T.F.; Jakobsen, K.S.; Jentoft, S. Evolutionary redesign of the Atlantic cod (Gadus morhua L.) Toll-like receptor repertoire by gene losses and expansions. Sci. Rep. 2016, 6, 25211. [Google Scholar] [CrossRef]
- Eldon, E.; Kooyer, S.; D’Evelyn, D.; Duman, M.; Lawinger, P.; Botas, J.; Bellen, H. The Drosophila 18 wheeler is required for morphogenesis and has striking similarities to Toll. Development 1994, 120, 885–899. [Google Scholar] [CrossRef] [PubMed]
- Tauszig, S.; Jouanguy, E.; Hoffmann, J.A.; Imler, J.L. Toll–related receptors and the control of antimicrobial peptide expression in Drosophila. Proc. Natl. Acad. Sci. USA 2000, 97, 10520–10525. [Google Scholar] [CrossRef]
- Ooi, J.Y.; Yagi, Y.; Hu, X.; Ip, Y.T. The Drosophila Toll-9 activates a constitutive antimicrobial defense. EMBO Rep. 2002, 3, 82–87. [Google Scholar] [CrossRef]
- Luna, C.; Wang, X.; Huang, Y.; Zhang, J.; Zheng, L. Characterization of four Toll related genes during development and immune responses in Anopheles gambiae. Insect Biochem. Mol. Biol. 2002, 32, 1171–1179. [Google Scholar] [CrossRef]
- Bass, C.; Puinean, A.M.; Zimmer, C.T.; Denholm, I.; Field, L.M.; Foster, S.P.; Gutbrod, O.; Nauen, R.; Slater, R.; Williamson, M.S. The evolution of insecticide resistance in the peach potato aphid, Myzus persicae. Insect Biochem. Mol. Biol. 2014, 51, 41–51. [Google Scholar] [CrossRef]
- Blackman, R.L.; Eastop, V.F. Aphids on the World’s Crops, an Identification and Information Guide, 2nd ed.; John Wiley & Sons Ltd: Chichester, UK, 2000; p. 476. [Google Scholar]
- Mathers, T.C.; Chen, Y.Z.; Kaithakottil, G.; Legeai, F.; Mugford, S.T.; Baa-Puyoulet, P.; Bretaudeau, A.; Clavijo, B.; Colella, S.; Collin, O.; et al. Rapid transcriptional plasticity of duplicated gene clusters enables a clonally reproducing aphid to colonise diverse plant species. Genome. Biol. 2017, 18, 27. [Google Scholar] [CrossRef]
- Ma, L.; Chen, F.; Wang, W.; Xu, L.; Lu, Z.Q. Identification of two clip domain serine proteases involved in the pea aphid’s defense against bacterial and fungal infection. Insect Sci. 2020, 27, 735–744. [Google Scholar] [CrossRef]
- Scharf, M.E. Termites as targets and models for biotechnology. Annu. Rev. Entomol. 2015, 60, 77–102. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real–time quantitative PCR and the 2−ΔΔCt method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- De Gregorio, E.; Spellman, P.T.; Tzou, P.; Rubin, G.M.; Lemaitre, B. The Toll and Imd pathways are the major regulators of the immune response in Drosophila. EMBO J. 2002, 21, 2568–2579. [Google Scholar] [CrossRef]
- Gerardo, N.M.; Altincicek, B.; Anselme, C.; Atamian, H.; Barribeau, S.M.; De Vos, M.; Duncan, E.J.; Evans, J.D.; Gabaldón, T.; Ghanim, M.; et al. Immunity and other defenses in pea aphids, Acyrthosiphon pisum. Genome. Biol. 2010, 11, R21. [Google Scholar] [CrossRef]
- Imamura, M.; Yamakawa, M. Molecular cloning and expression of a Toll receptor gene homologue from the silkworm, Bombyx mori. Biochim. Biophys. Acta. 2002, 1576, 246–254. [Google Scholar] [CrossRef]
- Cheng, T.C.; Zhang, Y.L.; Liu, C.; Xu, P.Z.; Gao, Z.H.; Xia, Q.Y.; Xiang, Z.H. Identification and analysis of Toll–related genes in the domesticated silkworm, Bombyx mori. Dev. Comp. Immunol. 2008, 32, 464–475. [Google Scholar] [CrossRef]
- Liu, L.; Wei, Y.S.; Wang, D. Identification of core genes of toll-like receptor pathway from Lymantria dispar and induced expression upon immune stimulant. Insects 2021, 12, 827. [Google Scholar] [CrossRef]
- Tauszig-Delamasure, S.; Bilak, H.; Capovilla, M.; Hoffmann, J.A.; Imler, J.L. Drosophila MyD88 is required for the response to fungal and Gram–positive bacterial infections. Nat. Immunol. 2002, 3, 91–97. [Google Scholar] [CrossRef]
- Bella, J.; Hindle, K.L.; McEwan, P.A.; Lovell, S.C. The leucine–rich repeat structure. Cell. Mol. Life Sci. 2008, 65, 2307–2333. [Google Scholar] [CrossRef]
- Zhang, S.S.; Jia, Y.H.; Li, J.Y.; Lin, J.H.; You, M.S.; Xia, X.F. Cloning and functional characterization of Toll–6 gene in the diamond back moth Plutella xylostella. J. Plant Prot. 2022, 49, 574–586. [Google Scholar]
- Rosas-Ballina, M.; Tracey, K.J. The neurology of the immune system: Neural reflexes regulate immunity. Neuron 2009, 64, 28–32. [Google Scholar] [CrossRef]
- Irazoqui, J.E.; Urbach, J.M.; Ausubel, F.M. Evolution of host innate defence: Insights from Caenorhabditis elegans and primitive invertebrates. Nat. Rev. Immunol. 2010, 10, 47–58. [Google Scholar] [CrossRef]
- Liao, W.L.; Bu, X.L.; Jiang, W.Y.; Liu, J.S. Influence of lipopolysaccharide on the expression of the toll-like receptor gene BmToll9-2 in larval Bombyx mori. Acta. Entomol. Sin. 2017, 60, 372–378. [Google Scholar]
- Hoffmann, J.A.; Reichhart, J.M. Drosophila innate immunity: An evolutionary perspective. Nat. Immunol. 2002, 3, 121–126. [Google Scholar] [CrossRef]
- Wen, Y.Y.; He, Z.; Xu, T.; Jiao, Y.; Liu, X.S.; Wang, Y.F.; Yu, X.Q. Ingestion of killed bacteria activates antimicrobial peptide genes in Drosophila melanogaster and protects flies from septic infection. Dev. Comp. Immunol. 2019, 95, 10–18. [Google Scholar] [CrossRef]
- Nishide, Y.; Kageyama, D.; Yokoi, K.; Jouraku, A.; Tanaka, H.; Futahashi, R.; Fukatsu, T. Functional crosstalk across IMD and Toll pathways: Insight into the evolution of incomplete immune cascades. P. Roy. Soc. B Biol. Sci. 2019, 286, 20182207. [Google Scholar] [CrossRef]
- Wu, S.; Zhang, X.F.; Chen, X.M.; Cao, P.S.; Beerntsen, B.T.; Ling, E.J. BmToll9, an Arthropod conservative Toll, is likely involved in the local gut immune response in the silkworm, Bombyx mori. Dev. Comp. Immunol. 2010, 34, 93–96. [Google Scholar] [CrossRef]
- Brennan, C.A.; Anderson, K.V. Drosophila: The genetics of innate immune recognition and response. Annu. Rev. Immunol. 2004, 22, 457–483. [Google Scholar] [CrossRef]
- Yan, Y.; Jia, M.H.; Le, Z.J.; Xu, K.K.; Li, C.; Yang, W.J. Four peptidoglycan recognition proteins are indispensable for antibacterial immunity in the cigarette beetle Lasioderma serricorne (Fabricius). Int. J. Biol. Macromol. 2022, 220, 1212–1220. [Google Scholar] [CrossRef]
- Altincicek, B.; Gross, J.; Vilcinskas, A. Wounding-mediated gene expression and accelerated viviparous reproduction of the pea aphid Acyrthosiphon pisum. Insect Mol. Biol. 2008, 17, 711–716. [Google Scholar] [CrossRef]

| Application of Primers | Primer Name | Forward Primer (5′–3′) | Reverse Primer (5′–3′) |
|---|---|---|---|
| Genes cloned | MpToll F1/R1 | TTCGTGTTCGCAGTTGTC | GCTTGGTCAGGCATAGTAA |
| MpToll F2/R2 | GGCGTCTCAATCAACTACT | TCAGTCTTGTTGTCATCCTT | |
| MpToll F3/R3 | GGATGACAACAAGACTGATT | GTGGAAGGGCATATCTCAA | |
| MpToll6 F1/R1 | GCGAGTATGTGCGTGTTCTT | CGGCTTATCGTTGACCACTG | |
| MpToll6 F2/R2 | TGCCGTAATGCCGTCCAA | CAGCCTGATCGCTTCTATGC | |
| MpToll6 F3/R3 | ATCGGCTCCGCAATATAACTTC | CTCGCAATCACACGCATCAA | |
| MpToll6 F4/R4 | GATGCGTGTGATTGCGAGAT | ACCGTCATCTTTCAGGCTGTA | |
| MpToll6 F5/R5 | GGACGTAAGTATTCGAGACCAA | GCAAATCACGCCCAAAGATG | |
| MpToll6-1 F1/R1 | AGTATTGCGAGTGTCCGTCTT | AGGTTCAGGTTGGCGAGTC | |
| MpToll6-1 F2/R2 | CGACAACAACATCTGGAACCT | CGAGCGTCTTGAGCATGTG | |
| MpToll6-1 F3/R3 | TCTTGGACAGGAACCGCATC | CTCGTACTCGCACAGGAACT | |
| MpToll6-1 F4/R4 | CCGCATTATGGACATGGACTC | TACGAACACGACGATGAACAG | |
| MpToll6-1 F5/R5 | TGCTGTGCTCGGACAAGAT | ACAGACAAGAAGAAGCAGATGG | |
| MpToll7 F1/R1 | ATTATTGTCGGCTACTCGG | ATCTCGGTGATGCTGTTG | |
| MpToll7 F2/R2 | GCACACGTCCAACTTAGA | TTGAGTCAGGTTACCGATAA | |
| MpToll7 F3/R3 | CGGATTGCGACTTGTAGA | AACGAGCTAAGCCTGTTG | |
| MpToll7 F4/R4 | CATCGTGGACTGTTCGTA | CACTCGGTCTGTATGAAGT | |
| MpToll7 F5/R5 | GCAACAAACACTTCTACGAGGA | GTAACGACGACTTGCGAGG | |
| MpTollo F1/R1 | TGCCCGTTCGTTGATGTCT | AAGTTGGTCAGGTGCGAGAA | |
| MpTollo F2/R2 | TTCTCGCACCTGACCAACTT | AATAATTGCCGTCCACCCTGA | |
| MpTollo F3/R3 | CATCTCATCAACCTCGCCAATA | TGGAACTGACGCTGTGGAA | |
| MpTollo F4/R4 | AAGAAGCGTCCGAAGAACAG | TATATGAATGGCAAGCGGTCTT | |
| qPCR (q) | qMpToll F/R | AAGGATGACAACAAGACTGA | GGCAACTTGGTGATGGAA |
| qMpToll6 F/R | CTTATTAGGCTGGTTGCGTTGA | CGAGGTTCCGAAGTGGTATGA | |
| qMpToll6-1 F/R | ACTGTTCCATTCCACTCGGTAT | AGGTGCTGGTCAACTCGTT | |
| qMpToll7 F/R | ATACCGAACAGCGTGGAAGT | TTAACTCGTTGGCGTACAAGTC | |
| qMpTollo F/R | CGGTCGTCACTCACAACAAC | CGGCAACTTGGCGATGTT | |
| qMpβ-actin F/R | TGGTATCGTCTTGGATTCTG | TTAGGTAGTCGGTGAGATCA | |
| qMpRps20 F/R | CTCCGAGATGATTGCCGATAT | GTCTTTGAACCTTCACCACAAG | |
| dsRNA synthesis (ds) | dsMpToll F/R | T7-CGTGCCATCAATCATCAACTCT | T7-AGGAATCCAATCGCCGACTAC |
| dsMpToll6 F/R | T7-GCCTTCGTCCAGTCAGTGT | T7-CAACGCAACCAGCCTAATAAGA | |
| dsMpToll6-1 F/R | T7-ACTGTTCCATTCCACTCGGTAT | T7-GATGCGGTTCCTGTCCAAGA | |
| dsMpToll7 F/R | T7-TGCCAGTCACAATCACATAACC | T7-GCGTCGTATTCACAACACTTG | |
| dsMpTollo F/R | T7-TTCCACAGCGTCAGTTCCA | T7-GCGAGCGTCAGATGAGTCA | |
| dsGFP F/R | T7-GCCAACACTTGTCACTACTT | T7-GGAGTATTTTGTTGATAATGGTCG |
| Gene | ORF Length (bp) | Protein Sequence (aa) | Molecular Weight (kDa) | Isoelectric Points | Leucine Ratio (%) | Instability Index |
|---|---|---|---|---|---|---|
| MpToll | 2952 | 983 | 113.69 | 6.42 | 15.3 (150) | 42.45 |
| MpToll6 | 3789 | 1262 | 142.50 | 5.54 | 12.4 (157) | 45.48 |
| MpToll6-1 | 3882 | 1293 | 146.24 | 5.89 | 13.9 (180) | 40.65 |
| MpToll7 | 4008 | 1335 | 151.83 | 6.80 | 12.7 (170) | 43.18 |
| MpTollo | 3747 | 1248 | 142.14 | 6.07 | 13.3 (166) | 36.35 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
He, L.; Zhang, C.; Yang, H.; Ding, B.; Yang, H.-Z.; Zhang, S.-W. Characterization and Functional Analysis of Toll Receptor Genes during Antibacterial Immunity in the Green Peach Aphid Myzus persicae (Sulzer). Insects 2023, 14, 275. https://doi.org/10.3390/insects14030275
He L, Zhang C, Yang H, Ding B, Yang H-Z, Zhang S-W. Characterization and Functional Analysis of Toll Receptor Genes during Antibacterial Immunity in the Green Peach Aphid Myzus persicae (Sulzer). Insects. 2023; 14(3):275. https://doi.org/10.3390/insects14030275
Chicago/Turabian StyleHe, Li, Chao Zhang, Hong Yang, Bo Ding, Han-Zhi Yang, and Sen-Wen Zhang. 2023. "Characterization and Functional Analysis of Toll Receptor Genes during Antibacterial Immunity in the Green Peach Aphid Myzus persicae (Sulzer)" Insects 14, no. 3: 275. https://doi.org/10.3390/insects14030275
APA StyleHe, L., Zhang, C., Yang, H., Ding, B., Yang, H.-Z., & Zhang, S.-W. (2023). Characterization and Functional Analysis of Toll Receptor Genes during Antibacterial Immunity in the Green Peach Aphid Myzus persicae (Sulzer). Insects, 14(3), 275. https://doi.org/10.3390/insects14030275






